Abstract
Biological networks play increasingly important roles in omics data integration and systems biology. Over the past decade, many excellent tools have been developed to support creation, analysis and visualization of biological networks. However, important limitations remain: most tools are standalone programs, the majority of them focus on protein-protein interaction (PPI) or metabolic networks, and visualizations often suffer from ‘hairball’ effects when networks become large. To help address these limitations, we developed OmicsNet - a novel web-based tool that allows users to easily create different types of molecular interaction networks and visually explore them in a three-dimensional (3D) space. Users can upload one or multiple lists of molecules of interest (genes/proteins, microRNAs, transcription factors or metabolites) to create and merge different types of biological networks. The 3D network visualization system was implemented using the powerful Web Graphics Library (WebGL) technology that works natively in most major browsers. OmicsNet supports force-directed layout, multi-layered perspective layout, as well as spherical layout to help visualize and navigate complex networks. A rich set of functions have been implemented to allow users to perform coloring, shading, topology analysis, and enrichment analysis. OmicsNet is freely available at http://www.omicsnet.ca.
INTRODUCTION
The growing applications of large-scale multi-omics studies in current life sciences have generated vast amounts of molecular measurements at DNA, RNA, protein and metabolite levels. Novel bioinformatics tools and computational methods are urgently needed to help researchers analyze these complex datasets to facilitate systems-level understanding. Two general approaches have emerged - the statistical approach and the network-based approach. The statistical approach aims to identify overall patterns or shared signatures across multiple datasets by employing various multivariate statistical methods (1,2), while the network-based approach views the biological system as interconnected networks of molecular entities, and is primarily concerned with creating and computing on such networks (3,4). Multivariate statistics are inherently complex. Although numerous methods have been developed to deal with multi-omics datasets, there is a general lack of well-established guidelines and strong use cases to promote their wide adoption and application (5). In contrast, the network-based approach is particularly appealing as networks can easily integrate new data into current knowledge framework and visually engage researchers to facilitate data understanding. Over the past decade, large-scale experiments have enabled comprehensive collection of high-quality molecular interaction data. Many excellent public databases and bioinformatics tools have been developed for storage, visualization, and analysis of such data (6–11). These expansive resources have made the network-based approach the preferred choice in current multi-omics data integration and systems biology.
The first step in the network-based approach is to create a subnetwork (or a few subnetworks) that connects significant molecules identified from individual omics data analysis. Protein–protein interactions (PPI) and metabolic reactions have been widely used for building such subnetworks. In general, there is a lack of easy-to-use bioinformatics tools that permits facile incorporation of important regulators such as microRNAs (miRNAs) or transcription factors (TFs) into biological networks. These two types of molecules are important players in gene regulations and are integral components in systems biology. High-quality public resources housing gene regulator data have become readily available in recent years. For instance, TarBase (12) and miRTarBase (13) are two comprehensive databases that host experimentally validated miRNA-target interactions. Meanwhile, the ENCODE (14), JASPAR(15) and TRRUST (16) databases have provided high-quality information on TFs and their potential target genes. Integrating these resources to allow users to easily include these important players into widely-supported PPI or metabolic networks would therefore enable deep insights for systems understanding.
After the creation of subnetworks that can, ideally, connect a significant portion of the molecules of interest, the next step is to analyze the subnetworks. Although graph theory is often used to help identify important patterns and links, a key strength of network analysis lies in organizing and visualizing the considerable knowledge about the interplay among biological molecules to help researchers make informed decisions or to develop new hypotheses (17). Therefore, an important goal of network visualization is to facilitate easy interpretation and absorption of large quantities of information without being overwhelmed by it. However, as networks become larger, it often leads to the well-known ‘hairball’ effect, which is caused by a large number of overlapping nodes and edges. Many empirical methods have been developed to address this issue such as trimming uninformative nodes, edge bundling or applying different layouts (18,19). One potential solution is to increase the visualization space from the conventional 2D to 3D space, thereby providing more viewing perspectives and reducing intersections between nodes and edges. In addition, the extra dimension can present critical information unique in multi-omics and time-series data to facilitate systems-level understanding (20,21).
Most current network visualization tools are standalone programs focusing primarily on 2D network visualization, such as Cytoscape (6) and Gephi (22). A few of them also support 3D visualizations such as BioLayout3D (23), iCAVE (24), NAViGaTOR (10), Arena3D (25) and 3DScapeCS (26). Over the past several years, there is a clear trend to move away from standalone applications towards integration of visualization within web browsers (27–29). To the best of our knowledge, no dedicated web-based tools are currently available to support 3D visualization of biological networks. There are some technical reasons behind this. Early 3D rendering was often implemented using Flash or Java 3D, both of which require plugins in order to work within a web browser. In addition, 3D rendering is inherently a computationally intensive task, and displaying large networks in 3D could easily exceed the computing capacity of early web browsers. The situation has significantly changed over the past few years. Modern web browsers are much more powerful. Browser-based applications with hardware acceleration using graphics processing units (GPUs) can deliver excellent user experience through their interactive, media-rich interfaces. The recent arrival of WebGL technology, now standard in all modern web browsers, has made it possible to implement interactive 3D graphics directly in a web browser. When properly implemented, WebGL can deliver higher performance as compared to other existing technologies such as canvas or scalable vector graphics (SVG) (30). Leveraging this new web technology to enable intuitive online 3D network visualization represent a promising direction to address the current challenges in large network visualization and multi-omics integration.
We introduce OmicsNet, a novel web-based tool for biological network creation and visual exploration in 3D space. OmicsNet was developed using the state-of-the-art WebGL technology to enable 3D network visual analytics, with built-in support for flexible creations of composite networks. The key features of OmisNet include:
Accepting lists of genes/proteins, transcription factors, miRNAs, metabolites, as well as network files (.txt, .sif or .graphml);
Supporting ten molecular interaction databases on protein-protein, miRNA-target, TF-target and enzyme-metabolite interactions, with multiple procedures for network customization;
Fully-featured 3D network visualization system supporting three layouts (force-directed layout, multi-layered perspective layout and spherical layout) and a wide array of 3D visual effects and interactions (shading, zooming, highlighting, rotating, drag-and-drop, etc.);
Comprehensive support for functional analysis based on GO, KEGG, Reactome and PANTHER (31–34), as well as network topology analysis including module detection, computing shortest paths and node centralities;
OmicsNet contains a comprehensive list of frequently asked questions (FAQs) and multiple tutorials on different use cases to help researchers navigate common analysis tasks. The public server is freely available at http://www.omicsnet.ca.
PROGRAM DESCRIPTION AND METHODS
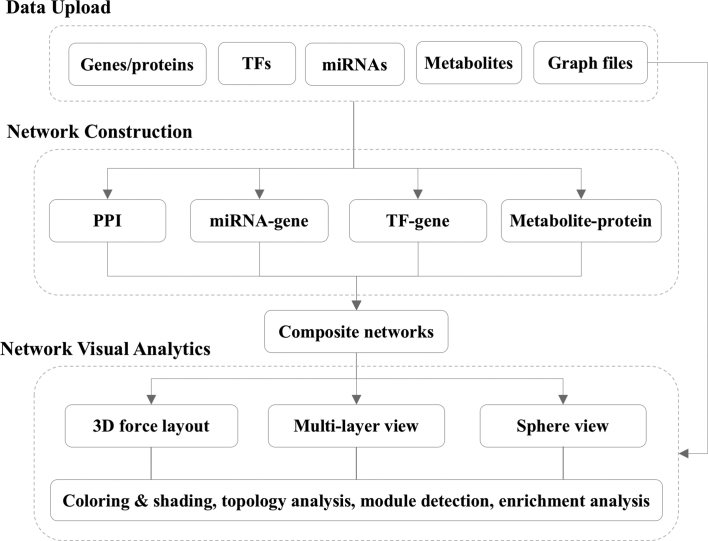
OmicsNet is mainly composed of three web pages corresponding to the three tasks - data input, network creation, and network visual analytics. Figure 1 shows the overall design and workflow of OmicsNet. Users can upload lists of genes/proteins, TFs, miRNAs or metabolites to search different molecular interaction databases. The results will be used to create different subnetworks, which can be explored in our 3D visualization system. Each component is furnished with various options to facilitate users’ tasks. The key features of each page are described in the sections below.
Figure 1.
The overall workflow of OmicsNet. Users can upload lists of genes/proteins, TFs, miRNAs or metabolites to search different molecular interaction databases. The results will be used to create composite networks, which can be explored in a powerful 3D visualization system with comprehensive built-in support for different layouts, topology analysis and functional analysis.
Creation of knowledgebase on molecular interactions
To support the construction of biological networks for different types of molecules, the first task is to create a comprehensive knowledgebase on molecular interactions. In addition to PPI and metabolic interactions, we have also included transcriptional and post-transcriptional regulations. Together they represent the four main types of molecular interactions in a simplified biological system. In total, data from ten different databases were collected, including three PPI databases (STRING (7), InnateDB (9), and IntAct (8)), two miRNA-target database (TarBase (12) and miRTarBase (13)), two metabolic databases (KEGG (32) and Recon2 (35)), and three TF-target databases (TRRUST (16), JASPAR (15), and ENCODE (36)). These publicly available databases are well maintained. We will perform annual check to synchronize our knowledgebase with the major releases of these databases.
Data upload and processing
The query input can be one or multiple lists of genes/proteins, miRNAs, TFs or metabolites. OmicsNet currently supports nine organisms (human, mouse, rat, cattle, chicken, zebra fish, fruit fly, Caenorhabditis elegans and Schistosoma mansoni). In addition to supporting creation of conventional PPI, miRNA-gene, TF-gene and metabolic networks, OmicsNet has been designed to support three general use cases for systems biology and multi-omics integration: (i) starting from a list of genes, proteins or metabolites to build PPI or metabolic networks and further include miRNAs or TFs that target these nodes; (ii) starting from a few miRNAs or TFs to identify their target genes and further add interactions between these target genes/proteins based on PPI information. Note it is not advisable to start from a long list of TFs or miRNAs as primary queries because they tend to have large numbers of interaction partners, making it impossible to identify any meaningful connections through visual inspection of the resulting networks; (iii) starting from multiple lists of molecules (genes, miRNAs, and TFs) to identify known interactions among them. Finally, users can also directly upload their own networks in several common graph file formats (.txt, .sif, or .graphml) for 3D visual exploration.
Network construction
After users have uploaded one or more lists of molecules of interest, they can proceed to the next page for network building. The interface allows users to select one or more (up to three) types of interactions (PPI, miRNA-gene, TF-gene or protein–metabolite) to be included in the network. For building composite network containing more than one interaction types, users need to specify the order of network creation (primary, secondary, or tertiary interactions). The primary interaction should be selected to build networks consisting of molecular entities of main interest and their immediate interactors. The secondary and tertiary interactions are mainly to ‘enrich’ the information contained in the primary network through: (a) adding new edges - when the PPI database is chosen as secondary, the process will introduce new edges between gene/protein nodes in the current network; or (b) adding new nodes - when the TF or miRNA database is chosen, the process will introduce new regulator nodes that target gene nodes in the current network. If multiple lists are uploaded, the lists corresponding to the secondary and/or tertiary interactions will serve as constraints to make the resulting networks more context-specific by filtering out nodes that are not in the input lists. In addition, we have implemented the ‘targeted node search’ function, which allows users to search for higher-order interactions for a selected node during the network visualization stage. The details will be described later in the corresponding section.
Once interaction types are chosen and submitted for network building, a table will be displayed indicating the number of edges and nodes of the resulting networks to help users make decisions regarding whether to perform network filtering or proceed to network visualization. The purpose of network filtering is to reduce the network size by excluding less-informative nodes based on their topological properties, such as degrees or betweenness. Users can also compute and extract a minimum network that connects all current seed nodes.
Network visual analytics
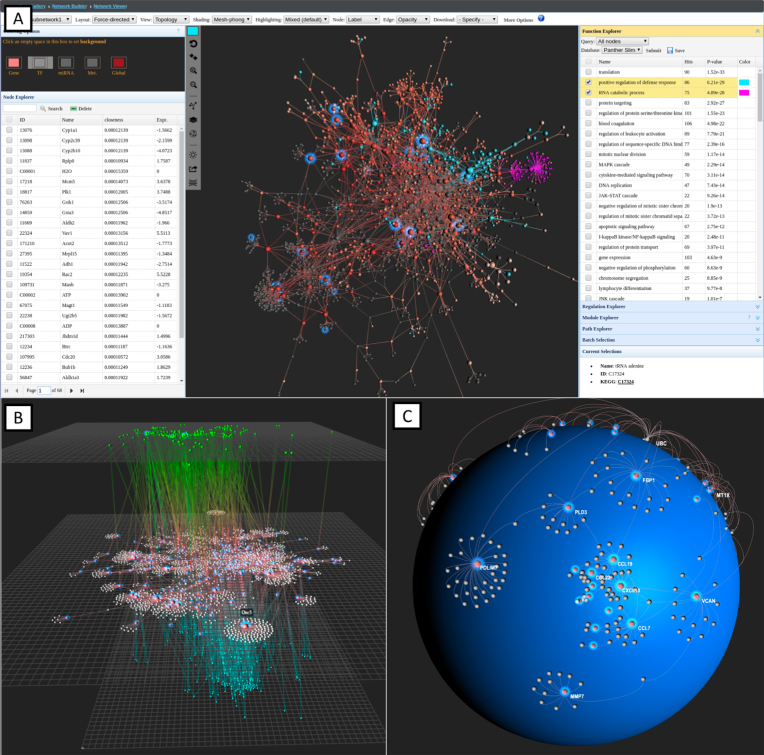
OmicsNet offers comprehensive options for network visualization, customization, topology analysis, and functional analysis. A screenshot of the Network Viewer page is shown in Figure 2A. The top tool bar contains various menu items for network viewing and customization, the left panel displays node-related information, the center panel shows the network, and the right panel consists of various functions for enrichment analysis and network topology analysis.
Figure 2.
Some screenshots of the Network Viewer showing the main features and different network layouts. (A) A force-directed subnetwork composed of ∼2000 nodes and ∼4000 edges. Seed nodes are indicated using halo effect, and nodes from two enriched pathways are highlighted in different colors. (B) A 2D perspective view of PPI subnetwork further enhanced with TFs and miRNAs targeting the key genes; (C) A spherical layout showing a module extracted from a large PPI network
Visual exploration through mouse controls
OmicsNet allows users to intuitively navigate 3D network using a mouse or trackpad. The basic mouse controls are described below:
Zoom in/out: scroll the mouse wheel in the middle. Node labels will show up automatically based on the zoom levels;
Rotate the current view: press the left mouse button and drag. The network will stay in the center;
Obtain node information: move the mouse over a node to show its label; click a node to display more detailed information about the node in the ‘Current selections’ panel on the bottom right;
Drag and drop: in the 3D force-directed layout, users can directly drag a single node or a group of highlighted nodes depending on the current scope selection. In the 2D perspective mode, users can drag and drop individual layers using the grey triangle located at one corner of the layer. Node dragging is not yet supported at the moment in the spherical layout.
Other advanced options: users can right-click on a node to search for interaction partners for this particular node against several databases (targeted node search will be discussed later), or select two nodes (source and target) and search for shortest paths between them.
Coloring
Coloring is probably the most important factor for effective visualizations. OmicsNet provides three places for users to adjust the colors of their networks. The ‘Coloring Options’ panel on the top-left corner allow users to set the background color, as well as to customize the node colors for different molecular types. The ‘Node’ option on the top tool bar provides a comprehensive list of coloring schemes based on different node topology measures or node expression values (if available). Some of the most commonly used functions are provided in the vertical toolbar located inside the network view. Located on the top is the color picker, which is used to set the current highlight color that will be applied in subsequent highlighting when users double click a node or click the halo icon (a circle with rays) to indicate the ‘seed’ nodes.
Shading
Shading is a unique feature in 3D visualization. When applied, the colors of a node surface will vary based on its angle and distance to the light source to produce more realistic 3D effects. To minimize memory load, the default network nodes are generated using premade texture mapped to point primitives to simulate 3D effect. OmicsNet supports six different shading options under the ‘Shading’ drop-down menu. Note the ‘Mesh-phong’ shading was implemented based on 3D mesh objects, which is more memory intensive thus only suitable for small and medium networks. For visualization of very large networks, it is recommended to turn off the shading effects for better performance.
Node highlighting
This is an important function to help bring out important nodes and connections. OmicsNet currently supports three options for node highlighting - mixed mode (default), halo effect and node color. In the default mixed mode, halo effect is used for node searching (when users click a node name in the node table), and for highlighting seed nodes (when users click the halo icon); while the node coloring is used for direct node highlighting (when user double click a node in the network) and for highlighting functions, modules, or shortest paths (when users click an item from the results of enrichment analysis, module detection, or shortest path finding). The highlighting color (including the color for halo effect) is controlled by the color palette located on the top-left corner of the center panel.
Network layout
Network layout (arrangements of nodes) plays a critical role in revealing important patterns during network visualization. Unlike the 2D layout where numerous algorithms have been implemented, very few ready-to-use algorithms are available for 3D network layout. OmicsNet offers the standard 3D force-directed layout as default. We have also spent significant efforts to implement two other layouts - a multi-layered 2D perspective layout and a 3D spherical layout. These three layouts are described below.
Force-directed layout. This algorithm was adapted from the standard 2D force-directed layout algorithm (37). It rearranges nodes in the current network using a physical model where all pairs of nodes repulse and adjacent nodes attract each other with edges acting as springs. It often results in an aesthetically pleasing graph with reasonable node distribution and clustering. An example is shown in Figure 2A. In some cases, the default force-directed layout in 3D may seem even more cluttered than the 2D view. There are several options to help partially resolve this issue including edge bundling, manual drag-and-drop of nodes to reduce overlap, decreasing edge opacity using the ‘Edge’ option in the top toolbar, or rotating the network to a different viewing angle.
Multi-layered perspective layout. When networks contain more than one node type (i.e. bipartite or tripartite graphs), it is often more intuitive to apply a multi-layered layout that takes advantage of the best of both 2D and 3D. This layout, first introduced by Arena3D (25), separates the network into an array of 2D networks using existing context information (i.e. types of molecules). This arrangement can greatly reduce the number of edge-crossings and emphasizes the source data type of each node. This feature is also available in iCAVE (24). An example is shown in Figure 2B. Users can use their mouse to move each layer by dragging the grey triangle at one corner to improve the layout. The type of layer (grid, plane, or none) can also be specified using the corresponding option under the ‘More Options’ menu.
Spherical layout. The spherical layout is inspired by flight paths around the globe, which is implemented by projecting a 2D force-directed network onto the surface of a sphere. This layout improves the visual experience in some cases by reducing visual occlusions and avoids information overload by showing only a part of the network. An example is shown in Figure 2C. Users can change both the color and opacity of the globe using the corresponding option under the ‘More Options’ menu.
Functional and topology analysis
OmicsNet supports functional enrichment analysis on genes displayed in the current network. It uses hypergeometric tests for over-representation analysis (38), and can be performed against GO, PANTHER GO-Slim, Reactome or KEGG pathways (31–34). OmicsNet supports three network topology analyses including node centrality analysis, module detection, and shortest path finding. Five different node centrality measures can be computed (degree, betweenness, closeness, eigenvalue, and transitivity), with degree being the default. To view different centrality measures, users can use the ‘Topology’ option under the ‘View’ menu or the ‘Color’ option under the ‘Node’ menu. Module analysis aims to find subsets of nodes that are more closely connected than expected by chance. Three module detection algorithms are supported in OmicsNet including InfoMap (39), Walktrap (40) and Label Propagation (41). Finally, users can use the ‘Path Explorer’ panel to search for the shortest paths between any two nodes of interest. Users can either enter the corresponding node IDs or right click the two nodes to define the source and target. Click a returned path will highlighted it in the current network.
Targeted node search
Due to practical reasons, the network creation interface does not allow users to introduce high-order interaction partners in batches (i.e. for all nodes). To address this limitation, we added the ‘Targeted node search’ to allow users to search higher-order interactions for a particular node displayed in the current network. To do this, users must right click a node of interest to show a drop-down menu containing different databases, then click to search a particular database. The detailed results will be displayed in the ‘Regulation Explorer’ panel on the right. Users can then use checkboxes to select one or more hits, and then click the ‘Add nodes’ button located directly above the result table. These new nodes will be added to the current network via connections to the target node.
Other features
The top menu bar contains most of the functions related to network viewing and customization. From the left side, the ‘Network’ menu allows users to access the other subnetworks created during network building; the ‘Layout’ menu contains the three different layout options; the ‘Shading’ menu allow users to select different shading effects or turn off the shading; the ‘Node’ and ‘Edge’ menus allow users to customize the node style (size, color and label) and edge style (opacity, color and bundling). Finally, the ‘More Options’ menu contains various advanced functions to customize the scope of selection for highlighting, dragging as highlighting styles. The network can be exported as a PNG image or graph files (.txt, .sif or .graphml) in the ‘Download’ menu.
Case study: Understanding complex immune regulations during helminth infection
Parasitic nematodes (helminths) are known to employ a wide array of immunomodulatory mechanisms in order to maintain their long-time survival in the host (42). To better understand the effects of helminth infection, we recently performed a meta-analysis of multiple gene expression datasets from helminth-infected mice, and revealed a core signature of genes that are differentially expressed across multiple independent studies (43). It is of great interest to further identify potential regulators (i.e. miRNAs or TFs) involved in the host immune response. To achieve this, we first built a PPI network using the InnateDB database from the signature gene list that maximally connects all seed genes and then further included miRNA-gene and TF-gene regulatory relationships using the miRNet and TRRUST databases. From the 2D perspective view, Sp1 and mir-9-5p clearly stands out as the key regulatory hub nodes in the composite network. Literature search indicates that both molecules play important roles in the immune system. Sp1 is a transcription factor involved in the regulation of Il-10 (44), a key effector in regulatory T cell response that mediates helminth-mediated immunoregulation (45), while the miRNA-9 family is involved in the regulation of the immune response (46,47). More detailed step-by-step analysis together with screenshots are available as Tutorial #4 on the OmicsNet website.
IMPLEMENTATION
OmicsNet was developed using a server-client design. The server side was implemented using the PrimeFaces component library (version 6.1) for the web framework, and R (version 3.4.3) for back-end computing. The client side was implemented based on JavaScript using the Three.js library (https://threejs.org/) as an interface to WebGL. WebGL can take advantage of the GPU acceleration by sending and executing code directly on the GPU to render graphics. This type of code is termed shader. To minimize the memory load and computational resources required, we used material rendered using low-level custom shader to represent nodes as opposed to memory intensive meshes. Additionally, to minimize the instances of time-consuming data passing to CPU, we store the geometry data of nodes and edges in buffers before sending them. Our empirical testing shows that OmicsNet can display large networks with ∼10,000 nodes. A key limiting factor in terms of performance is the high interactivity of the current implementation. Supporting features such as drag-and-drop and dynamic updating visual properties (color, size, etc.) of nodes/edges requires a large amount of event listeners which will negatively impact the performance in the cases of larger networks. We are developing a specialized version for 3D viewing only (zoom and rotate) that will allow visualization of up to one million nodes with same size and color. We intend to add this option in the near future. Meanwhile, we recommend users to keep network size between 200 and 2000 for practical reasons. OmicsNet also supports retina display by automatically adjusts the pixels of rendered networks depending on the user's screen resolution. Since most of the network visualization functions come from browser-side JavaScript functions, its performance is dependent on the user's browser and graphics card. The public server is hosted on a Google Cloud Engine with 30GB of RAM and eight virtual CPUs with 2.6 GHz each. OmicsNet has been tested in most major web browsers such as Chrome 50+, Firefox 47+, Safari 10.1+ and Edge 12+ with WebGL enabled.
Comparison with other tools
OmicsNet is a 3D network visualization and integrative analysis tool containing a comprehensive built-in molecular interaction knowledgebase that supports an array of different organisms. To the best of our knowledge, it is currently the only web-based application dedicated for visualizing biological network in 3D space. Table 1 compares OmicsNet with several well-known stand-alone tools that support 3D biological network visualization, including 3DScapeCS, BioLayout3D, Arena3D, and NAViGaTOR. Compared to these tools, OmicsNet distinguishes itself as being the only web-based tool with comprehensive built-in support for generation of different types of molecular interaction networks and a fully-featured 3D visualization system.
Table 1. Comparison of OmicsNet with other network visualization tools.
| Tools | OmicsNet | 3DScapeCS | BioLayout3D | Arena3D | NAViGaTOR |
|---|---|---|---|---|---|
| Platform | Web | Cytoscape plug-in | Standalone | Standalone | Standalone |
| Inputs | One or more lists of genes, proteins, TFs, miRNAs, metabolites; Or graph files (.txt, .sif, graphml) | List of genes, multiple graph files; time-series data | List of genes, Multiple graph files, expression matrix | Multiple graph files; time-series data | List of genes, multiple graph files |
| Network Construction and Integration | |||||
| Built-in database support | Yes | - | Yes | - | Yes |
| Network integration | Up to three types of interactions | - | - | - | - |
| Network Visualization and Analysis | |||||
| 3D visualization | Yes | Yes | Yes | Yes | Yes |
| Perspective layout | Yes | - | - | Yes | - |
| Spherical layout | Yes | - | - | - | - |
| Node drag-drop | Yes | - | 2D only | - | Yes |
| Enrichment Analysis | GO, KEGG, Reactome, PANTHER | - | - | - | - |
| Module Detection | Yes | - | - | - | - |
The URL for each tool is given below the table (note, evaluation for 3DScapeCS is based on functions offered by the plug-in itself).
• OmicsNet: http://omicsnet.ca/.
• 3DScapeCS: http://scape3d.sourceforge.net/.
• BioLayout3D: https://kajeka.com/graphia-professional/.
• Arena3D: http://arena3d.org/.
• NAViGaTOR: http://ophid.utoronto.ca/navigator/.
Current limitations and future perspectives
While the standard 3D visualization increases the viewing space and provides greater freedom in navigation, new issues are introduced such as edge occlusion and lack of perceptual reproducibility due to excessive numbers of viewing perspectives. To address these issues, we have implemented two enhanced layout options by dividing the nodes into multiple layers based on node types (the multi-layered perspective view), and by projecting the network on a globe's surface to mask network complexity while maintaining connectivity and ease of navigation (the sphere view). To further reduce edge occlusion, we implemented a force-directed edge bundling. In the future, we will implement additional network layouts and editing options to improve both the performance and visualization experience. Meanwhile, we also intend to increase its interoperability with community network visualization tools such as Cytoscape (6) and Gephi (22). At the moment, OmicsNet does not support directed or weighted edges, both features are important for many biological network visualization and interpretation. This will be our focus in the next updates. Other features to be added is the support of time-series, dynamic networks and general functionalities to perform differential network analysis. Indeed, as tremendous progresses have been made in the field of personalized medicine, there is an increasing need in the processing and visualization of -omics data from a single source over a period of time (48). This remains a huge challenge in the field and novel features such as integrating animation and other additional dimensions could facilitate its visualization and analysis (49). Another extension of the current work is to explore the effects of virtual reality (VR) through browsers using the WebVR API. This is already achievable with either Firefox or Chrome using a VR device such as the Oculus Rift.
CONCLUSION
Driven by the growing numbers of studies on multi-omics data integration and systems biology, there are strong demands for user-friendly web-based tools to allow researchers to easily create, integrate and visualize different types of biological networks. To address this need, we have developed OmicsNet to support intuitive network construction from a single or multiple lists of molecules. To facilitate data visualization experience, OmicsNet leverages the powerful WebGL technology to enable native 3D rendering of complex biological networks within modern web browsers. Three graph layouts have been implemented to provide different perspectives of the same network. The interface allows users to easily customize their visualizations through coloring, shading, highlighting, drag-and-drop, etc. In addition, users can also perform targeted node search, functional enrichment analysis, module detection, and shortest path computing. OmicsNet therefore fills an important gap by providing an easy-to-use web-based tool for 3D network visual analytics.
FUNDING
Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant; Genome Canada; Canada Research Chairs Program (CRC). Funding for open access charge: CRC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ritchie M.D., Holzinger E.R., Li R., Pendergrass S.A., Kim D.. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 2015; 16:85–97. [DOI] [PubMed] [Google Scholar]
- 2. Meng C., Zeleznik O.A., Thallinger G.G., Kuster B., Gholami A.M., Culhane A.C.. Dimension reduction techniques for the integrative analysis of multi-omics data. Brief. Bioinformatics. 2016; 17:628–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kitano H. Systems biology: a brief overview. Science. 2002; 295:1662–1664. [DOI] [PubMed] [Google Scholar]
- 4. Robinson J.L., Nielsen J.. Integrative analysis of human omics data using biomolecular networks. Mol. Biosyst. 2016; 12:2953–2964. [DOI] [PubMed] [Google Scholar]
- 5. Chong J., Xia J.. Computational approaches for integrative analysis of the metabolome and microbiome. Metabolites. 2017; 7:E62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P.. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., Del-Toro N.. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2013; 42:D358–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breuer K., Foroushani A.K., Laird M.R., Chen C., Sribnaia A., Lo R., Winsor G.L., Hancock R.E., Brinkman F.S., Lynn D.J.. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 2013; 41:D1228–D1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown K.R., Otasek D., Ali M., McGuffin M.J., Xie W., Devani B., Toch I.L.v, Jurisica I.. NAViGaTOR: network analysis, visualization and graphing Toronto. Bioinformatics. 2009; 25:3327–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu Z., Snitkin E.S., DeLisi C.. VisANT: an integrative framework for networks in systems biology. Brief. Bioinformatics. 2008; 9:317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karagkouni D., Paraskevopoulou M.D., Chatzopoulos S., Vlachos I.S., Tastsoglou S., Kanellos I., Papadimitriou D., Kavakiotis I., Maniou S., Skoufos G. et al. . DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018; 46:D239–D245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., Huang W.C., Sun T.H., Tu S.J., Lee W.H. et al. . miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018; 46:D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kellis M., Wold B., Snyder M.P., Bernstein B.E., Kundaje A., Marinov G.K., Ward L.D., Birney E., Crawford G.E., Dekker J. et al. . Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:6131–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R., Bessy A., Cheneby J., Kulkarni S.R., Tan G. et al. . JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018; 46:D260–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han H., Cho J.W., Lee S., Yun A., Kim H., Bae D., Yang S., Kim C.Y., Lee M., Kim E. et al. . TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018; 46:D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barabasi A.L., Gulbahce N., Loscalzo J.. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011; 12:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barsky A., Gardy J.L., Hancock R.E.W., Munzner T.. Cerebral: a Cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation. Bioinformatics. 2007; 23:1040–1042. [DOI] [PubMed] [Google Scholar]
- 19. Holten D., Van Wijk J.J.. Computer Graphics Forum. 2009; 28:Wiley Online Library; 983–990. [Google Scholar]
- 20. De Domenico M., Porter M.A., Arenas A.. MuxViz: a tool for multilayer analysis and visualization of networks. J. Complex Netw. 2015; 3:159–176. [Google Scholar]
- 21. Secrier M., Pavlopoulos G.A., Aerts J., Schneider R.. Arena3D: visualizing time-driven phenotypic differences in biological systems. BMC Bioinformatics. 2012; 13:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacomy M., Venturini T., Heymann S., Bastian M.. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS One. 2014; 9:e98679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Theocharidis A., Van Dongen S., Enright A.J., Freeman T.C.. Network visualization and analysis of gene expression data using BioLayout Express3D. Nat. Proto. 2009; 4:1535–1550. [DOI] [PubMed] [Google Scholar]
- 24. Liluashvili V., Kalayci S., Fluder E., Wilson M., Gabow A., Gümüş Z.H.. iCAVE: an open source tool for visualizing biomolecular networks in 3D, stereoscopic 3D and immersive 3D. GigaScience. 2017; 6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pavlopoulos G.A., O’Donoghue S.I., Satagopam V.P., Soldatos T.G., Pafilis E., Schneider R.. Arena3D: visualization of biological networks in 3D. BMC Syst. Biol. 2008; 2:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Q., Tang B., Song L., Ren B., Liang Q., Xie F., Zhuo Y., Liu X., Zhang L.. 3DScapeCS: application of three dimensional, parallel, dynamic network visualization in Cytoscape. BMC Bioinformatics. 2013; 14:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xia J., Gill E.E., Hancock R.E.. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015; 10:823–844. [DOI] [PubMed] [Google Scholar]
- 28. Salavert F., Garcia-Alonso L., Sanchez R., Alonso R., Bleda M., Medina I., Dopazo J.. Web-based network analysis and visualization using CellMaps. Bioinformatics (Oxford, England). 2016; 32:3041–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan Y., Siklenka K., Arora S.K., Ribeiro P., Kimmins S., Xia J.. miRNet - dissecting miRNA–target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016; 44:W135–W141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kee D.E., Salowitz L., Chang R.. Comparing interactive web-based visualization rendering techniques. In Poster Proc. IEEE Conf. InfoVis. 2012. [Google Scholar]
- 31. Reference Genome Group of the Gene Ontology, C. The Gene Ontology's Reference Genome Project: a unified framework for functional annotation across species. PLoS Comput. Biol. 2009; 5:e1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B. et al. . The reactome pathway knowledgebase. Nucleic Acids Res. 2018; 46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D.. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016; 44:D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Swainston N., Smallbone K., Hefzi H., Dobson P.D., Brewer J., Hanscho M., Zielinski D.C., Ang K.S., Gardiner N.J., Gutierrez J.M.. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics. 2016; 12:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Consortium E.P. The ENCODE (ENCyclopedia of DNA elements) project. Science. 2004; 306:636–640. [DOI] [PubMed] [Google Scholar]
- 37. Fruchterman T.M., Reingold E.M.. Graph drawing by force‐directed placement. Software: Pract. Exp. 1991; 21:1129–1164. [Google Scholar]
- 38. Cho R.J., Huang M., Campbell M.J., Dong H., Steinmetz L., Sapinoso L., Hampton G., Elledge S.J., Davis R.W., Lockhart D.J.. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 2001; 27:48. [DOI] [PubMed] [Google Scholar]
- 39. Rosvall M., Bergstrom C.T.. An information-theoretic framework for resolving community structure in complex networks. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pons P., Latapy M.. Computing communities in large networks using random walks. Lect. Notes Comput. Sci. 2005; 3733:284–293. [Google Scholar]
- 41. Raghavan U.N., Albert R., Kumara S.. Near linear time algorithm to detect community structures in large-scale networks. Phys. Rev. E Stat. Nonlin. Soft. Matter Phys. 2007; 76:036106. [DOI] [PubMed] [Google Scholar]
- 42. Maizels R.M., Yazdanbakhsh M.. Immune regulation by helminth parasites: cellular and molecular mechanisms. Nat. Rev. Immunol. 2003; 3:733–744. [DOI] [PubMed] [Google Scholar]
- 43. Zhou G., Stevenson M.M., Geary T.G., Xia J.. Comprehensive transcriptome meta-analysis to characterize host immune responses in helminth infections. PLoS Neglect. Trop. Dis. 2016; 10:e0004624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang X., Edwards J.P., Mosser D.M.. Dynamic and transient remodeling of the macrophage IL-10 promoter during transcription. J. Immunol. 2006; 177:1282–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sher A., Gazzinelli R.T., Oswald I.P., Clerici M., Kullberg M., Pearce E.J., Berzofsky J.A., Mosmann T.R., James S.L., MorseIII H.. Role of T‐Cell derived cytokines in the downregulation of immune responses in parasitic and retroviral infection. Immunol. Rev. 1992; 127:183–204. [DOI] [PubMed] [Google Scholar]
- 46. Bazzoni F., Rossato M., Fabbri M., Gaudiosi D., Mirolo M., Mori L., Tamassia N., Mantovani A., Cassatella M.A., Locati M.. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:5282–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nie K., Gomez M., Landgraf P., Garcia J.-F., Liu Y., Tan L.H., Chadburn A., Tuschl T., Knowles D.M., Tam W.. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am. J. Pathol. 2008; 173:242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schork N.J. Personalized medicine: time for one-person trials. Nature. 2015; 520:609–611. [DOI] [PubMed] [Google Scholar]
- 49. Pavlopoulos G.A., Malliarakis D., Papanikolaou N., Theodosiou T., Enright A.J., Iliopoulos I.. Visualizing genome and systems biology: technologies, tools, implementation techniques and trends, past, present and future. GigaScience. 2015; 4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]