Abstract
The structural states of proteins include ordered globular domains as well as intrinsically disordered protein regions that exist as highly flexible conformational ensembles in isolation. Various computational tools have been developed to discriminate ordered and disordered segments based on the amino acid sequence. However, properties of IDRs can also depend on various conditions, including binding to globular protein partners or environmental factors, such as redox potential. These cases provide further challenges for the computational characterization of disordered segments. In this work we present IUPred2A, a combined web interface that allows to generate energy estimation based predictions for ordered and disordered residues by IUPred2 and for disordered binding regions by ANCHOR2. The updated web server retains the robustness of the original programs but offers several new features. While only minor bug fixes are implemented for IUPred, the next version of ANCHOR is significantly improved through a new architecture and parameters optimized on novel datasets. In addition, redox-sensitive regions can also be highlighted through a novel experimental feature. The web server offers graphical and text outputs, a RESTful interface, access to software download and extensive help, and can be accessed at a new location: http://iupred2a.elte.hu.
INTRODUCTION
Intrinsically disordered proteins and protein regions (IDPs/IDRs) carry out important biological functions without relying on a single well-defined conformation, defying the traditional structure-function paradigm (1). Such regions are best characterized as ensembles of highly fluctuating conformations in isolation but their detailed properties are delicately tailored for their specific function (2). The activities of IDPs can directly emerge from their flexible nature, exhibiting entropic chain functions or serving as linkers between ordered domains. Disordered proteins can also mediate protein-protein interactions by recognizing specific partners and undergo a disorder-to-order transition by adopting a more structured conformation. Such disordered binding regions or MoRFs (molecular recognition features) commonly occur in modular proteins involved in signaling and regulation (3,4). The specific properties of these compact functional modules, such as their plasticity and flexibility, enable their regulation depending on cellular cues through various mechanisms including post-translational modifications (PTMs) or competitive binding (5). While the majority of known disordered binding regions lose their flexibility upon interaction (with the exception of fuzzy complexes (6,7)), an order-to-disorder transition is the key for the function of another group of proteins. These conditionally disordered proteins are folded in isolation but their functional state requires a local or global unfolding to a more disordered state. The transition can be induced by interactions with other macromolecules or changes in environmental factors, such as pH, temperature or redox potential (8). One example for such conditional disorder is presented by Hsp33 from Escherichia coli. This redox-sensing chaperone becomes active upon oxidative stress, which induces a transition to a more disordered state exposing the substrate binding surface of the protein (9).
The growing number of examples of experimentally verified disordered segments are collected into dedicated databases, such as the DisProt database, which currently holds 2,167 such disordered regions from 803 proteins (10). However, these entries only provide a small sample of IDPs/IDRs that are widespread in all domains of life but are most prevalent in eukaryotic organisms (11–14). At this scale, protein disorder can only be studied through computational approaches. The distinct sequence properties of IDPs compared to that of globular proteins enable the discrimination of these two groups at the amino acid sequence level at reasonable accuracies. So far, over 50 prediction methods have been developed using a wide arsenal of approaches, including simple amino acid propensity scales, simplified biophysical models, machine learning techniques and meta-servers (15–17). IUPred is one of the commonly used methods for predicting protein disorder and it is based on capturing the basic biophysical properties of IDPs (18,19). The basic assumption of this method is that intrinsically disordered proteins have a specific amino acid composition that does not allow the formation of enough favorable inter-residue interactions to stabilize a well-defined structural state (20,21). In IUPred, the interaction capacity of each residue is captured by an energy estimation scheme. While there are other methods that can achieve higher accuracies on particular datasets, IUPred still provides robust predictions with a favorable trade-off between speed and accuracy (22–24). As a result, IUPred is frequently used in itself or in combination with other tools to provide information about disorder (25).
The next challenge following the prediction of protein disorder is the characterization of the functional properties of IDPs/IDRs. Towards this end, most efforts focused on predicting regions of disordered proteins that are involved in protein-protein interactions, although methods that aim to predict regions binding to DNA and RNA, or to recognize linker regions have also been developed (26,27). The first publicly available method developed to recognize disordered binding regions was ANCHOR (28,29). Similarly to IUPred, this method relies on the energy estimation approach to characterize the disordered tendency and binding capacity of protein segments. Apart from ANCHOR, machine learning methods, in particular support vector machines (SVM) have also been developed for the prediction of disordered binding regions. MoRFpred and fMoRFpred utilize SVM models in their predictions incorporating sequence conservation data and amino-acid physicochemical properties, in addition to predictions of intrinsic disorder, relative solvent accessibility and residue flexibility (30,31). MFSPSSMpred and DISOPRED3 predict MoRFs based on an SVM with a radial basis function kernel, and using sequence-derived features and evolutionary profiles as inputs (32,33). MoRFchibi also employs SVMs, but uses a dual architecture to efficiently discriminate short MoRF regions from their flanking regions and to recognize similarity to already known instances (34).
The precision of the computational identification of disordered binding regions is usually evaluated against predicting such regions within globular proteins. However, these prediction methods should also have a discriminatory power against disordered regions in general. The main challenge is that currently we do not have a clear idea about the prevalence of disordered binding regions in proteins in general. One well-characterized example, p53 shows a nearly complete coverage by overlapping binding regions within its N- and C-terminal disordered segments (35). Other examples suggest that this could be a common scenario for many IDPs/IDRs, however, methods are often evaluated on proteins with a single known disordered binding site. A further limitation for accurate method development originates from a limited set of well-characterized examples used for training and testing. As a result, larger datasets were resorted to PDB complexes formed between short and longer segments, assuming that the short segments are usually associated with disorder (30). However, this approach resulted in noisy datasets without experimental verification. In this regard, a major new development was the launch of the DIBS database, which collects protein complexes where one partner was shown experimentally to be both disordered in isolation and being involved in disorder-to-order transition (36). This database currently contains 773 entries, providing a reliable platform for further method development for recognizing disordered binding regions.
Conditionally disordered regions provide further computational challenges for the characterization of IDPs (8). An important category in this class corresponds to redox potential regulated proteins that play important roles in oxidant signalling and protein biogenesis events (37). Fascinating examples, such as Hsp33(9), COX17(38) or CP12 (39) indicate that redox sensing can be coupled to disorder-to-order or order-to-disorder transitions. While the limited number of such cases currently prevents systematic analyses, we found that the biophysical model of IUPred is already equipped to highlight redox-sensitive regions in proteins.
Recently, we have relocated our web-server IUPred to a new location (25). This gave us access to further improvements. Here, we describe the IUPred2A web server, which provides a combined interface to collect predictions for disordered regions via an improved version of IUPred, disordered binding segments via a new version of ANCHOR, and can highlight redox-sensitive regions in proteins based on the energy estimation method. These predictions can be accessed through an HTML server, a RESTful web server and as a downloadable software.
METHODS
IUPred2
IUPred uses an energy estimation method at its core. This approach utilizes a low-resolution statistical potential to characterize the tendencies of amino acid pairs to form contacts, observed in a collection of globular protein structures (40). When the structure is known, the statistical potential allows the calculation of the energy for each residue based on its interactions with other contacting residues in the structure. The sum of these residue-level energy terms can be used to quantify the total stabilizing energy contribution of intrachain interactions in a given protein structure. To open up a way to estimate these energies directly from the amino acid sequence without a known structure, a novel method was developed (18). In this model, the energy of each residue in the amino acid sequence is estimated based on the following formula:
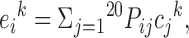 |
where 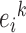 is the energy of the residue in position k of type i, Pij is the ijth element of the energy predictor matrix, and cj is the jth element of amino acid composition vector, specifying the ratio of amino acid type j in the sequence neighbourhood of position k. P is a 20 × 20 energy predictor matrix that connects the amino acid composition vector to the energy of the given residue. Its parameters were optimized on a set of globular proteins to minimize the difference between the energy calculated from the known structures using the statistical potential and the energy estimated from the amino acid sequence. Based on the energy estimation, residues that have favorable energies are predicted as ordered and residues with unfavorable energies are predicted disordered. The energies calculated for each residue in the amino acid sequence are smoothed with the window size (w0) and are transformed into a score between 0 and 1, so they can be interpreted as quasi-probabilities of a given residue being disordered.
is the energy of the residue in position k of type i, Pij is the ijth element of the energy predictor matrix, and cj is the jth element of amino acid composition vector, specifying the ratio of amino acid type j in the sequence neighbourhood of position k. P is a 20 × 20 energy predictor matrix that connects the amino acid composition vector to the energy of the given residue. Its parameters were optimized on a set of globular proteins to minimize the difference between the energy calculated from the known structures using the statistical potential and the energy estimated from the amino acid sequence. Based on the energy estimation, residues that have favorable energies are predicted as ordered and residues with unfavorable energies are predicted disordered. The energies calculated for each residue in the amino acid sequence are smoothed with the window size (w0) and are transformed into a score between 0 and 1, so they can be interpreted as quasi-probabilities of a given residue being disordered.
The resulting method, IUPred (19) is able to recognize regions of proteins that are not compatible with ordered regions based on their inability to form enough favorable intrachain interactions. As the method relies on a low-resolution biophysical model of protein folding, its parameters are easily interpretable. Furthermore, calculations involve only simple arithmetics and as a result IUPred not only makes reliable and highly robust predictions, but is currently one of the fastest available disorder prediction algorithms, making it especially suited for large-scale studies.
In the current version, IUPred2, the force field and the architecture of the method were left unchanged. However, integration into several resources, such as MobiDBlite (41), MobiDB 3.0 (42) and InterPro (43) made it necessary to implement several minor bug-fixes. IUPred2 was tested on both the original testing sets of disordered and globular structures (18), and the newest version of DisProt (10) as a positive testing set, and a custom-built negative testing set of single domain ordered proteins with known structures (see Supplementary material). The efficiencies of IUPred2 and the original IUPred are consistent with earlier independent testing results (22,24), and are virtually the same. This is evidenced by the high similarities between the two receiver operating characteristic (ROC) curves of the two algorithms on both pairs of testing datasets (see Supplementary material for the ROC curves), with the areas under the curves being nearly identical (AUC = 0.855 and 0.856 for IUPred2 and IUPred on the new testing sets, and AUC = 0.924 and 0.926 on the original testing sets). From a practical point of view, these efficiencies correspond to true positive rates of 59.6% and 68.72% when using IUPred2 with 5% and 10% false positive rates, respectively, on the new testing sets.
ANCHOR2
Similarly to IUPred, ANCHOR also utilizes the energy estimation approach, for the identification of disordered binding sites. Besides the general disorder tendency, two additional terms were also incorporated into the method that estimate the energy associated with interaction with a globular protein and with the local disordered sequence environment (28). These tendencies were combined using a linear combination and were transformed to yield a normalized score between 0 and 1 representing the probability of a given residue being part of a disordered binding region. In the presented IUPred2A server, ANCHOR was substantially reworked to give better predictions.
Concept and architecture of ANCHOR2
Retaining the original idea behind ANCHOR, the new ANCHOR2 methods also employs a simple biophysics-based model to describe disordered binding regions. In this framework, residues belonging to disordered binding sites have to fulfill two distinct criteria: (i) they have to be able to form favourable interactions with the binding surface of an ordered protein and (ii) they should be embedded in a generally disordered sequence environment. These two criteria are formulized as follows:
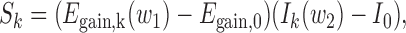 |
where Sk is the score assigned to residue k; Egain,k(w1) = Eloc,k(w1) – Eint,k is the energy the residue gains by making interactions with an averaged ordered interacting surface (represented by the composition vector Eint) instead of its own sequential environment (represented by the composition vector Eloc,k(w1), calculated in a w1 half-window sequential neighborhood of residue k); Ik(w2) is the averaged IUPred score in the w2 half-window sequential neighborhood of residue k; Egain,0 and I0 are parameters that determine the minimum energy gain and minimum average disorder tendency a residue has to possess in order to become a disordered binding site. The sign of Egain is chosen in a way that high positive values mark true binding residues (as usually expected from prediction methods), which is different from the standard choice for true free energy. Keeping this in mind, the architecture of ANCHOR2 has a clear biophysical meaning and contains only four parameters (w1, w2, Egain,0 and I0) that need to be optimized during training.
Training and benchmarking
ANCHOR2 was trained and tested using the disordered binding regions in the DIBS database (36) filtered for 30% sequence identity as the positive set, using only short binding regions below 30 residues yielding a total of 374 protein regions. Four distinctively different datasets were used as negative (see Supplementary material). The first negative dataset (ordered monomers) comprises sequence regions (also filtered for 30% sequence identity) that encode single structural domains with determined monomeric structures in the PDB (4,549 protein regions). The second dataset contains 389 flexible linker regions, used previously in the assessment of DISOPRED3(33). These two datasets can be considered as verified in a sense that they are unlikely to contain currently unknown disordered binding regions. The third dataset (decoy sequences) were collected as ∼15,000 protein segments taken randomly from the human proteome, excluding extracellular proteins, transmembrane regions and known structural Pfam domains to increase the expected ratio of disordered regions. The fourth negative dataset contains 1,042 known disordered protein regions from the DisProt database (10) that do not overlap with entries in DIBS. These two datasets cannot be assumed to be devoid of currently unknown disordered binding regions (unverified datasets). However, for parameter optimization and testing, the positive dataset, the ordered monomer set and the decoy set were split, and two thirds of all three were used in training and the remaining one third was used in testing.
During training the four adjustable parameters w1, w2, Egain,0 and I0 were tuned to their optimal values. The Egain term of the score basically describes the distinction between disordered binding regions and other sequence regions in general (non-binding disordered segments in particular). In accord, w1 was set to achieve the highest information gain (similarly to the protocol employed in (44)) calculated in the separation of the positive and decoy training sets (see Supplementary material). While the decoy set can in theory contain any number of disordered binding regions, due to the random assignment we expect their numbers to be fairly low. In contrast to the energy gain term, the I term of the score primarily describes the separation between disordered binding regions and ordered proteins. Thus, w2 was set to achieve the highest information gain in the distinction between the positive and the ordered monomer training sets (see Supplementary material). As a final step, Egain,0 and I0 were also set to best discriminate the elements of the positive and the two negative training sets.
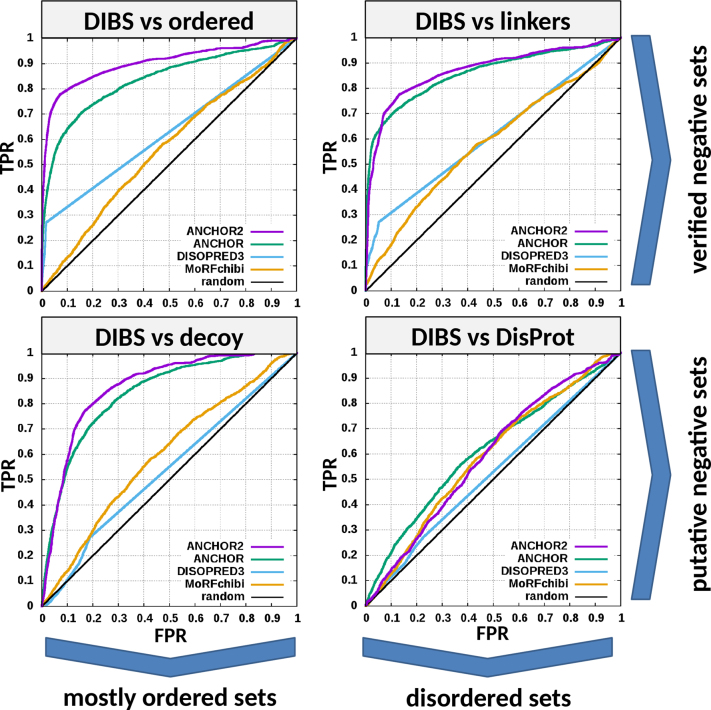
Testing of ANCHOR2 was done by calculating residue-based ROC curves evaluating the ability of the method to separate the testing positive dataset from any of the four negative testing datasets. To better gain insights into the strengths and weaknesses of ANCHOR2, three other methods capable of predicting disordered binding regions: the original ANCHOR, DISOPRED3 and MoRFchibi, were also evaluated on the same datasets. The obtained ROC curves for all four negative testing sets are shown in Figure 1, while the calculated AUC values for all methods for all datasets are shown in Table 1. The obtained efficiencies of the four methods outline the clear differences between their applicability. Both DISOPRED3 and MoRFchibi are machine learning based methods and were trained to have very low false positive prediction rates in both ordered and disordered protein regions. However, this comes at the expense of recognizing disordered binding regions that are not similar to currently known ones. ANCHOR and ANCHOR2 on the other hand incorporate a direct description of protein disorder in their predictions and thus excel at giving an extremely low false positive rate on ordered protein regions. They are also remarkable at distinguishing flexible linkers, but predict a higher ratio of disordered binding sites in generic disordered protein datasets, such as DisProt. While this may involve over-prediction, it is worth noting that the exact number of true disordered binding regions in DisProt sequences are not known and thus it is hard to determine the optimal behaviour of disorder binding site predictions on these data.
Figure 1.
ROC curves of four methods predicting disordered binding regions/MoRFs on the four different negative testing datasets. The upper row shows testing on verified negative data containing virtually no disordered binding regions. Negative sets of the bottom row might contain an unknown number of disordered binding regions, albeit with a significantly lower frequency compared to the positive set.
Table 1. Area under the curve (AUC) values calculated from the ROC curves in Figure 1.
| Methods | |||||
|---|---|---|---|---|---|
| ANCHOR2 | ANCHOR | DISOPRED3 | MoRFchibi | ||
| Datasets | ordered monomers | 0.901 | 0.835 | 0.627 | 0.561 |
| linkers | 0.870 | 0.859 | 0.612 | 0.581 | |
| decoy | 0.865 | 0.840 | 0.536 | 0.595 | |
| DisProt | 0.590 | 0.610 | 0.522 | 0.588 | |
AUC values can range from 0.5 for random predictions to 1 for perfect predictions. The highest AUC values for each negative dataset are highlighted in bold.
As a final step, the prediction score of ANCHOR was normalized to fall between 0 and 1 in such a way that the ratio of binding residues stayed below 50% even in the DisProt database, where it was the highest among the negative datasets. Using this threshold, the ratio of residues predicted to be binding in the positive and negative datasets is shown in Table 2. While this reduces the apparent efficiency of ANCHOR2 as compared to the scaling used in the original ANCHOR (the 0.5 cutoff corresponded to 5% false positive prediction on ordered protein segments), ANCHOR2 is still able to correctly predict nearly 64% of residues in known binding regions (true positive rate), with over 72% of known binding regions harboring at least one correctly predicted residue (segment-level true positive rate).
Table 2. Prediction rates of ANCHOR2 on training and testing datasets.
| Dataset name | Dataset type | Fraction of residues predicted to be disordered binding regions by ANCHOR2 |
|---|---|---|
| DIBS training | Verified positive | 57.31% (66.40% at segment level) |
| DIBS testing | 63.83% (72.58% at segment level) | |
| Ordered monomers training | Verified negative | 2.38% |
| Ordered monomers testing | 2.44% | |
| Linker regions | Verified negative | 6.03% |
| Decoy training | Putative negative | 10.69% |
| Decoy testing | 11.55% | |
| DisProt | Putative negative | 50% |
Datasets were evaluated using 0.5 cutoff to discriminate between disordered binding regions and non-binding residues.
Redox-state dependent prediction of protein disorder
In another group of conditionally disordered proteins, changes of the oxidation status are coupled to disorder-to-order or order-to-disorder transitions (37). One example for this behaviour is provided by the human small copper chaperone Cox17. This protein can be viewed as a prototype for proteins that are synthesized on cytosolic ribosomes and diffuse as intrinsically disordered proteins to the mitochondrial intermembrane space, where they become oxidized and fold into their functional conformations (38). The activity of Hsp33 also depends on oxidative conditions, however, for this protein the functional state is disordered. Under non-stress conditions, Hsp33 is a compactly folded zinc-binding protein with negligible activity. Oxidative stress causes the formation of two intramolecular disulfide bonds and the release of Zn2+ ions. This leads to the unfolding of the zinc-binding domain, exposing the substrate binding surface of the chaperone that is necessary for its activity (45).
The key sensors built into these redox-regulated proteins are cysteine residues which can undergo reversible thiol oxidation in response to the oxidation status of the molecular environment. Under reducing conditions cysteine residues can behave as polar amino acids, most similar to serine, without contributing much to protein stability. However, they can also play essential roles in stabilizing the folded conformation by coordinating Zn2+ ions under reducing conditions, or by forming disulfide bonds that are commonly used by extracellular proteins that experience oxidative conditions (46). In our energy estimation scheme, the strong stabilizing feature of cysteine residues can be adequately captured, with the most extreme energy terms corresponding to interactions mediated by cysteine residues. In order to capture the other end of the spectrum, cysteine residues can be changed to serine in the amino acid sequence. Thus, we generate two disorder prediction profiles, one corresponding to the state that is achieved through cysteine stabilization (redox-plus) and one without cysteine stabilization (redox-minus), modeled by a cysteine/serine swap. In many cases the two profiles would not differ significantly. However, our assumption is that in the case of conditionally disordered redox proteins the two profiles would be separated and would highlight redox-sensitive regions based on their different disorder tendencies. These regions are defined when the redox-minus line predicts disorder for a minimal region of 10 residues, while no disorder is predicted for the same region by the redox-plus profile. This core region is then extended in both directions to the point where the separation in the disorder score between the two lines falls below 0.15. Thus identified redox-sensitive regions are merged if their sequence separation is less than 10 residues (for details see Supplementary material). While this feature of IUPred2A cannot be tested rigorously, examples provided in later sections and on the server help pages indicate that the prediction of redox-sensitive regions can be used to explore this phenomenon at the large-scale. Our preliminary data suggests that redox sensitive regions can be quite common in the human proteome: the few experimentally characterized examples indicate that how this redox sensitivity is used in biological context can be more complex and can be fully understood only based on further experiments.
SERVER DESCRIPTION
Input
To ease the transition of users of the original IUPred server, the user interface of IUPred2A inherits a lot from its predecessor, enabling fast and straightforward usage. The main page features entry boxes, which accept a FASTA formatted or plain protein sequence, or any valid UniProt accession/ID. The sequences of corresponding UniProt entries are accessed through an SQL database containing information about the specified input, or extract the information directly from UniProt, in case of an SQL database failure. In addition, a multi-FASTA formatted file with a maximum size of 200 megabytes can also be uploaded. The new web-server also incorporates RESTful services using custom links for searches. For IUPred2 predictions, three types of predictions can be chosen depending on the type of structural regions the user wants to analyse: short stretches of disorder (such as flexible loops or linkers), long disordered regions (such as disordered domains), or structured domains. These options are directly inherited from the previous IUPred implementation (19,25). In addition, IUPred2A features optional context dependent prediction options, using either ANCHOR2 for the identification of disordered binding sites, or the redox-sensitive feature to uncover redox potential dependent disorder. Once the proper inputs are filled, the server calculates the results on a Django 2.0 based back-end. Each prediction is calculated on-the-fly server side, utilizing the latest MPI technology for maximum efficiency. To ease the load on the server, multi-FASTA uploads are treated separately and are queued until the server has enough free capacity.
Output
The latest version of Bokeh (0.12.14) is responsible for the visualization of the results that is directly integrated into the Django framework. The graphical output presents the requested predictions. By default, it contains disorder predictions from IUPred2 and binding site predictions from ANCHOR2, but the individual predictions can be turned on and off on the plot. Alternatively, the redox-sensitive regions are highlighted. Integration with the UniProt resource enables the display of various additional information about the requested protein (when available), such as PFAM annotations (47), low-throughput post-translational modifications (including phosphorylation, methylation and acetylation sites) from PhosphoSitePlus (48), related structures from the PDB (49) and experimentally verified disordered regions from three different databases: generic disorder from DisProt (10) and disordered binding regions from DIBS (36) and MFIB (50). Besides the visual output, both text based and JSON formatted outputs are downloadable for each prediction. Despite the intensive use of cutting-edge web technologies, IUPred2A supports all HTML5 and WebP compatible browsers.
Supporting features
To further enhance the usability of IUPred2A, the site features the description of the method, together with various examples that highlight its functionality and aid the correct interpretation of the results. Furthermore, IUPred2A also supports the local use of IUPred2 and ANCHOR2, as both methods are available for download as Python3 codes.
EXAMPLES
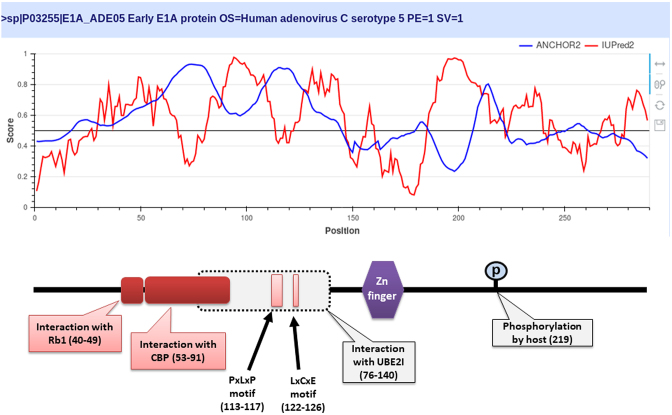
ANCHOR2 can correctly recognize many disordered binding regions that machine learning methods are likely to overlook due to their very conservative estimates of the occurrence of these functional modules. This is demonstrated through the example of the oncogenic Human adenovirus C early E1A protein (Figure 2). E1A is a largely disordered protein (51), which is essential for forcing the host cell into S phase via modulation of the Rb1/E2F1 pathway (52) and the inhibition of apoptosis via modulation of p53 degradation (53). These host-pathogen interactions are mediated by several binding events. Rb1 and CBP are targeted by two N-terminal tandem binding sites with determined complex structures deposited in the PDB. These known disordered binding regions are identified by ANCHOR2 as two distinct neighbouring peaks in the output score. While no other E1A-human protein complexes are currently known in structural detail, E1A harbors two additional known motifs capable of forming host-specific interactions. Both motifs, together with the putative binding site for the deubiquitinase UBE2I are correctly recognized by ANCHOR2 as a separate peak in the prediction score. A distinct peak C-terminal of the structured zinc-finger has no known binding partners; however it entails a serine residue that was shown to be phosphorylated by host kinases (54), hinting at an additional important binding region with currently limited characterization.
Figure 2.
The output of IUPred2 and ANCHOR2 for the oncogenic Human adenovirus C early E1A protein. Top: IUPred2 and ANCHOR2 scores are shown in red and blue. Bottom: schematic architecture of E1A. Disordered binding regions with known complex structure are shown in deep red boxes. Light red boxes correspond to known linear motifs. Grey box marks the region sufficient for interaction with UBE2I.
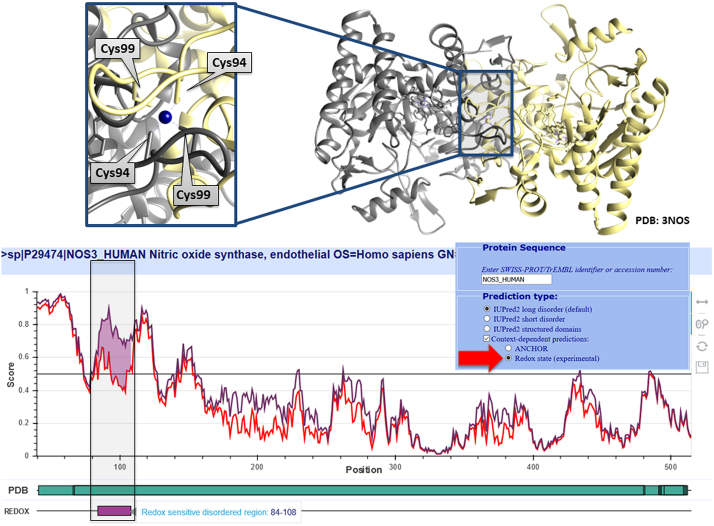
In the case of disordered binding regions, the transition between the disordered and the folded state is induced by the presence of a protein partner. However, in certain cases both the structural state and molecular interactions can be influenced by redox potential. A prime example of such behaviour is presented by the endothelial nitric oxide synthase (NOS3). Dimerization of this protein is essential for its oxidoreductase activity. The dimer interface is formed through a Zn2+-cysteine complex, where Cys94 and Cys99 from each subunit coordinate the Zn2+. These cysteines appeared susceptible to redox modifications which promote a disulfide bond formation within each monomer and subsequent release of Zn2+. This results in the disruption of the dimer and a transition to the monomeric state, parallelled by the disruption of the enzyme activity (46,55). Figure 3 shows the prediction for NOS3 generated using the experimental redox-state option of IUPred2, correctly capturing the redox-sensitive region involved in this structural transition.
Figure 3.
The output of the redox-state dependent IUPred2 predictor for the N-terminal region of NOS3. Top: the coordination of Zn2+ by cysteines 94 and 99 from both chains in the dimeric NOS3 structure. Bottom: the output of IUPred2 using the redox state modeling option, where the estimated sensitivity of the disorder tendency is marked in purple. The plot is zoomed into the N-terminal region that can be seen in the dimeric complex (PDB: 3NOS).
CONCLUSION
The current paper presents the new IUPred2A server that serves as a unified platform for both generic and context-dependent prediction of protein disorder. IUPred2A combines and supersedes our general disorder prediction method IUPred and disordered binding region prediction method ANCHOR. While IUPred2 features only slight improvements over its predecessor, ANCHOR2 was completely re-trained and re-tested built on a new architecture, bringing a significant improvement over the original version. In addition, IUPred2A also incorporates a new experimental feature that targets the identification of protein regions capable of redox-state dependent transition between disordered and ordered states. These methods are available through a completely rewritten server at a new location. The IUPred2A server retains all options for data input from previous versions, but also significantly expands its functionality by introducing RESTful services, and automated data integration from a range of databases with information about protein structure. Furthermore, completely rewritten codes for IUPred2 and ANCHOR2 are available for download to aid local large-scale analyses.
Concurrent machine learning algorithms typically excel at correctly predicting protein regions with a substantial similarity to training examples. However, owing to their biophysics-based models, IUPred2 and ANCHOR2 are expected to be able to correctly recognize protein regions that share limited to no resemblance to currently known disordered regions or binding sites. This, together with the fact that both IUPred and ANCHOR present virtually the fastest methods with high accuracies in their respective fields (56), make them outstandingly suited for de novo identification of binding- and non-binding disordered protein regions in large-scale studies.
While the computational identification of protein disorder in general has already been targeted by several methods, the possible context dependence of structural features has been generally overlooked from a prediction standpoint. IUPred2A presents the first attempt at the unified description of the context-dependence of protein disorder by being able to describe the lack of structure and its dependence on the presence of a partner protein or a change in redox environment. As IUPred2A is rooted in a biophysical model of molecular interactions, it holds the potential for the future extension of its architecture to successfully incorporate the effects of other structure-modifying environmental factors, such as pH or post-translational modifications.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Domenico Cozzetto and David T. Jones for kindly providing the flexible linker dataset used for benchmarking DISOPRED3. The constructive remarks of László Dobson concerning IUPred2A functionality are gratefully acknowledged.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Hungarian Academy of Sciences [LP2014-18 ‘Lendület’]; Országos Tudományos Kutatási Alapprogramok [K108798]. Funding for open access charge: OTKA K108798.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wright P.E., Dyson H.J.. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999; 293:321–331. [DOI] [PubMed] [Google Scholar]
- 2. van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T. et al. . Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014; 114:6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mészáros B., Tompa P., Simon I., Dosztányi Z.. Molecular principles of the interactions of disordered proteins. J. Mol. Biol. 2007; 372:549–561. [DOI] [PubMed] [Google Scholar]
- 4. Vacic V., Oldfield C.J., Mohan A., Radivojac P., Cortese M.S., Uversky V.N., Dunker A.K.. Characterization of molecular recognition features, MoRFs, and their binding partners. J. Proteome Res. 2007; 6:2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Roey K., Gibson T.J., Davey N.E.. Motif switches: decision-making in cell regulation. Curr. Opin. Struct. Biol. 2012; 22:378–385. [DOI] [PubMed] [Google Scholar]
- 6. Borgia A., Borgia M.B., Bugge K., Kissling V.M., Heidarsson P.O., Fernandes C.B., Sottini A., Soranno A., Buholzer K.J., Nettels D. et al. . Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018; 555:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miskei M., Antal C., Fuxreiter M.. FuzDB: database of fuzzy complexes, a tool to develop stochastic structure-function relationships for protein complexes and higher-order assemblies. Nucleic Acids Res. 2017; 45:D228–D235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jakob U., Kriwacki R., Uversky V.N.. Conditionally and transiently disordered proteins: awakening cryptic disorder to regulate protein function. Chem. Rev. 2014; 114:6779–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reichmann D., Xu Y., Cremers C.M., Ilbert M., Mittelman R., Fitzgerald M.C., Jakob U.. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell. 2012; 148:947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piovesan D., Tabaro F., Mičetić I., Necci M., Quaglia F., Oldfield C.J., Aspromonte M.C., Davey N.E., Davidović R., Dosztányi Z. et al. . DisProt 7.0: a major update of the database of disordered proteins. Nucleic Acids Res. 2017; 45:D219–D227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ward J.J., Sodhi J.S., McGuffin L.J., Buxton B.F., Jones D.T.. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004; 337:635–645. [DOI] [PubMed] [Google Scholar]
- 12. Lobanov M.Y., Galzitskaya O.V.. How common is disorder? Occurrence of disordered residues in four domains of life. Int. J. Mol. Sci. 2015; 16:19490–19507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peng Z., Yan J., Fan X., Mizianty M.J., Xue B., Wang K., Hu G., Uversky V.N., Kurgan L.. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell. Mol. Life Sci. 2015; 72:137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xue B., Dunker A.K., Uversky V.N.. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 2012; 30:137–149. [DOI] [PubMed] [Google Scholar]
- 15. He B., Wang K., Liu Y., Xue B., Uversky V.N., Dunker A.K.. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009; 19:929–949. [DOI] [PubMed] [Google Scholar]
- 16. Dosztányi Z., Mészáros B., Simon I.. Bioinformatical approaches to characterize intrinsically disordered/unstructured proteins. Brief. Bioinform. 2010; 11:225–243. [DOI] [PubMed] [Google Scholar]
- 17. Meng F., Uversky V.N., Kurgan L.. Comprehensive review of methods for prediction of intrinsic disorder and its molecular functions. Cell. Mol. Life Sci. 2017; 74:3069–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dosztányi Z., Csizmók V., Tompa P., Simon I.. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005; 347:827–839. [DOI] [PubMed] [Google Scholar]
- 19. Dosztányi Z., Csizmok V., Tompa P., Simon I.. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005; 21:3433–3434. [DOI] [PubMed] [Google Scholar]
- 20. Garbuzynskiy S.O., Lobanov M.Y., Galzitskaya O.V.. To be folded or to be unfolded. Protein Sci. 2004; 13:2871–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dosztányi Z., Csizmók V., Tompa P., Simon I.. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005; 347:827–839. [DOI] [PubMed] [Google Scholar]
- 22. Peng Z.-L., Kurgan L.. Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr. Protein Pept. Sci. 2012; 13:6–18. [DOI] [PubMed] [Google Scholar]
- 23. Walsh I., Giollo M., Di Domenico T., Ferrari C., Zimmermann O., Tosatto S.C.E.. Comprehensive large-scale assessment of intrinsic protein disorder. Bioinformatics. 2015; 31:201–208. [DOI] [PubMed] [Google Scholar]
- 24. Necci M., Piovesan D., Dosztányi Z., Tompa P., Tosatto S.C.E.. A comprehensive assessment of long intrinsic protein disorder from the DisProt database. Bioinformatics. 2018; 34:445–452. [DOI] [PubMed] [Google Scholar]
- 25. Dosztányi Z. Prediction of protein disorder based on IUPred. Protein Sci. 2018; 27:331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng Z., Wang C., Uversky V.N., Kurgan L.. Prediction of disordered RNA, DNA, and protein binding regions using DisoRDPbind. Methods Mol. Biol. 2017; 1484:187–203. [DOI] [PubMed] [Google Scholar]
- 27. Meng F., Kurgan L.. DFLpred: High-throughput prediction of disordered flexible linker regions in protein sequences. Bioinformatics. 2016; 32:i341–i350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mészáros B., Simon I., Dosztányi Z.. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009; 5:e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dosztányi Z., Mészáros B., Simon I.. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009; 25:2745–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Disfani F.M., Hsu W.-L., Mizianty M.J., Oldfield C.J., Xue B., Dunker A.K., Uversky V.N., Kurgan L.. MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics. 2012; 28:i75–i83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yan J., Dunker A.K., Uversky V.N., Kurgan L.. Molecular recognition features (MoRFs) in three domains of life. Mol. Biosyst. 2016; 12:697–710. [DOI] [PubMed] [Google Scholar]
- 32. Fang C., Noguchi T., Tominaga D., Yamana H.. MFSPSSMpred: identifying short disorder-to-order binding regions in disordered proteins based on contextual local evolutionary conservation. BMC Bioinformatics. 2013; 14:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones D.T., Cozzetto D.. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015; 31:857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malhis N., Gsponer J.. Computational identification of MoRFs in protein sequences. Bioinformatics. 2015; 31:1738–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gibson T.J. Cell regulation: determined to signal discrete cooperation. Trends Biochem. Sci. 2009; 34:471–482. [DOI] [PubMed] [Google Scholar]
- 36. Schad E., Fichó E., Pancsa R., Simon I., Dosztányi Z., Mészáros B.. DIBS: a repository of disordered binding sites mediating interactions with ordered proteins. Bioinformatics. 2018; 34:535–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reichmann D., Jakob U.. The roles of conditional disorder in redox proteins. Curr. Opin. Struct. Biol. 2013; 23:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fraga H., Pujols J., Gil-Garcia M., Roque A., Bernardo-Seisdedos G., Santambrogio C., Bech-Serra J.-J., Canals F., Bernadó P., Grandori R. et al. . Disulfide driven folding for a conditionally disordered protein. Sci. Rep. 2017; 7:16994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gontero B., Maberly S.C.. An intrinsically disordered protein, CP12: jack of all trades and master of the Calvin cycle. Biochem. Soc. Trans. 2012; 40:995–999. [DOI] [PubMed] [Google Scholar]
- 40. Thomas P.D., Dill K.A.. An iterative method for extracting energy-like quantities from protein structures. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:11628–11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Necci M., Piovesan D., Dosztányi Z., Tosatto S.C.E.. MobiDB-lite: fast and highly specific consensus prediction of intrinsic disorder in proteins. Bioinformatics. 2017; 33:1402–1404. [DOI] [PubMed] [Google Scholar]
- 42. Piovesan D., Tabaro F., Paladin L., Necci M., Micetic I., Camilloni C., Davey N., Dosztányi Z., Mészáros B., Monzon A.M. et al. . MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 2018; 46:D471–D476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.-Y., Dosztányi Z., El-Gebali S., Fraser M. et al. . InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017; 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Erdős G., Szaniszló T., Pajkos M., Hajdu-Soltész B., Kiss B., Pál G., Nyitray L., Dosztányi Z.. Novel linear motif filtering protocol reveals the role of the LC8 dynein light chain in the Hippo pathway. PLoS Comput. Biol. 2017; 13:e1005885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reichmann D., Xu Y., Cremers C.M., Ilbert M., Mittelman R., Fitzgerald M.C., Jakob U.. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell. 2012; 148:947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pace N.J., Weerapana E.. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014; 4:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. . The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E.. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015; 43:D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The protein data bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fichó E., Reményi I., Simon I., Mészáros B.. MFIB: a repository of protein complexes with mutual folding induced by binding. Bioinformatics. 2017; 33:3682–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ferreon A.C.M., Ferreon J.C., Wright P.E., Deniz A.A.. Modulation of allostery by protein intrinsic disorder. Nature. 2013; 498:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Egan C., Bayley S.T., Branton P.E.. Binding of the Rb1 protein to E1A products is required for adenovirus transformation. Oncogene. 1989; 4:383–388. [PubMed] [Google Scholar]
- 53. Lowe S.W., Ruley H.E.. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993; 7:535–545. [DOI] [PubMed] [Google Scholar]
- 54. Tremblay M.L., McGlade C.J., Gerber G.E., Branton P.E.. Identification of the phosphorylation sites in early region 1A proteins of adenovirus type 5 by amino acid sequencing of peptide fragments. J. Biol. Chem. 1988; 263:6375–6383. [PubMed] [Google Scholar]
- 55. Zou M.-H., Shi C., Cohen R.A.. Oxidation of the zinc-thiolate complex and uncoupling of endothelial nitric oxide synthase by peroxynitrite. J. Clin. Invest. 2002; 109:817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Malhis N., Wong E.T.C., Nassar R., Gsponer J.. Computational identification of MoRFs in protein sequences using hierarchical application of Bayes rule. PLoS One. 2015; 10:e0141603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.