Abstract
Motivation
As cancer genomics initiatives move toward comprehensive identification of genetic alterations in cancer, attention is now turning to understanding how interactions among these genes lead to the acquisition of tumor hallmarks. Emerging pharmacological and clinical data suggest a highly promising role of cancer-specific protein–protein interactions (PPIs) as druggable cancer targets. However, large-scale experimental identification of cancer-related PPIs remains challenging, and currently available resources to explore oncogenic PPI networks are limited.
Results
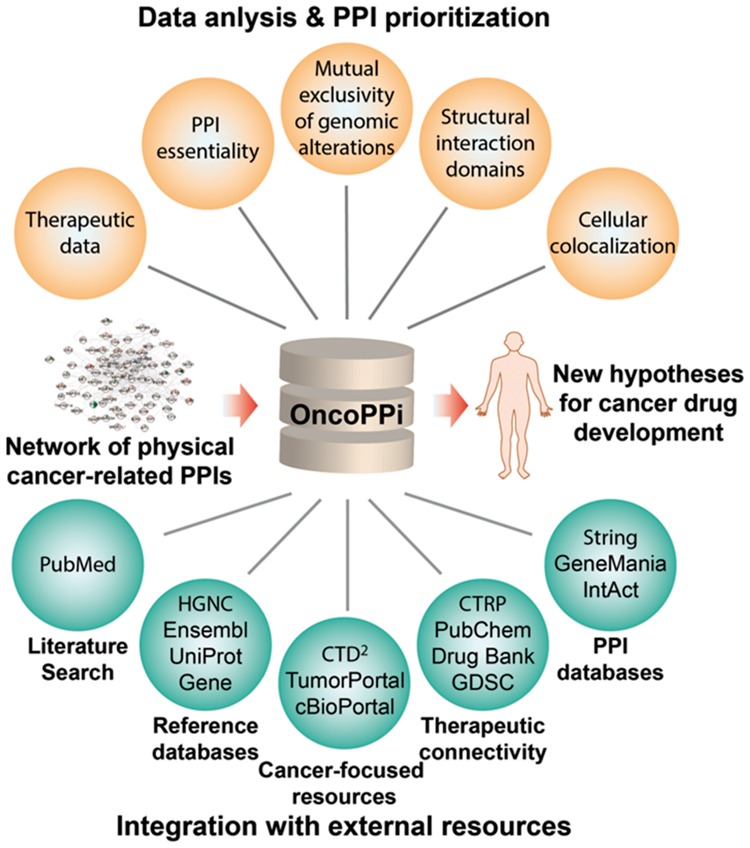
Recently, we have developed a PPI high-throughput screening platform to detect PPIs between cancer-associated proteins in the context of cancer cells. Here, we present the OncoPPi Portal, an interactive web resource that allows investigators to access, manipulate and interpret a high-quality cancer-focused network of PPIs experimentally detected in cancer cell lines. To facilitate prioritization of PPIs for further biological studies, this resource combines network connectivity analysis, mutual exclusivity analysis of genomic alterations, cellular co-localization of interacting proteins and domain–domain interactions. Estimates of PPI essentiality allow users to evaluate the functional impact of PPI disruption on cancer cell proliferation. Furthermore, connecting the OncoPPi network with the approved drugs and compounds in clinical trials enables discovery of new tumor dependencies to inform strategies to interrogate undruggable targets like tumor suppressors. The OncoPPi Portal serves as a resource for the cancer research community to facilitate discovery of cancer targets and therapeutic development.
Availability and implementation
The OncoPPi Portal is available at http://oncoppi.emory.edu.
1 Introduction
Protein–protein interactions (PPIs) play pivotal roles in biological processes in cells. Recent advances in technology and proteomics have enabled large-scale, high-throughput screening (HTS) studies of PPIs (Huttlin et al., 2015; Rolland et al., 2014; Vo et al., 2016). These studies, performed mostly in normal human HEK293T cells or yeast, have led to the discovery of thousands of PPIs involved in hundreds of metabolic and signaling pathways (Kanehisa et al., 2017; Ritz et al., 2016). A rapidly growing number of PPIs detected with various methods under different physiological conditions are now available in large PPI databases, providing a general landscape of human interactome (Chatr-Aryamontri et al., 2015; Huttlin et al., 2017; Orchard et al., 2014; Szklarczyk et al., 2015; Warde-Farley et al., 2010). However, in cancer cells, gene mutations, deletions or amplifications disturb the normal balance between death and survival signals. The genomic alterations promote the acquisition of cancer hallmarks (Hanahan and Weinberg, 2011; Vogelstein et al., 2013) through a re-wired network of oncogenic PPIs (Ivanov et al., 2013). Emerging pharmacological and clinical data suggest a highly promising role for cancer-specific PPIs as druggable cancer targets (Ivanov et al., 2013; Nero et al., 2014; Scott et al., 2016). Thus, identification of such cancer-associated PPIs could lead to new biological models for oncogenic signaling and provide new avenues for cancer therapeutic development. A number of powerful approaches have been developed to predict oncogenic PPIs based on mRNA expression data analysis (Potts et al., 2013; Wang et al., 2009), analysis of mutual exclusivity of genomic alterations (Babur et al., 2015; Ciriello et al., 2012), or through the analysis of genomic dependencies in loss-of-function screens (Cowley et al., 2014; Rosenbluh et al., 2012). However, experimental detection of cancer-associated PPIs in a cancer cell environment remains a challenge, and resources focused on physical oncogenic PPIs are limited (Alanis-Lobato et al., 2017; Meng et al., 2015; Van Coillie et al., 2016).
To facilitate discovery of PPIs involved in regulation of tumorigenesis we have recently developed a PPI HTS platform to detect the PPIs between cancer-associated proteins in the context of cancer cells (Li et al., 2017). Characterization of ∼3500 PPIs tested for a set of lung-cancer related proteins resulted in a network of high-confidence direct PPIs, termed OncoPPi (version 1) (Li et al., 2017). Overlap of OncoPPi with PPIs described in public databases revealed that more than 85% of the OncoPPi interactions are novel. Validation of newly discovered PPIs with conventional affinity pull-down assays indicated that at least 80% of OncoPPi PPIs can be confirmed as true-positive interactions (Li et al., 2017). Moreover, the network is enriched with PPIs that share known interacting structural domains, cellular co-localization, and show mutual exclusivity of genomic alterations that further support the HTS data.
To enable streamlined and integrated analysis of PPI datasets, here we present the OncoPPi Portal, a web-based resource that integrates the network of experimentally detected cancer-associated PPIs with cancer genomics, pharmacological and protein structural data. OncoPPi Portal is a unique, multifunctional web resource that facilitates analysis of PPI datasets to uncover new oncogenic programs and support cancer research.
2 Materials and methods
2.1 OncoPPi gene library
A set of 83 lung cancer-associated genes was curated based on the analysis of genomic alterations in lung cancer patients and literature searches (Li et al., 2017). It includes the major oncogenes (e.g. EGFR, KRAS, BRAF, MYC, PIK3CA, ERBB2) and tumor suppressors (e.g. TP53, STK11, CDKN2A, SMARCA4 or RB1). In addition to major tumor drivers, the protein library is populated with key regulators of oncogenic pathways, such as 14-3-3, MAPK14 (p38) or Beclin1. Structurally, the various classes of proteins (such as transmembrane receptors, kinases, GTPases, transcription factors or adaptor proteins) with different cellular localization are included in the library.
2.2 Protein–protein interaction dataset
The network of lung cancer-associated PPIs was generated based on the recent Time-Resolved Förster Energy Transfer (TR-FRET) screening performed in lung cancer H1299 cells using the cell lysates derived from systematic, pairwise transfection of GST- and Venus-fusion expression vectors containing the genes included in the OncoPPi gene library. The combination of GST- and Venus-tagged proteins allows to generate a strong TR-FRET signal of PPI that can be detected in the multi-well plate format as described by Li et al. (2017). Each PPI was tested in triplicate, with both fusion tags along with the corresponding empty vector negative controls. The whole PPI screening was repeated in three independent experiments (Li et al., 2017). The positive and negative experimental data for a total of 3486 PPI pairs is available through the OncoPPi Portal.
2.3 Statistical parameters
2.3.1 Fold-over control
Positive interactions were identified as described previously (Li et al., 2017). For each experiment the average TR-FRET signals for the PPI (SPPIG1V2, SPPIG2V1), GST empty vector control (SVecG1, G2) and Venus empty vector control (SVecV1, V2) were calculated over the triplicates for each of two tested fusions (GST, Venus). Then, for each fusion the fold-over control (FOC) values were calculated for each fusion with the following equation:
The maximum of the FOC values obtained for the two fusions (GST, Venus) was considered as the final FOC value for a given PPI in a given experiment. Then, the final FOC values were calculated by averaging individual FOC values for triplicate experiments. These values were considered as the final FOC value for a given PPI.
2.3.2 Statistical significance
A statistical significance of positive PPIs was estimated in terms of P-values calculated with the permutation test. The raw TR-FRET PPI signals detected for both fusions in triplicate in three independent experiments (a total of 18 data points), and the corresponding signals of empty vector controls (36 data points total) were used to build the PPI and Control groups, respectively. For each PPI the permutation test was performed according to the following procedure. First, for a given PPI pair, all PPI signals and control signals obtained in all experiments were ranked. The sum of the ranks of the PPI signals was calculated and was used as the test statistic for the permutation test. The permutation tests were repeated 10 000 times. The P-value was calculated with the following equation: P = (Ns + 1)/10 001, where Ns is the number of cases where total ranks of shuffled labels exceed or are equal to that of true label. The P-values adjusted for the multiple comparisons (q-values) were calculated with the Benjamini-Hochberg procedure. A set of PPIs with FOC ≥ 1.5 and q-values < 0.01, as well as PPIs that had been previously detected in other experiments (Chatr-Aryamontri et al., 2015; Orchard et al., 2014; Szklarczyk et al., 2015; Warde-Farley et al., 2010) that had FOC >1.2 and P-values < 0.05 formed the OncoPPi network (version 1) of high-confident cancer-associated PPIs (Li et al., 2017).
2.4 Integration of domain–domain interactions
The protein library was annotated with structural domain information extracted from the Pfam database (Finn et al., 2016). Specifically, for each protein all domain names (e.g. the Kinase domain) and corresponding Pfam IDs (e.g. PF00069) associated with the protein UniProt IDs were collected. Then, for each domain the domain–domain interactions (DDI) observed in crystallized protein complexes were extracted from the 3DID database based on the Pfam IDs (Mosca et al., 2014). Thus, for each domain of each protein a list of the interacting domains was generated. For example, based on the 3DID data, the Kinase domain was co-crystallized with 73 other structural domains. Then, for each domain of each protein the identified DDIs were superimposed with the DDIs identified for the protein binding partners. The protein–protein pairs with the common interacting domain pairs formed a subset of PPIs supported by structural data of domain–domain interactions.
2.5 Mutual exclusivity analysis
Analysis of mutual exclusivity (ME) of genomic alterations was performed in MatLab package (MathWorks, Inc., Natick, MA, USA) using the Cancer Genomics Data Server tool box (Cerami et al., 2012; Gao et al., 2013). The complete tumor samples from lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) TCGA Provisional datasets (downloaded 1/2016) were used to analyze the mutual exclusivity of genomic alterations in lung cancer patient samples (Cancer Genome Atlas Research, 2014; Hammerman et al., 2012). Mutations, DNA amplifications and deletions were taken into account. Mutual exclusivity was evaluated in terms of log odds ratio (OD) values calculated as described previously (Gao et al., 2013). The alterations of two genes were considered as mutually exclusive if Log(OD) < 0.
2.6 Predicted cellular co-localization
Recently, the large-scale image based analysis of cellular localization of more than 12 000 human proteins has been reported (Thul et al., 2017). These data were combined with the cellular localization previously reported in the Gene Ontology (GO) database (Ashburner et al., 2000) for the 83 proteins tested in our PPI screening. Then, cellular localization data were overlapped with the set of all PPIs tested in the screening. Two proteins were considered as potentially co-localized if they shared the same cellular compartment.
2.7 MEDICI PPI essentiality data
The PPI essentiality values for over 7900 PPIs in 206 single cell lines were calculated using the Mining Essentiality Data to Identify Critical Interactions (MEDICI) algorithm as described by Harati et al. (2017). If multiple cell lines are selected, the averaged essentiality values are calculated for the given set of cell lines.
2.8 Protein–drug connectivity
The OncoPPi network was integrated with the approved drugs. The drug names and corresponding target proteins were extracted from the Drug Bank version 5.0.9 (Wishart et al., 2006). The set of Pharmacologically Active approved drugs with the mechanism of action directly related to the specified target has been used. A total of 42 protein–drug associations have been determined.
2.9 External resources
To simplify and accelerate the data mining and exploration of detailed information associated with the OncoPPi proteins, each protein included in the network is directly linked with several external databases, including the general protein annotation servers: Human Genome Organization (HUGO) Gene Nomenclature Committee (HGCN), Ensembl, UniProt, Gene; literature database PubMed; cancer-focused resources: Cancer Target Discovery and Development (CTD2) Dasboard, TumorPortal, cBioPortal; pharmacological databases: Cancer Therapeutic Response Portal (CTRP), PubChem, DrugBank, Genomics of Drug Sensitivity in Cancer (GDSC) server; general PPI databases String, GeneMania and IntAct.
2.10 Network visualization
The PPI networks are visualized using the Cytoscape.js library (Franz et al., 2016).
2.11 Implementation
The PPI Portal application resides on a Linux server that provides Apache 2 for web services, MySQL for relational database management, and the PHP for server-side scripting services. The portal application itself utilizes those backend services along with a variety of other web technologies that include PHP, JavaScript, jQuery, Cytoscape.js and qTip2. Since the significant changes in cancer genomics data, such as the ME, protein cellular localization and DDI data are relatively infrequent, the automated ‘on-fly’ mining of corresponding datasets is not efficient, and will unnecessarily decrease the speed and accessibility of the OncoPPi Portal. Therefore, the corresponding datasets are curated manually, and updated quarterly. The protein–drug connectivity data is updated simultaneously with new DrugBank releases.
3 Results
3.1 OncoPPi portal overview
The OncoPPi Portal, available at http://oncoppi.emory.edu, provides a user-friendly interface to explore a network of cancer-associated PPIs to facilitate discovery of new targets for cancer therapy (Fig. 1). The core of the Portal is the network of PPIs experimentally detected in cancer cells (Li et al., 2017). The PPI network is integrated with the results of mutual exclusivity of genomic alterations, cellular co-localization of interacting proteins and domain–domain interactions analyses. Direct connection of the network with external cancer-focused resources and general protein databases simplifies and accelerates data mining of PPI and protein annotations. The OncoPPI Portal also integrates predicted functional impact of PPI disruption on cancer cell viability by incorporating a graphical interface for our recently developed Mining Essentiality Data to Identify Critical Interactions (MEDICI) algorithm (Harati et al., 2017). Furthermore, to enable discovery of new cancer dependencies for target discovery, the OncoPPi protein–drug connectivity network has been constructed for a set of approved drugs (Wishart et al., 2006).
Fig. 1.
OncoPPi Portal overview. The OncoPPi Portal provides a web-based interface to explore, visualize and export the network of experimentally determined cancer-associated PPIs. To facilitate prioritization of PPIs for further biological studies, the PPI network is integrated with genomics, pharmacological and structural data. A direct connection of PPIs and individual proteins with external resources enable detailed protein annotations and efficient data mining. Together, the OncoPPi Portal provides a framework to generate new hypotheses and biological models for cancer target discovery
3.2 The OncoPPi network of cancer-associated PPIs
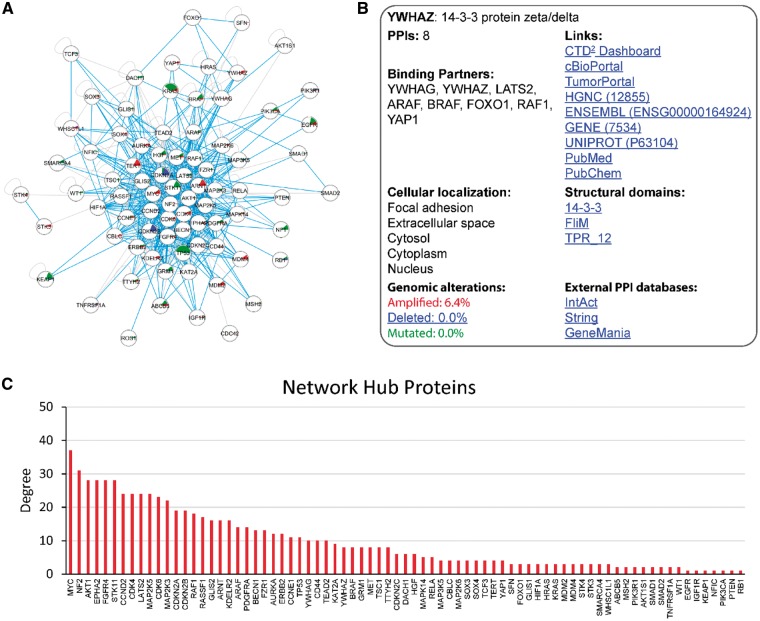
The OncoPPi Portal enables visualization and browsing of annotated PPI networks. In its first iteration, the OncoPPI Portal contains the recently published OncoPPI v1 experimental dataset resulting from a TR-FRET-based PPI screening performed in the lung cancer cells (Fig. 2A) (Li et al., 2017). Currently, a total of 3486 PPIs tested in the high-throughput PPI screening are available through the Portal along with the corresponding FOC values, permutation test P-values and the q-values (see Materials and methods section). The Statistical Properties panel allows the user to adjust these parameters to increase or decrease the thresholds for defining positive PPIs. To simplify navigation through the PPI network, all proteins included in the network are listed in the PPI Hub panel. PPIs for specific proteins can be visualized by selecting of the corresponding protein name from the list. The predefined OncoPPi network of 397 high-confidence PPIs (Fig. 2A) is also included in the PPI Hub panel. In addition, the Search panel allows the user to query the network for single or multiple proteins. A selection of multiple proteins results in a subnetwork of PPIs identified for this particular set of proteins that meet the specified statistical parameters and filters.
Fig. 2.
Visualization and analysis of the OncoPPi network. (A) The OncoPPi Portal allows the user to explore the network of PPIs experimentally determined in cancer cells. The PPIs supported by mutual exclusivity of genomic alterations are highlighted with blue lines. The frequency of gene mutations, amplification or deletions in lung cancer patients are indicated inside the corresponding circles with green, red and blue sectors, respectively. (B) The Protein Information panel provides a detailed information about the selected protein. It includes the standard gene name, complete protein name, the number of protein binding partners detected with the given statistical thresholds, and a complete list of the binding proteins. In addition, the cellular localization, structural domains linked with the Pfam server, and percent of genomic alterations in lung adenocarcinoma patients are provided. To facilitate further data mining and protein annotations, the Protein information panel links every protein with the major general PPI and cancer-specific external resources. (C) Distribution of the hub protein degree in the OncoPPi network (Color version of this figure is available at Bioinformatics online.)
3.3 Annotation and prioritization of cancer-associated PPIs
3.3.1 Analysis of mutual exclusivity of genomic alterations
Analysis of large genomic datasets, such as TCGA (Tomczak et al., 2015), revealed that genes involved in the same biological pathways often demonstrate mutual exclusivity (ME) of genomic alterations in cancer patient samples (Babur et al., 2015; Ciriello et al., 2012). Therefore, ME analysis has become a standard approach to identify functionally connected genes, and to link those genes with oncogenic signaling pathways. We calculated the ME of genomic alterations observed in lung adenocarcinoma and lung squamous cell carcinoma patients for all 3486 PPIs tested in the original HT PPI screening (Li et al., 2017). A total of 2516 PPIs with ME were identified, and 257 PPIs out of the 397 (65%) in the OncoPPi network were predicted to be functionally connected based on the ME analysis. PPIs that demonstrate ME of genomic alterations are highlighted with the blue lines. Also by checking the corresponding option in the Settings panel a user can hide the PPIs not supported by the ME analysis.
3.3.2 Cellular co-localization of interacting proteins
In order to form a complex, interacting proteins are expected to share a common compartment in the cell. Although cellular localization data are limited by currently available experimental studies performed under specific physiological conditions, the determined protein cellular localization can provide additional support for the observed PPIs, and may serve as an important factor for PPI prioritization for further studies. Therefore, each protein in the OncoPPi library was annotated with its cellular localization compartments (Fig. 2B) based on the analysis of GO annotations (Ashburner et al., 2000; Carbon et al., 2017) and recently reported large-scale image-based profiling of cellular localization of more than 12 000 human proteins (Thul et al., 2017). We have found that 353 out of 397 PPIs (89%) included in the OncoPPi network are supported by potential cellular localization. The PPIs between the proteins with predicted cellular co-localization can be visualized in the network using the ‘Co-localized proteins’ option in the Settings panel.
3.3.3 Analysis of domain–domain interactions
Often, specificity and binding affinity of PPIs is defined by the presence of specific structural domains and binding motifs in the protein structures (Chakrabarti and Janin, 2002; Mosca et al., 2014; Sillitoe et al.). Such structural domains, derived from the co-crystallized protein–protein or protein–peptide complexes are manually curated, well-annotated, and available through public databases. We integrated Pfam (Finn et al., 2016) and 3DID (Mosca et al., 2014) data into the OncoPPI Portal. The analysis of domain–domain interactions (DDIs) has revealed that 180 out of 397 OncoPPi interacting proteins (45%) also share the known interacting domains. The PPIs with known DDIs can be highlighted by checking the DDI option in the Settings panel. Overlap of domain–domain interactions (DDIs) with the PPI network provides additional structural support for novel PPIs. For example, the newly discovered STK11/RASSF1 PPI is supported by known interaction between STK11 Protein kinase domain and RASSF1 Mst1_SARAH domain. Furthermore, analysis of DDIs may guide identification of the PPI interface surface and a target site for small molecule PPI inhibitors.
3.3.4 Integration of external resources
A key step in prioritization of proteins and PPIs for detailed biological studies is the gathering and analyses of different types of information, including biochemical, biophysical, genomic and clinical data. However, different types of data are accessible through different, often poorly connected, sources and databases, and in many cases the data availability is limited. Together, it makes a comprehensive data analysis time consuming and difficult. To facilitate data mining for prioritization of cancer-associated PPIs and individual proteins for further experiments, every protein in the OncoPPi library is annotated and connected with a number of external resources. This detailed information is available in the ‘Protein information’ window that appears when a user right-clicks on a protein in the network (Fig. 2B). In addition to the complete protein name and standard gene symbol, all binding partners detected for the given protein in the currently selected network are listed in the Information panel. The PPIs reported for a given protein in external PPI databases can be directly accessed through links to the protein pages in String, Intact and Genemania datasets (Fig. 2B) (Orchard et al., 2014; Szklarczyk et al., 2015; Warde-Farley et al., 2010). Often, genes frequently altered in cancer patients play a critical role in regulation of oncogenic signaling and tumorigenesis (Chang et al., 2016; Lawrence et al., 2014). The OncoPPi network was established for a set of lung cancer-associated genes (Li et al., 2017). Therefore, for every gene in the library, a percentage of lung adenocarcinoma patient samples with gene DNA amplifications, deletions or mutations is provided in the Information panel, and the frequency of these alterations is visualized with red, blue and green sectors inside the protein node circle in the network. Additionally, the major cellular localization sites and known structural domains for each protein are shown and linked with the Pfam webserver.
To provide direct access to general genomic and proteomic annotations, every protein is linked to the HGNC, ENSEMBL, GENE and UniProt databases (NCBI R.C. 2017; The UniProt Consortium 2017; Aken et al., 2016; Gray et al., 2015). Furthermore, the recently published literature related to a selected protein can be accessed through a direct link to PubMed (NCBI R.C. 2017) (Fig. 2B).
The role of a protein as a biological target, its druggability, and available small molecule modulators of its activity can be gathered through the PubChem database (Kim et al., 2016). Moreover, every protein is associated with several major cancer-focused resources. In particular, the CTD2 Network (Schreiber et al., 2010) provides a broad spectrum of the most recent experimental cancer biology data generated by the network of leading groups in cancer biology, clinical oncology, genomic and bioinformatics fields. In addition, detailed information on genomic alterations in different tumor types and their impact on cancer patient survival can be accessed and explored though direct links to cBioPortal (Cerami et al., 2012; Gao et al., 2013) and TumorPortal (Lawrence et al., 2014). Together, the Protein Information panel serves as a dashboard for the cancer-focused data mining and annotations for the oncogenic signaling networks.
3.3.5 The network hub proteins
Statistical analyses of large-scale PPI networks combined with gene knockdown experiments have revealed that cells are more sensitive to the deletion of proteins that are involved in many PPIs (hub proteins), and deletion of hub proteins is likely to be more lethal than deletion of non-hub proteins (He and Zhang, 2006; Jeong et al. 2001). Therefore, identification of hub proteins in the cancer-associated PPI network may lead to discovery of novel major regulators of oncogenic signaling, as potential drug targets. With this goal, we integrated the analysis of protein connectivity implemented as an interactive bar graph shown under the PPI network (Fig. 2C). Specifically, the X-axis of the graph shows the proteins included in the given network, and the Y-axis shows the number of protein binding partners (node degree) identified with the selected statistical parameters. The protein order along the X-axis and data shown on the graph changes dynamically when the network and statistical thresholds are changed (e.g. FOC or P-values) (Fig. 2C). Furthermore, all the binding partners of a hub protein and their interconnections can be shown by clicking on the corresponding bar on the graph. The graphical representation of protein connectivity allows immediate visualization and ranking of the major hub proteins, and identification of their binding partners for further analysis.
Adjustments of statistical parameters combined with different filters allows determination of sets of promising PPIs. For example, the application of all three filters (ME, DDI and co-localization) to the OncoPPi dataset results in a subnetwork of 91 PPIs. The analysis of this subnetwork reveals many well-known oncogenic PPIs, including KRAS/BRAF, TP53/MDM2, ARNT/HIF1A or MET/HGF. On the other hand, a set of novel PPIs supported by structural and genomic data can be extracted from this subnetwork. For example, we found that in addition to the well-characterized interaction between mitogen-activated protein kinase 14 (MAPK14) and the mitogen-activated protein kinase kinase 3 (MAP2K3), MAP2K3 also interacts with growth factors FGFR4 and EPHA2, while serine-threonine kinase 11 (STK11) forms a complex with the cyclin-dependent kinase 4 (CDK4). Indeed, these interactions have been validated in secondary glutathione S-transferase (GST) pull-down assays (Ivanov et al., 2017; Li et al., 2017).
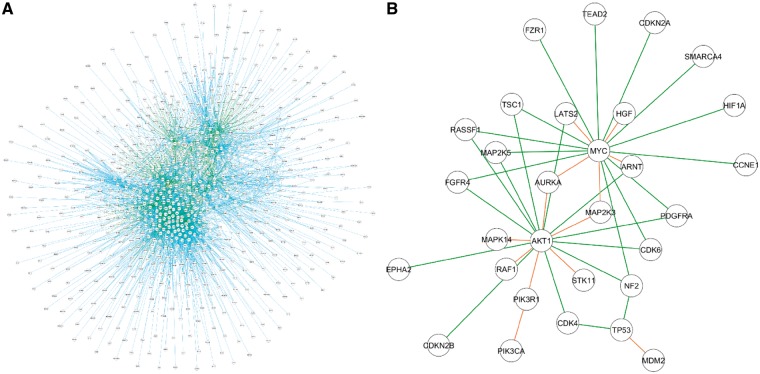
3.3.6 PPI essentiality
Recently, we have reported the MEDICI algorithm designed to predict the role of individual PPIs in cancer cell death and survival (Harati et al., 2017). The MEDICI approach couples gene knockdown studies with network models of protein interaction pathways to quantify and rank PPI essentiality in specific cancer cell lines. To simplify the practical application and accessibility of the MEDICI algorithm, we have developed a graphical user interface implemented on the PPI Essentiality page of the Portal. The main part of the PPI Essentiality page is the PPI superpathway network constructed by combining curated PPI pathways available through the Molecular Signature Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb) (Fig. 3A). Currently, the essentiality values calculated in 206 cell lines from multiple tissue types for a total 7906 PPIs derived for a set of 1548 proteins are available for the analysis. The essentiality values range from 0 (less essential) to 1 (most essential PPIs). Accordingly, the subsets of PPIs with a desired level of essentiality can be visualized by adjusting the Essentiality Threshold in the Statistical Properties panel. The essentiality values can be calculated either for all 206 available cell lines or for a specific set of cell lines as well as for the individual cell lines. The cell lines can be filtered based on the corresponding tumor type or through the Search option. Similar to the OncoPPi Network page, all proteins included in the dataset are listed in the PPI Hubs panel. In addition, the proteins can be filtered based on their involvement in defined 196 biological pathways identified by the Pathway Interaction Database (Schaefer et al., 2009).
Fig. 3.
Visualization and analysis of predicted PPI essentiality. (A) The network shows protein–protein pairs subjected for the MEDICI calculations to predict their essentiality for lung cancer cells. A total of 7906 PPIs are available for the analysis. The proteins are connected by lines colored based on the essentiality vales: E = 0—grey, 0 < E ≤ 0.2—black, 0.2 < E ≤ 0.4 – blue, 0.4 < E ≤ 0.6 – green, 0.6 < E ≤ 0.8 – orange, 0.8 < E ≤ 1 – red. (B) A subnetwork of PPIs included in the OncoPPi network and predicted as critical for lung cancer cell viability. A number of well-known oncogenic PPIs, e.g. RAF1-AKT, MDM2-TP53 or PIK3CA-PIK3R1 dimerization are predicted as essential for the cancer cells along with a set of novel PPIs, such as MYC-MAP2K3 or RASSF1-AKT1 (Color version of this figure is available at Bioinformatics online.)
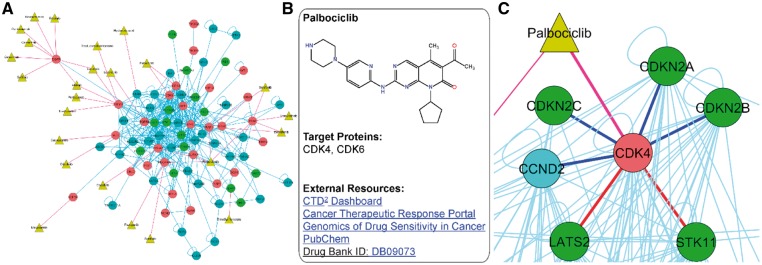
3.3.7 Therapeutic connectivity
To identify PPIs detected for validated cancer drug target proteins and to link PPIs with available therapeutic data, the OncoPPi network is also annotated with the approved drugs (Fig. 4). A total of 42 protein–drug interactions have been identified for the 32 approved drugs and 14 proteins included in OncoPPi network. The resulting OncoPPi protein–drug connectivity network is available through the Therapeutic Connectivity page of the OncoPPi Portal (Fig. 4A). To provide immediate access to the chemical, biological and pharmacological data available for the compounds, each drug is linked with five external databases, including the CTD2 Network Dashboard (Schreiber et al., 2010), Cancer Therapeutic Response Portal (Rees et al., 2015), Genomics of Drug Sensitivity in Cancer project (Yang et al., 2013), PubChem (Kim et al., 2016) and the Drug Bank (Wishart et al., 2006) (Fig. 4B). Examples of the target proteins identified for the approved drugs include key oncogenic proteins, such as BRAF, MET, FGFR4, ERBB2 or CDK4. Upon binding to a target protein, small molecule compounds can regulate PPIs through inhibition of enzymatic activity or induction of conformational changes of target proteins, leading to alterations in oncogenic signal transduction. We found more than 150 PPIs that could be potentially regulated by 32 approved drugs identified for the OncoPPi set of proteins. For example, it can be hypothesized that inhibition of CDK4 kinase by palbociclib (Owsley et al., 2016) can affect some of the 24 PPIs observed for CDK4 in the OncoPPi network. Interestingly, the network analysis revealed that CDK4 binds to several tumor suppressor proteins, including LATS2 and STK11, which are widely mutated in lung adenocarcinoma (Fig. 4C). Based on these observations, we have found that indeed CDK4 binds to STK11 in lung cancer cells at endogenous levels and that the CDK4 inhibitor, palbociclib, disrupts the CDK4/STK11 PPI (Li et al., 2017). Furthermore, we found that STK11 silenced cells showed increased sensitivity to palbociclib compared to the parental H1792 lung cancer cells (Li et al., 2017). Together, analysis of the OncoPPi network combined with the protein–drug connectivity data revealed that STK11-loss may lead to enhanced-dependency of cancer cells on CDK4. Based on these data, we can speculate that LATS2 mutations may also lead to an enhanced sensitivity of cancer cells to CDK4 inhibition, and the CDK4/LATS2 PPI can also be regulated by palbociclib and other CDK4 inhibitors. Thus, exploration and analysis of OncoPPi protein–drug connectivity provides a valuable resource to discover new mechanisms of tumor dependencies.
Fig. 4.
OncoPPi drug-connectivity network. (A) The drug-connectivity network constructed for the approved drugs and the corresponding target proteins included in the OncoPPi network. Protein–drug connections are shown with red lines, blue lines indicate the PPIs. The drugs are highlighted as yellow triangles, and proteins are shown as circles. Red circles indicate known oncogenes, and green circles highlight tumor suppressor proteins. (B) The Drug Information panel shows the chemical structure of the drug compound along with the corresponding target proteins. The direct links to external resources facilitate the mining of clinical and biological data available for the given compound. (C) Analysis of protein–drug connectivity allows the user to hypothesize a role of palbociclib in regulation of CDK4-STK11 and CDK4-LATS2 PPIs (shown with red lines). On the other hand, STK11 and LATS2 mutations in lung cancer may lead to increased sensitivity to CDK4 inhibitors. Well known CDK4 PPIs with CCND2, CDKN2A, B and C are shown with blue lines (Color version of this figure is available at Bioinformatics online.)
3.4 Data export
There are two ways to export data from the OncoPPi Portal for further analysis. The Save Image icon allows a user to save the network image as a portable network graphics (png) file. The Export PPI Data icon allows the user to export all data associated with selected (shown) PPIs as a comma-separated file (csv). The data includes statistical characteristics of the PPI, namely the FOC, P-value and q-value, indication of mutual exclusivity of alterations of corresponding genes in lung adenocarcinoma or lung squamous cell carcinoma, predicted interacting domains, co-localized proteins and essentiality values along with the corresponding cell lines for the PPI Essentiality module.
3.5 Prioritizing PPI targets with the OncoPPi Portal
To illustrate a practical application of the OncoPPi Portal for prioritization of PPIs for biological studies we first analyzed the OncoPPi network connectivity using the Network Hub protein bar graph for the OncoPPi v1 network (Fig. 2C). As indicated on the graph, the MYC oncogene (Meyer and Penn, 2008) is the largest hub of the OncoPPi network with 37 binding partners, including MYC homodimer. After clicking on the red MYC bar in the graph, the MYC network hub appears in the graphical window. We next apply the ‘PPIs with mutual exclusivity’ filter available in the Statistical Properties panel to identify experimentally observed MYC binding partners that also demonstrate mutual exclusivity in TCGA lung adenomcarcinoma datasets. We found that genomic alterations of 18 genes are mutually exclusive with genomic alterations of MYC in lung adenocarcinoma or lung squamous cell carcinoma: ARAF, ARNT, AURKA, CCNE1, CDKN2A, CDKN2B, DACH1, FZR1, GLIS2, HIF1A, KAT2A, LATS2, MAP2K3, MAP2K5, PDGFRA, SMARCA4, WHSC1L1, WT1. These genes can be highlighted by on the MYC node. When the ‘Co-localized proteins’ filter is added, all of these proteins also appear to be co-localized with MYC based on the GO annotations. A query of String, Intact and GeneMania PPI databases revealed that CDKN2A, FZR1, KAT2A and SMARCA4 are known binding partners of MYC. In contrast, 14 other proteins have not been previously reported as direct MYC binding partners. These data are readily available in a table format through the Data Export option, or through the direct links to corresponding databases accessible from the MYC Information window. Then, the MEDICI algorithm was utilized to evaluate an impact of novel MYC PPIs on the viability of the lung cancer cells. The MYC PPIs with essentiality values (E) were averaged across all lung tumor cell lines, and the PPIs with E > 0.5 were selected. The analysis suggested that among novel MYC PPIs lung cancer cells are sensitive to a disruption of MYC interactions with ARNT, AURKA, CCNE1, HIF1A, LATS2, MAP2K3, MAP2K5, PDGFRA and WT1. This allows us to hypothesize a novel role for these PPIs in control of MYC-driven tumorigenesis. Indeed, we were able to confirm all of the prioritized interactions, except MYC/PDGFRA, with our recently developed nanoPCA assay that was performed in live H1299 lung cancer cells (Mo et al., 2017). Interestingly, among all OncoPPi PPIs, the MAP2K3/MYC interaction is characterized by one of the highest PPI Essentiality values of 0.916 calculated for the H1299 lung cancer cells. In agreement with the predicted importance of MYC/MAP2K3 PPI, we have observed that MAP2K3 not only binds to MYC endogenously but upregulates MYC protein stability and transcriptional activity in cancer cells (Ivanov et al., 2017). Thus, analysis of OncoPPi network connectivity, integration of ME analysis and evaluation of PPI essentiality allowed a prioritization of PPIs as potential modulators of MYC-driven program for further biological studies.
4 Discussion
During the past decades, a number of powerful databases and internet resources have been developed to provide detailed characterization and annotation of cancer-related genes based on their genomic alterations in cancer patients and individual cell lines (Cerami et al., 2012; Gao et al., 2013; Lawrence et al., 2014). In addition, intensive bioinformatics analyses of mRNA expression and cell line sensitivity for individual gene knock-outs have been utilized to predict gene functional connectivity in cancer cells. However, identification and prioritization of oncogenic PPIs for detailed functional studies remains a challenge.
In this study, we have presented the OncoPPi Portal, a new resource available to the public that bridges cancer genomics, clinical and pharmacological data with a network of experimentally determined direct interactions between cancer-associated proteins. A user-friendly interface and flexible query options allows the exploration, visualization and export of the physical PPIs annotated with the analysis of mutual exclusivity of genomic alterations, cellular co-localization and interacting structural domains. The implementation of the graphical interface for the MEDICI algorithm allows the user to evaluate the PPI essentiality for the cell lines of different tumor types. The specific focus of the OncoPPi Portal on direct cancer-related PPIs distinguishes this resource from general PPI databases. On the other hand, the Portal links the PPIs and individual proteins with major PPI servers, and leading cancer-focused resources, including the CTD2 Dashboard (Schreiber et al., 2010), cBioPortal (Cerami et al., 2012; Gao et al., 2013) and TumorPortal (Lawrence et al., 2014). A practical application of the Portal is illustrated by analysis of PPIs experimentally determined for the MYC oncogene. A prioritization of MYC interactions based on the OncoPPi network connectivity, novelty, ME, co-localization and PPI essentiality data allowed us to select PPIs with a potential role in regulation of MYC-driven tumorigenesis, including the MAP2K3/MYC interaction. Recently, we have demonstrated that MAP2K3 binds MYC exogenously and endogenously in cancer cells leading to enhanced MYC stability and activity (Ivanov et al., 2017).
The basis of the current version of the OncoPPi Portal is the OncoPPi v1 network of lung cancer-related PPIs (Li et al., 2017), providing a foundation for expansion and growth. The OncoPPi is a dynamic resource, expanding in several directions, including the gene library, the number of tested cancer-associated PPIs, PPI screening technologies and the tumor models for the PPI screening. The next versions of the Portal will also include the options for users to import external PPI sets for the analysis. We believe that the OncoPPi Portal will serve as a valuable resource to explore, annotate and prioritize cancer-associated PPIs for cancer biology and drug discovery studies.
Funding
The results published here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. This research was supported in part by the National Cancer Institute of the NIH (Cancer Target Discovery and Development Network grants U01CA168449 and U01CA217875, H.F.), Emory University Research Committee grant (A.A.I.) and Winship Cancer Institute (NIH 5P30CA138292).
Conflict of Interest: none declared.
References
- Aken B.L. et al. (2016) The Ensembl gene annotation system. Database (Oxford), doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed]
- Alanis-Lobato G. et al. (2017) HIPPIE v2.0: enhancing meaningfulness and reliability of protein–protein interaction networks. Nucleic Acids Res., 45, D408–D414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babur O. et al. (2015) Systematic identification of cancer driving signaling pathways based on mutual exclusivity of genomic alterations. Genome Biol., 16, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature, 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E. et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery, 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S. et al. (2017) Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res., 45, D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti P., Janin J.. (2002). Dissecting protein–protein recognition sites. Proteins, 47, 334–343. [DOI] [PubMed] [Google Scholar]
- Chang M.T. et al. (2016). Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol., 34, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A. et al. (2015) The BioGRID interaction database: 2015 update. Nucleic Acids Res., 43, D470–D478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G. et al. (2012) Mutual exclusivity analysis identifies oncogenic network modules. Genome Res., 22, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley G.S. et al. (2014) Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data, 1, 140035.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D. et al. (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res., 44, D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz M. et al. (2016) Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics (Oxford, England), 32, 309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6, pl1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K.A. et al. (2015) Genenames.org: the HGNC resources in 2015. Nucleic Acids Res., 43, D1079–D1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerman P.S. et al. (2012) Comprehensive genomic characterization of squamous cell lung cancers. Nature, 489, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Harati S. et al. (2017) MEDICI: mining essentiality data to identify critical interactions for cancer drug target discovery and development. PloS One, 12, e0170339.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Zhang J. (2006) Why do hubs tend to be essential in protein networks? PLoS Genet., 2, e88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L. et al. (2017) Architecture of the human interactome defines protein communities and disease networks. Nature, 545, 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L. et al. (2015) The BioPlex network: a systematic exploration of the human interactome. Cell, 162, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A.A. et al. (2017) OncoPPi-informed discovery of mitogen-activated protein kinase kinase 3 as a novel binding partner of c-Myc. Oncogene, 36, 5852–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A.A. et al. (2013) Targeting protein–protein interactions as an anticancer strategy. Trends Pharmacol. Sci., 34, 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H. et al. (2001) Lethality and centrality in protein networks. Nature, 411, 41–42. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. et al. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res., 45, D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. et al. (2016) PubChem substance and compound databases. Nucleic Acids Res., 44, D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S. et al. (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature, 505, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. et al. (2017) The OncoPPi network of cancer-focused protein–protein interactions to inform biological insights and therapeutic strategies. Nat. Commun., 8, 14356 doi:10.1038/ncomms14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X. et al. (2015) CancerNet: a database for decoding multilevel molecular interactions across diverse cancer types. Oncogenesis, 4, e177.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N., Penn L.Z. (2008) Reflecting on 25 years with MYC. Nat. Rev. Cancer, 8, 976–990. [DOI] [PubMed] [Google Scholar]
- Mo X.L. et al. (2017) AKT1, LKB1, and YAP1 revealed as MYC interactors with NanoLuc-based protein-fragment complementation assay. Mol. Pharmacol., 91, 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca R. et al. (2014) 3did: a catalog of domain-based interactions of known three-dimensional structure. Nucleic Acids Res., 42, D374–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI R.C. (2017) Database resources of the national center for biotechnology information. Nucleic Acids Res., 45, D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nero T.L. et al. (2014) Oncogenic protein interfaces: small molecules, big challenges. Nat. Rev. Cancer, 14, 248–262. [DOI] [PubMed] [Google Scholar]
- Orchard S. et al. (2014) The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res., 42, D358–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsley J. et al. (2016) Palbociclib: CDK4/6 inhibition in the treatment of ER-positive breast cancer. Drugs Today (Barc), 52, 119–129. [DOI] [PubMed] [Google Scholar]
- Potts M.B. et al. (2013) Using functional signature ontology (FUSION) to identify mechanisms of action for natural products. Sci. Signal., 6, ra90.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees M.G. et al. (2015) Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol., 12, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz A. et al. (2016) Pathways on demand: automated reconstruction of human signaling networks. NPJ Syst. Biol. Appl., 2, 16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T. et al. (2014) A proteome-scale map of the human interactome network. Cell, 159, 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J. et al. (2012) beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell, 151, 1457–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer C.F. et al. (2009) PID: the Pathway Interaction Database. Nucleic Acids Res., 37, D674–D679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S.L. et al. (2010) Towards patient-based cancer therapeutics. Nat. Biotechnol., 28, 904–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D.E. et al. (2016) Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nat. Rev. Drug Discov., 15, 533–550. [DOI] [PubMed] [Google Scholar]
- Sillitoe I. et al. New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. [DOI] [PMC free article] [PubMed]
- Szklarczyk D. et al. (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res., 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul P.J. et al. (2017) A subcellular map of the human proteome. Science, 356, doi: 10.1126/science.aal3321. [DOI] [PubMed]
- The UniProt Consortium. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res., 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczak K. et al. (2015) The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (Poznan, Poland), 1A, A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Coillie S. et al. (2016) OncoBinder facilitates interpretation of proteomic interaction data by capturing coactivation pairs in cancer. Oncotarget, 7, 17608–17615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo T.V. et al. (2016) A proteome-wide fission yeast interactome reveals network evolution principles from yeasts to human. Cell, 164, 310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B. et al. (2013) Cancer genome landscapes. Science, 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. (2009) Genome-wide identification of post-translational modulators of transcription factor activity in human B cells. Nat. Biotechnol., 27, 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D. et al. (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res., 38, W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S. et al. (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res., 34, D668–D672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. et al. (2013) Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res., 41, D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]