Abstract
We present AAI-profiler, a web server for exploratory analysis and quality control in comparative genomics. AAI-profiler summarizes proteome-wide sequence search results to identify novel species, assess the need for taxonomic reclassification and detect multi-isolate and contaminated samples. AAI-profiler visualises results using a scatterplot that shows the Average Amino-acid Identity (AAI) from the query proteome to all similar species in the sequence database. Taxonomic groups are indicated by colour and marker styles, making outliers easy to spot. AAI-profiler uses SANSparallel to perform high-performance homology searches, making proteome-wide analysis possible. We demonstrate the efficacy of AAI-profiler in the discovery of a close relationship between two bacterial symbionts of an omnivorous pirate bug (Orius) and a thrip (Frankliniella occidentalis), an important pest in agriculture. The symbionts represent novel species within the genus Rosenbergiella so far described only in floral nectar. AAI-profiler is easy to use, the analysis presented only required two mouse clicks and was completed in a few minutes. AAI-profiler is available at http://ekhidna2.biocenter.helsinki.fi/AAI.
INTRODUCTION
Whole-genome shotgun sequencing has propelled the re-evaluation of taxonomic classifications and the emergence of single-cell genomics is vastly expanding knowledge about biodiversity (1). In all these application domains, direct comparison of sequence data is a quicker way to get an overview of taxonomic and phylogenetic relationships than searching the original literature on taxonomic classification. Unfortunately, metadata in sequence databases can be out-of-date, using old synonyms or be entirely misclassified. Correct metadata is important because many inference methods test the congruence of sequence trees with the species tree (taxonomy) assuming that species assignments of the sequences are correct. Such applications include tree reconciliation to identify speciation, gene duplication events (2) and lateral gene transfer events (3), lowest common ancestor (LCA) approaches for taxonomic profiling in metagenomics (4) and assignment of the last common ancestor taxon to a cluster of sequences (5).
Pairwise overall genomic relatedness indices (OGRIs (6)) have gained popularity in species discovery and delineation in recent years. OGRIs overcome some of the limitations of gene-based computational tests such as the limited resolution of 16S rRNA gene sequences and missing data in Multilocus Sequence Analysis (MLSA) (7). Measures of overall genomic relatedness include the Karlin genomic signatures, Average Nucleotide Identity (ANI), Average Amino Acid Identity (AAI), supertrees, and in silico Genome-to-Genome Distance Hybridization (GGDH) (8). In particular, a switch is anticipated from the classic polyphasic to a genomic microbial taxonomy to avoid phenotypic characterization using time-consuming laboratory tests (8–11).
Here, we introduce AAI-profiler, a user-friendly web server which computes AAI between a query proteome and all target species in the Uniprot database. In contrast to several existing tools (12–15) that only compare either two species, a small predefined set of species or are directed to metagenomics, AAI-profiler takes the proteome of just one species as input and automatically searches the protein database for species with similar proteins. AAI-profiler is similar to the MiGA web site (http://microbial-genomes.org/) that implements a suite of metagenome and genome comparative analyses including AAI distance searches. Using SANSparallel (16) instead of BLAST, AAI-profiler has a faster response time but the most important difference is that the Uniprot database searched by AAI-profiler has a larger representation of species (809 540 unique labels) than the prokaryotic reference genome collections searched by MiGA (1927 references in NCBI RefSeq and 11 566 references in NCBI Prok).
AAI-profiler is powered by SANSparallel (16), a fast homology search tool. The homology search detects neighbours down to ∼50% sequence identity, which is sufficient to identify neighbouring bacterial genera or mammalian families. The query species is represented by its proteome (protein sequences in FASTA format). Comparing amino acid sequences rather than nucleotide sequences allows AAI-profiler to be practically applied to eukaryotes. Eukaryotic genomes are hundreds to thousands of times longer than bacterial genomes but, given their lower gene density, a eukaryote's proteome is typically only ten times larger than that of a bacterium. For example, the Escherichia coli and ferret genomes are about 5 Mb and 2.4 Gb long and encode about 5000 and 48 000 proteins, respectively (NCBI genomes, https://www.ncbi.nlm.nih.gov/genome). Using SANSparallel, AAI-profiler is capable of processing a bacterial proteome in a few minutes and a eukaryotic proteome in an hour.
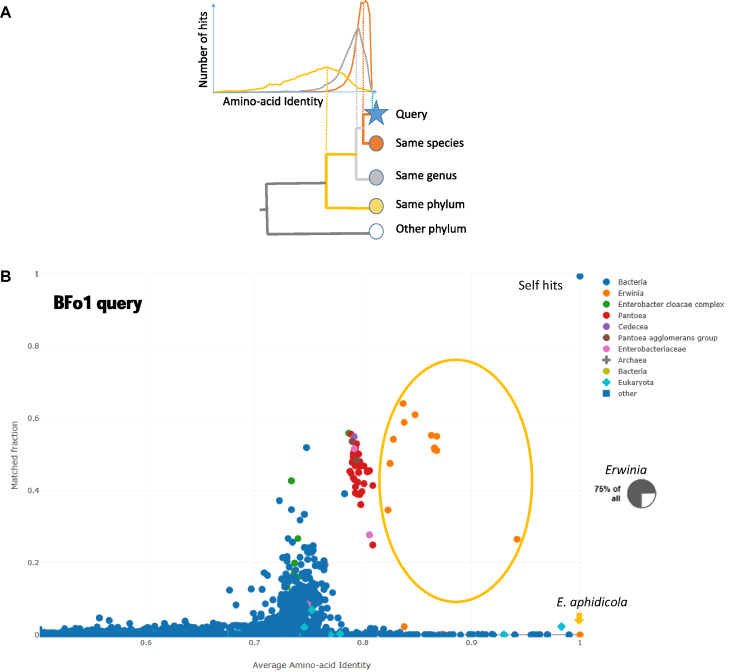
The results of AAI-profiler are presented in a two-dimensional scatterplot. The horizontal axis is AAI, where several sources put the species boundary ∼95% AAI (11). Sister species and sister genera have successively lower AAI values (Figure 1A). The vertical axis is coverage, i.e. the proportion of matched protein pairs. Species with completely sequenced genomes give more robust AAI estimates than species with small samples, because different protein families evolve at different rates. The data points in the scatterplot are coloured according to taxonomic groupings, making it easy to visually spot exceptions to the expected monophyletic pattern. One expects that taxa are monophyletic and therefore distances within a taxon should be smaller than distances between species from different taxa. Thus, the expected pattern shows the species and genus of the query proteome in a uniformly coloured cluster at the highest AAI values (Figure 1B). In many cases, however, we see incongruently labeled species interspersed within this ‘self’ cluster. At high coverage of the query proteome (high ‘matched fraction’), these exceptions are due to misclassified or mislabeled samples. At lower coverage, they signal contamination or, possibly, horizontal gene transfer.
Figure 1.
(A) The principle of AAI-profiler, using colour to indicate database species related to the Query species at species level (orange), genus level (light gray), or phylum level (yellow). Top: Distributions of pairwise sequence identities between proteins of the Query proteome and their best match in a species in the database. Bottom: Cladogram showing nested taxonomic groupings. Different genes evolve at different rates, broadening the distributions of taxa which are more distantly related to the Query species. The vertical lines indicate that, for a given Query proteome, the proteome-wide average of the pairwise sequence identities (Average Amino-acid Identity, AAI) correlates with taxonomic distance. (B) AAI-profiler scatterplot for bacteria symbiont BFo1 of Frankinella occidentalis. Selected bacterial genera are highlighted by different colours. The dot nearest coordinates (1,1) is the query species. Species of the genus Erwinia occupy the range AAI > 0.9. The inset shows a pie chart from the taxonomic profile view: the majority of query proteins have a closest match in Erwinia species.
MATERIALS AND METHODS
System architecture
AAI-profiler makes use of three servers: the web server for handling user requests, SANSparallel for protein homology searches (16) and DictServer for associating taxonomic metadata to the search results. SANSparallel and DictServer are maintained by our group (see http://ekhidna2.biocenter.helsinki.fi/sanspanz/) with monthly updates of the Uniprot sequence and taxonomy databases downloaded from http://uniprot.org. The computations in AAI-profiler are done by a Python script. All plots are generated using Plotly (https://plot.ly/javascript/) and KronaTools (https://github.com/marbl/Krona/wiki) (17).
AAI computation
We compute one-sided and bidirectional AAI profiles for a query proteome (protein sequences in FASTA format). We use SANSparallel to retrieve homologous proteins from Uniprot. Species information is retrieved from the OS tag in the Uniprot headers. Taxonomic metadata is retrieved from the DictServer. For each query protein, we retain the match to all database species with the highest bitscore. One-sided AAI profiles are based on a many-to-one mapping from query proteins to the target proteins of a database species. We define multiplicity as the number of query proteins having a match in the target species divided by the number of distinct target proteins. For example, if some protein family has expanded in the query species to a large number of paralogs, multiplicity can be larger than one. The effect can be notable if (pseudo)proteins encoded by transposable elements are included in the query proteome. Bidirectional AAI profiles are based on a one-to-one mapping, where we exclude the match of a query protein to a database protein if a higher scoring match exists for either sequence. The multiplicity of bidirectional hits is one by definition. The counts of matches per species are tallied in sequence identity bins which have a width of 1%. Sequence identity is computed per aligned positions in the alignment returned by SANSparallel. AAI is the average of sequence identities of all matched pairs between the query proteome and a database species, where each query protein has a weight of one. Query proteins with no matches (as reported by SANSparallel) have zero weight.
Scatterplots
The server shows scatterplots similar to Figure 1B. The horizontal axis shows AAI between the query and database species. The average is computed over the best match per database species over those query proteins, for which SANSparallel reports a match. The vertical axis shows the fraction of query proteins that have a match in the species. Species with higher AAI (to the right) are more closely related to the query than species with lower AAI (further to the left). If the query proteome is present in Uniprot, you see a dot near the top right corner (1.0, 1.0). Related species form a cloud to the left-and-down. More distantly related taxa have low AAI and low coverage because match counts are based on ∼100 nearest hits in the database. At the bottom, there is a band of matches from species for which only individual proteins have been sequenced. Data points are coloured according to genus (bacteria) or order (eukaryota). Eukaryotic species are marked as diamonds, bacteria as circles, archaea as crosses and anything else (viruses, metagenomes, unclassified samples) as squares.
AAI histograms
The server shows histograms of the distribution of AAI values of the top ranked species (top part of Figure 1A). Species are ranked based on the product of AAI and coverage, i.e. the sum of sequence identity values over all matched query proteins. Comparing species with fully sequenced genomes, the mode shifts to lower AAI values and the peak gets wider as the species diverge. Sometimes you see a low ranked species with a narrow peak at higher AAI values; its position in the ranking is then due to a low total count of matches.
Taxonomic profiles
The main purpose of AAI-profiler is to show the species neighbours of the query proteome. As a side product, we produce a taxonomic profile akin to metagenomics analysis. The taxonomic profile is based on mapping each query protein to the closest neighbour in the database, and binning the queries based on this target species. The target species counts are weighted by the percent-identity of the match to the query. The profile shows the frequencies of target species and can be inspected at different levels of the taxonomic hierarchy. When the query species is already included in the database, it will dominate the taxonomic profile, so we additionally generate a second taxonomic profile excluding hits to the top ranked species.
Availability
AAI-profiler is available as a web server at http://ekhidna2.biocenter.helsinki.fi/AAI. The scripts can also be downloaded and run locally using remote databases.
RESULTS
Here, we discuss examples of AAI-profiler analyses. We use published data and corroborate many findings described in the scientific literature. The AAI-profiler generated plots for all examples are viewable online at http://ekhidna2.biocenter.helsinki.fi/AAI/examples/.
Taxonomic identification (The good, …)
Facey et al. (18) took a comprehensive comparative genomics approach to investigate two prominent bacterial symbionts (BFo1 and BFo2) isolated from geographically separated populations of western flower thrips (Frankliniella occidentalis), an important pest insect in agriculture. They concluded that BFo1 is a close relative to Erwinia aphidicola and that BFo2 represents a highly novel species that may be related to known Pantoea. These conclusions are confirmed by AAI-profiler analysis (which only takes two mouse clicks). What is more, we identify the true identity of BFo2.
BFo1 clearly clusters with the genus Erwinia (Figure 1B). The closest match is to Erwinia aphidicola, which has only a few proteins in the database. Other Erwinia species (orange cloud) match with higher AAI values than other genera, as expected for a monophyletic clade. The small pie chart shows the fraction of query proteins whose nearest non-self-neighbours come from Erwinia.
Highlighting misclassification (… the bad, …)
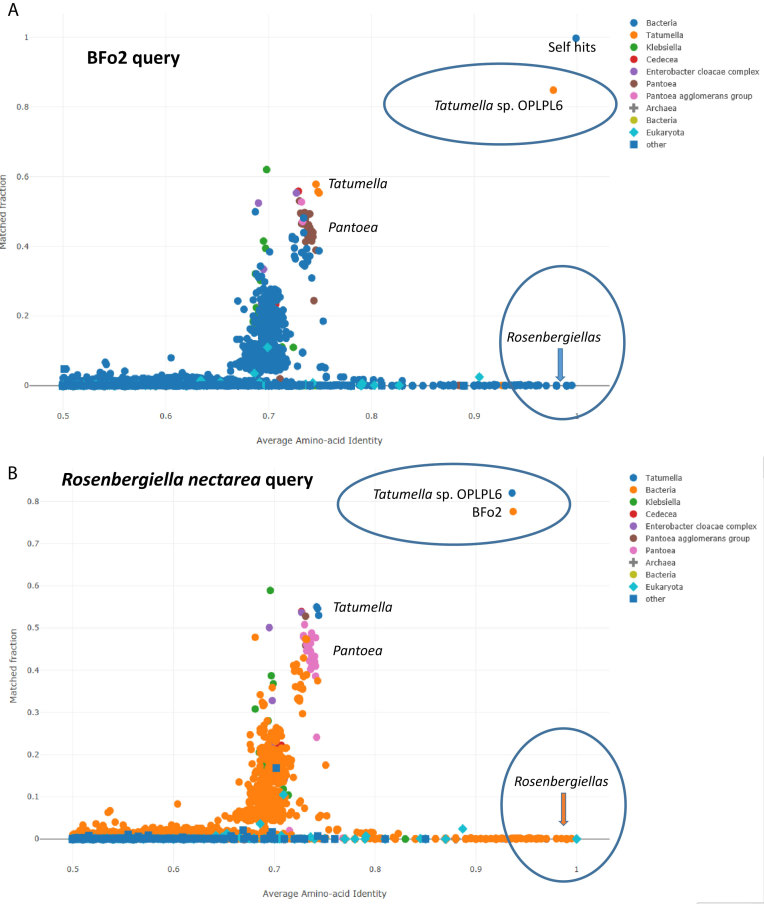
The second symbiont of Frankliniella occidentalis, BFo2, matches a tentatively assigned Tatumella sp. OPLPL6 (19) at 98% AAI (Figure 2A). However, known Tatumella species and other Erwiniaceae pile up around 75% AAI, suggesting a phylum-level relationship to Tatumella (13). The closest matches near the horizontal axis include four Rosenbergiella (20–21) species (Figure 2A). At the time of writing, the Uniprot database contained only 19 proteins from four Rosenbergiella species. Fortunately, the genome of Rosenbergiella nectarea has been sequenced and its predicted proteome is available from NCBI (https://www.ncbi.nlm.nih.gov/genome/?term=Rosenbergiella). A reverse search using the proteome of R. nectarea as query shows that the nearest matches are bacteria symbiont BFo2 of F. occidentalis and Tatumella sp. OPLPL6 at 94% AAI (Figure 2B). The high AAI to R. nectarea identifies strains OPLPL6 and BFo2 as belonging to the genus Rosenbergiella. To further validate the proposed reclassification, we used the MAFFT server (22) to generate multiple sequence alignments and phylogenetic trees of the rpoB, gyrB and atpD proteins of R. epipactidis and 100 homologues retrieved with SANSparallel. The four Rosenbergiella species, OPLPL6 and BFo2 formed a monophyletic clade within Erwiniaceae in all trees with 99%, 95% and 87% bootstrap support, respectively (data not shown).
Figure 2.
(A) AAI-profiler scatterplot for bacteria symbiont BFo2 of Frankinella occidentalis. The dot nearest coordinates (1,1) is the query species. (B) AAI-profiler scatterplot of Rosenbergiella nectarea. Only a few Rosenbergiella proteins are included in the Uniprot database. The query proteome was obtained from NCBI genomes. Rosenbergiella, OPLPL6 and BFo2 form a closely related group which is distinct from other genera within Erwiniaceae around 75% AAI.
Contamination in bacterial pan proteomes (… and the ugly)
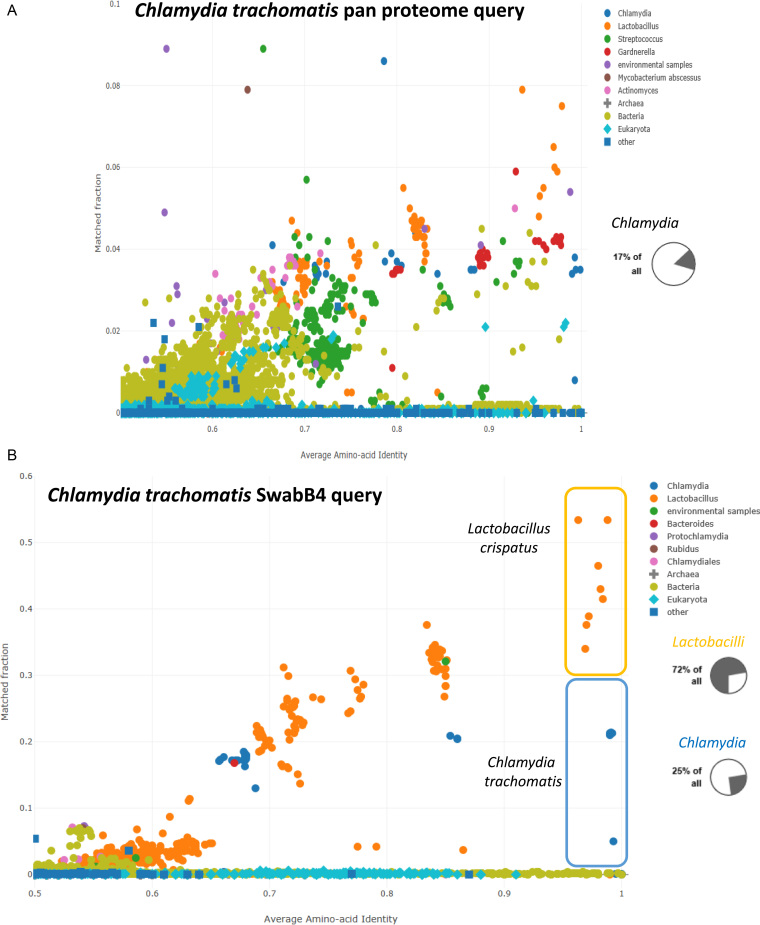
There are a small number of species which occur repeatedly in an unexpected position in AAI-profiler scatterplots. One of these cases is Chlamydia trachomatis. For example, the taxonomic profile of Lactobacillus crispatus 125-2-CHN shows 10% Chlamydia trachomatis. Tracking down the origin of this label required a bit of detective work. Chlamydia are taxonomically classified in a separate order Chlamydiales. Many strains of these pathogens have been sequenced. The AAI profiles of several Chlamydia strains show no trace of Lactobacilli. It turns out that Uniprot's organism source (OS = ) metadata can use the species name as the label for several strains. Thus, the information about which strain was sequenced is hidden from AAI-profiler. In the December 2017 release of Uniprot, the Chlamydia trachomatis (CHLTH) pan proteome was composed of 16 strains and contained 25 858 proteins. The whole pan proteome has an extremely diverse composition (Figure 3A). The taxonomic profile showed a mixture of only 17% Chlamydiales (self-hits to Chlamydia trachomatis were excluded), 38% Firmicutes, 25% Actinobacteria, 8% Bacteroidetes, 6% Tenericutes and 3% Fungi. The protein counts of the component strains range from 884 to 7320. One of these strains, SwabB4 (http://www.uniprot.org/proteomes/UP000044845), has 3922 predicted proteins and is a mixture of 72% Lactobacillus and 25% Chlamydia (Figure 3B). Most likely, the genomic samples of other strains that were incorporated into the pan proteome are multi-isolates like SwabB4. Clearly, a check of genome quality with AAI-profiler would be beneficial to ensure high quality derived data in databases.
Figure 3.
The Chlamydia trachomatis pan proteome illustrates data contamination due to mislabeled multi-isolate samples. (A) The AAI-profiler scatterplot of the Chlamydia pan proteome shows a complex mixture of several species. The pie chart (inset from taxonomic profile view) shows that a minor fraction of query proteins in the Chlamydia trachomatis pan proteome have a nearest match in Chlamydia. (B) The AAI-profiler scatterplot of Chlamydia trachomatis strain SwabB4 shows a superposition of two species (Lactobacillus crispatus [orange dots] and Chlamydia trachomatis [blue dots]). The pie charts (inset from taxonomic profile view) show the proportion of the two genera in the sample.
DISCUSSION
The main uses of AAI-profiler are in exploratory analysis and quality control in selecting data sets for comparative genomics. AAI-profiler reports sequence-based distances from the query proteome to other species. One strength of AAI-profiler is that it reveals inconsistencies in database metadata, unlike taxonomic profiling, and that it requires only one query proteome, unlike servers that generate pairwise AAI histograms or distance matrices from a user-defined set. At present, AAI-profiler is a visualization tool but the AAI concept can also be used to train classifiers for the identification of taxonomic affiliation (13).
By performing a proteome-wide search, AAI-profiler can pick up neighbour species with high AAI despite their being sparsely represented in the database. Such cases are easily missed by phylogenetic analyses of selected gene or protein families. A case in point is the analysis of BFo2. AAI-profiler analysis revealed a close relationship between an unclassified (BFo2) and a misclassified (Tatumella sp. OPLPL6) symbiont of two insect species and placed them in the genus Rosenbergiella, which has several well-characterized species with type strains (20–21). The two insect symbionts and Rosenbergiella are linked ecologically by a food chain. OPLPL6 (19) colonizes insects of the genus Orius (minute pirate bug), which feed mostly on smaller insects, including thrips, but will also feed on pollen and vascular sap. BFo2 (18) was isolated from the gut of Frankliniella occidentalis (western flower thrip), which lays its eggs in plants, often flowers. Finally, several Rosenbergiella species have been isolated from floral nectar, and have spread worldwide using insect vectors (21).
Many inconsistencies revealed by AAI-profiler have been noted before by sequence database curators, but they are buried in notes and remarks in an unsystematic way. AAI-profiler performs the search on the whole proteome and presents a summary report. On the NCBI website, many species with whole-genome sequences of multiple strains have links to a Genome Tree report. For some, but not all, species with multiple strains, NCBI genomes shows a dendrogram based on genomic BLAST and presents clade identifiers and a precomputed Genome neighbour report. Uniprot has clustered all proteins into unstructured sets (5). To our knowledge, the scatterplot visualization of sequence neighbours is unique to AAI-profiler. A similar looking plot of a large number of pairwise species comparisons has been used to propose absolute AAI cut-off values for taxonomic classification (23). In our experience with AAI-profiler, species and genus boundaries can be tighter or more relaxed in different branches of the taxonomy. Relative to a given query, however, AAI distances increase monotonically in a nested hierarchy and this test of monophyly is easily assessed with AAI-profiler.
DATA AVAILABILITY
AAI-profiler is an open source script available from our website (http://ekhidna2.biocenter.helsinki.fi/AAI/#download).
ACKNOWLEDGEMENTS
We thank P. Koirala, J. Taskinen and J. Tommila for discussion.
FUNDING
Academy of Finland [292589]; University of Helsinki Institute of Life Sciences (HiLife). Funding for open access charge: HiLife.
Conflict of interest statement. None declared.
REFERENCES
- 1. Parks D.H., Rinke C., Chuvochina M., Chaumeil P.-A., Woodcroft B.J., Evans P.N., Hugenholtz P., Tyson G.W.. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017; 2:1533–1542. [DOI] [PubMed] [Google Scholar]
- 2. Zmasek C.M., Eddy S.R.. A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics. 2001; 17:821–828. [DOI] [PubMed] [Google Scholar]
- 3. Ravenhall M., Škunca N., Lassalle F., Dessimoz C.. Inferring horizontal gene transfer. PLOS Comput. Biol. 2015; 11:e1004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huson D.H., Auch A.F., Qi J., Schuster S.C.. MEGAN analysis of metagenomic data. Genome Res. 2007; 17:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H.. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015; 31:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chun J., Rainey F.A.. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 2014; 64:316–324. [DOI] [PubMed] [Google Scholar]
- 7. Konstantinidis K.T., Tiedje J.M.. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol. 2007; 10:504–509. [DOI] [PubMed] [Google Scholar]
- 8. Thompson C.C., Chimetto L., Edwards R.A., Swings J., Stackebrandt E., Thompson F.L.. Microbial genomic taxonomy. BMC Genomics. 2013; 14:913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Richter M., Rossello-Mora R.. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vandamme P., Peeters C.. Time to revisit polyphasic taxonomy. Antonie Van Leeuwenhoek. 2014; 106:57–65. [DOI] [PubMed] [Google Scholar]
- 11. Konstantinidis K.T., Tiedje J.M.. Towards a Genome-Based taxonomy for prokaryotes. J. Bacteriol. 2005; 187:6258–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Richter M., Rosselló-Móra R., Glöckner F.O., Peplies J.. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016; 32:929–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo C., Rodriquez-R L.M., Konstantinidis K.T.. MyTaxa: an advanced taxonomic classifier for genomic and metagenomics sequences. Nucleic Acids Res. 2014; 42:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Konstantinidis K.T., Tiedje J.M.. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu J., Blom J., Glaeser S.P., Jaenicke S., Juhre T., Rupp T., Schwengers O., Spanig S., Goesmann A.. A review of bioinformatics platforms for comparative genomics. Recent developments of the EDGAR 2.0 platform and its utility for taxonomic and phylogenetic studies. J. Biotechnol. 2017; 261:2–9. [DOI] [PubMed] [Google Scholar]
- 16. Somervuo P., Holm L.. SANSparallel: Interactive homology search against Uniprot. Nucleic Acids Res. 2015; 43:W24–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ondov B.D., Bergman N.H., Phillippy A.M.. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011; 12:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Facey P.D., Meric G., Hitchings M.D., Pachebat J.A., Hegarty M.J., Chen X., Morgan L.V., Hoeppner J.E., Whitten M.M., Kirk W.D. et al. . Draft genomes, phylogenetic reconstruction, and comparative genomics of two novel cohabiting bacterial symbionts isolated from frankliniella occidentalis. Genome Biol. Evol. 2015; 7:2188–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen X., Hitchings M.D., Mendoza J.E., Balanza V., Facey P.D., Dyson P.J., Bielza P., Del Sol R.. Comparative genomics of facultative bacterial symbionts isolated from european orius species reveals an ancestral symbiotic association. Front. Microbiol. 2017; 8:1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lenaerts M., Alvarez-Perez S., de Vega C., Van Assche A., Johnson S.D., Willems K.A., Herrera C.M., Jacquemyn H., Lievens B.. Rosenbergiella australoborealis sp.nov., Rosenbergiella collisarenosi sp.nov. and Rosenbergiella epipactidis sp.nov., three novel bacterial species isolated from floral nectar. Syst. Appl. Microbiol. 2014; 37:402–411. [DOI] [PubMed] [Google Scholar]
- 21. Halpern M., Fridman S., Atamna-Ismaeel N., Izhaki I.. Rosenbergiella nectarea gen. nov., sp. nov., in the family Enterobacteriaceae, isolated from floral nectar. Int J. Syst. Evol. Microbiol. 2013; 63:4259–4265. [DOI] [PubMed] [Google Scholar]
- 22. Katoh K., Rozewicki J., Yamada K.D.. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief.Bioinformatics. 2017; doi:10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodriquez-R L.M., Konstantinidis K.T.. Bypassing cultivation to identify bacterial species. Microbe. 2014; 9:111–118. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
AAI-profiler is an open source script available from our website (http://ekhidna2.biocenter.helsinki.fi/AAI/#download).