Abstract
The identification and interpretation of genomic variants play a key role in the diagnosis of genetic diseases and related research. These tasks increasingly rely on accessing relevant manually curated information from domain databases (e.g. SwissProt or ClinVar). However, due to the sheer volume of medical literature and high cost of expert curation, curated variant information in existing databases are often incomplete and out-of-date. In addition, the same genetic variant can be mentioned in publications with various names (e.g. ‘A146T’ versus ‘c.436G>A’ versus ‘rs121913527’). A search in PubMed using only one name usually cannot retrieve all relevant articles for the variant of interest. Hence, to help scientists, healthcare professionals, and database curators find the most up-to-date published variant research, we have developed LitVar for the search and retrieval of standardized variant information. In addition, LitVar uses advanced text mining techniques to compute and extract relationships between variants and other associated entities such as diseases and chemicals/drugs. LitVar is publicly available at https://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/LitVar.
INTRODUCTION
Most biomedical knowledge is available as unstructured information in scholarly publications (1). While multiple databases provide structured knowledge on variations (2–5), they heavily rely on manual curation, and thus need advanced variation-oriented text-mining tools to improve the annotation process (6). The task of linking omics data (fields of study in biology ending in -omics such as genomics and proteomics) with scientific literature is further complicated by multiple synonyms and abbreviations used by researchers to refer to one variant, gene, disease or chemical in publications (7). Hence, finding comprehensive and contextualized information about a specific genomic variation becomes an arduous task, as researchers and healthcare professionals rely on curated databases or keyword-based search engines (8,9) that are not suitable for the variety of formats and complexity in which a variation can be cited in literature. Consequently, a variant-oriented semantic search system, which improves the quality (sensitivity and specificity) of search results, is greatly needed. To date, a handful of automatic tools have attempted to address this issue. For instance, command-line automatic variation detection tools such as EMU (10), MutationFinder (11) or Nala (12) can recognize variation mentions in text and return the results in wNm format (e.g. ‘A146T’), while SETH (13) and tmVar (6) can further map the extracted mentions to the specific dbSNP identifiers (e.g. rs121913527). A number of web applications have also been developed to provide an improved search for variants, but they generally accept only specific variant identifiers (14), or are limited to variant information found in abstracts (15,16). GeneView (17) is a recent system which allows semantic search of variants (with or without gene information), but its search results do not include context information and are not always normalized to specific variants (Supplementary Table S1).
Here, we present LitVar, a novel tool that combines robust and advanced text mining, with data integrations from PubMed, PubMed Central Open Access Subset (hereafter called ‘PMC-OA’), dbSNP (5), and ClinVar (4) for the accurate search of variants and related information from unstructured human-related biomedical literature. Compared to PubMed, LitVar offers multiple advantages in variant searching. First, LitVar uses tmVar (6,18), a high-performance variant name recognition tool, supporting both abstracts and full-text articles (Supplementary Table S2) to normalize different names of the same variant into a unique and standardized form. This enables all matching articles to be returned regardless of the specific queried variant name (e.g. identical results will be returned for ‘A146T’, ‘c.436G>A’ or rs121913527). Second, LitVar combines variant-related literature from PubMed abstracts (>27 million) and PMC-OA full-text articles (>1.8 million) and provides a unified access to both literature resources. This is particularly important as abstracts have much lower biomedical concept coverage compared to full-text articles (19,20). Third, LitVar employs a state-of-the-art entity recognition toolset (6,21–23) as its backend processing method, such that users can explore related chemical and disease information for variants of interest. In addition, users can filter results by publication type (e.g. ‘Review’ or ‘Letter’), publication year (e.g. ‘Last Year’ or ‘Last 2 years’), specific journals, and different elements of a publication (e.g. abstract or table content). Finally, LitVar allows users to download search results and subscribe to Really Simple Syndication (RSS) feeds of the latest literature updates. In addition to providing a user-friendly and interactive interface for human users, LitVar also supports a set of RESTful APIs for computational analysis and open programmatic access to its standardized and normalized variant data.
SYSTEM DESCRIPTION
Literature data process—entity recognition/normalization and relation extraction
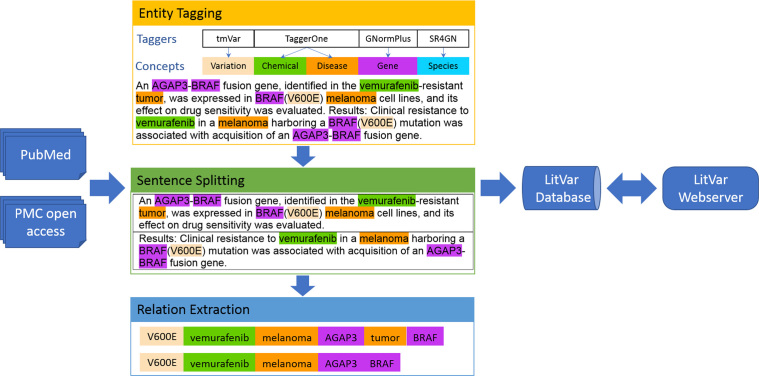
LitVar employs several state-of-the-art text mining and information extraction components in its data processing as shown in Figure 1. First, we processed the entire set of PubMed abstracts and PMC-OA full-text articles in the BioC XML format (24) to extract all variations and their associated entities (i.e. gene, disease, chemical, and species) using a suite of entity taggers, including tmVar for variants, GNormPlus for genes (22), TaggerOne for chemicals and diseases (21) and SR4GN for species (23). Following the lead of GeneView (17), we reported the performance of our taggers on previous benchmarked datasets in the supplementary document (Supplementary Table S3). We then normalized all detected entities to corresponding database identifiers (e.g. MeSH identifiers for chemical and diseases). When possible, we map variants in different forms into dbSNP identifiers (RSIDs). Otherwise, we normalize them into standard HGVS formats. After entity tagging, non-human papers were removed in order to be consistent with dbSNP, and a sentence splitter (25) was applied to segment remaining articles into individual sentences. Finally, we extracted relations between entities based on sentence co-occurrence. Our LitVar data is being updated every month.
Figure 1.
Pre-processing literature data for LitVar. Multiple scripts import publications, detect and normalize biological entities, retrieve relations and continuously update the database.
Query processing
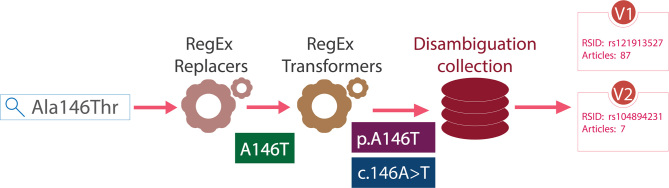
LitVar analyses user queries through a three-step normalization process (Figure 2). First, we use regular expressions to replace amino-acid codes to single-letter codes when applicable. For example, ‘Ala146Thr’ is replaced by ‘A146T.’ Second, we identify the main components of a variant mention (such as the sequence position and mutation type) and rewrite them in HGVS (Human Genome Variation Society) expression. For example, ‘A146T’ is transformed into ‘p.A146T’. Finally, the HGVS expression is used to match LitVar entries with the same name and return results sorted by the number of associated publications. The best match (i.e. the variant with the most publications) is used for the default search results, while other matching variants are also returned to the user for further review.
Figure 2.
LitVar normalizes user queries in real-time.
System implementation details
In LitVar, we aggregate text-mined entities and snippets from PubMed and PMC-OA and store them in a MongoDB database. Our Django web server then processes the requests of both the web application (based on AngularJS, one of the most popular web frameworks) and RESTful API clients. We have chosen both a JSON-like document-oriented database and JSON-oriented front-end to significantly reduce data transformations between storage and visualization of the content. The choice of client-side rendering also allows for better response to user interaction, thus improving the user experience. Currently, LitVar supports most popular web browsers, including the latest versions of Chrome, Safari, Firefox, IE11 and Edge.
RESULTS
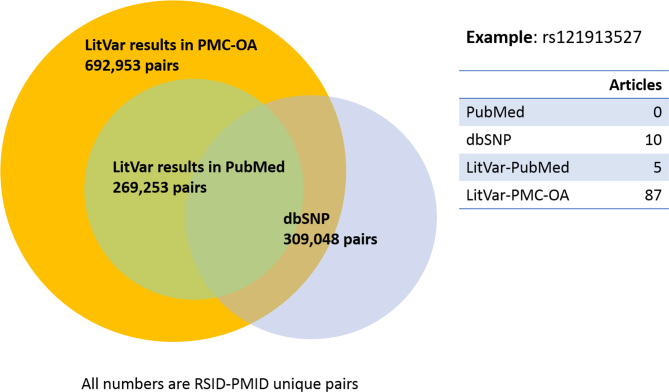
As of March 2018, there are 1 968 872 unique variants in LitVar, of which 852 489 are linked to RSIDs while the rest are expressed in standard HGVS forms. Figure 3 shows that there are 309 048 RSID-PMID links in dbSNP, while LitVar can detect 269 253 and 692 953 links by text-mining the entire PubMed and PMC-OA. On average, LitVar returns twice as many publications as in dbSNP, because of its ability to include many synonymous names. For example, in the case of ‘rs121913527’ as a search query, no results are found in PubMed, 10 results are found in dbSNP, while 87 articles are found in LitVar. As can be seen in Figure 3, some RSID-PMIDs exist only in dbSNP as LitVar is limited to the OA subset of PMC and does not currently process supplementary materials.
Figure 3.
The distribution of variants in PMC-OA, PubMed and dbSNP. All numbers are RSID-PMID unique pairs. Data accessed on 5 April 2018.
FUNCTIONALITY AND USAGE
Website
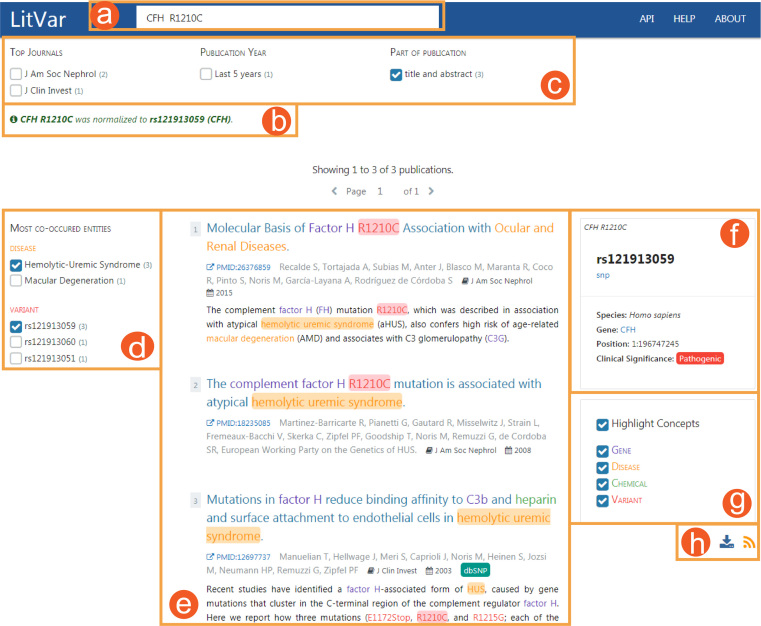
LitVar can be accessed through an easy-to-use graphical web interface, as shown in Figure 4. After a user enters a query in the search bar (Figure 4a), LitVar normalizes the query to find the best matching variant in its database, along with alternative disambiguation results (Figure 4b). Next, LitVar returns a list of publications containing this variant, ordered by publication date. This process has two main features. First, for each result, LitVar returns one or more snippets highlighting the searched variant as well as other entities (e.g. diseases, chemicals, and other variants) which appear the most often in the same sentence as the queried variant (Figure 4d). They are detected in publications during pre-processing and linked to each sentence in our database. This is particularly useful to detect potential relations (e.g. potential implication of a variant in several diseases). In addition to this entity-level filtering, users can also filter publications based on ‘Top Journals’, ‘Publication Year’, ‘Publication Type’ and ‘Part of publication’ matching the query (Figure 4c). Second, in addition to displaying publications associated with the relevant variant (Figure 4e), LitVar also displays a ‘Knowledge Panel’ (Figure 4f) to help users with the most important information about the queried variant, such as its clinical significance (this information is integrated from ClinVar).
Figure 4.
LitVar user interface. Multiple clearly delimited zones allow users to perform search and visualize results. This includes the (a) search, (b) disambiguation, (c) filters, (d) entity facets, (e) list of matching publications, (f) knowledge panel, (g) highlight customization panel and (h) automatic notification by RSS feed and download button.
Programmatic access via RESTful API
In addition to the interactive user interface, LitVar allows users to perform computational analyses through two types of RESTful APIs. The disambiguation API allows programmatic access to the LitVar disambiguation engine, which analyses a free-text query and returns a list of matching variants via VarIDs (a unique variation ID was created for LitVar because some variations could not be linked to an existing RSID after standardization). The second search API allows one to retrieve a list of PMIDs linked to any given variant specified by its VarID.
USE CASES
Below we demonstrate examples of how LitVar may be used under real-world circumstances.
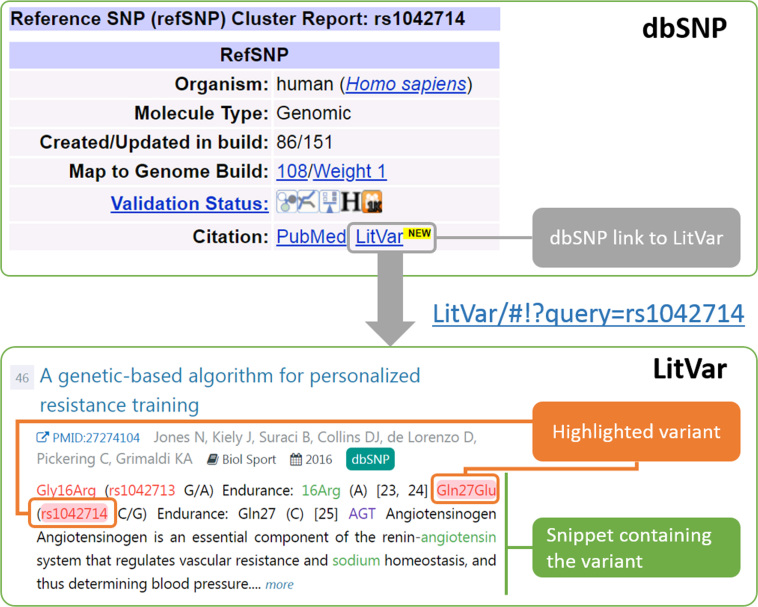
Case 1: citation link from dbSNP
When browsing variant information in dbSNP, researchers can click the citation link, to find and read relevant publications in PubMed. As mentioned earlier, the literature link in dbSNP shows often incomplete results. Hence, a second link to LitVar was added. For example, when searching information about a specific variant on dbSNP website, such as rs1042714, users can click on the LitVar link to review related publications when applicable. The newly added link not only allows users to view more publications (466) than with the link to PubMed (134) but also to display the context in which the variant appears in the publications (Figure 5). Furthermore, we display a small icon if a result in LitVar is the same as in dbSNP.
Figure 5.
LitVar snippets. LitVar displays the queried variation in the context of the sentences in which it appears in the publication. The queried variation has a red background, while other bioconcepts (other variants, genes, diseases, chemicals) are represented by specific colors.
To continue their investigation, the researcher can further restrict the search to articles published in the last two years (51 publications) or use the entity facets on the left sidebar to investigate the link between this variant and a disease such as ‘Hypertension’.
Case 2: variant-specific search
A mutation mention is highly ambiguous as it can refer to different variants located on different genes. Conversely, a single variant can be described in many ways in the literature. For example, c.37G>C, p.Gly13Arg, p.G13R and, ‘glycine to arginine substitution at position 13’ located on gene NRAS, all refer to the same RSID: rs121434595. Hence, searching variants in PubMed suffers both problems of precision and recall. A search with the unique identifier (RSID) in dbSNP, addresses this issue, but as shown in Figure 3, many RSID-PMID links are missing in dbSNP.
For instance, rare variants in the complement factor H (CFH) gene, are associated with age-related macular degeneration (AMD). We start by searching for ‘CFH R1210C’ on LitVar. The best hit, rs121913059, is a highly penetrant rare variant with 48 results in LitVar, compared to seven results in PubMed (with the same query) or five results in dbSNP (with RSID).
Furthermore, in the LitVar search results page, the highlighted snippets allow to easily select an abstract worth further investigation, for example ‘THE PATHOPHYSIOLOGY OF GEOGRAPHIC ATROPHY SECONDARY TO AGE-RELATED MACULAR DEGENERATION AND THE COMPLEMENT PATHWAY AS A THERAPEUTIC TARGET’. This relevant article is only found in LitVar results (i.e. absent in PubMed or dbSNP search results).
CONCLUSIONS
In summary, LitVar improves access to variant-specific information in the biomedical literature. LitVar not only processed the entire set of PubMed abstracts, but also applicable PMC-OA full-text articles. Furthermore, it allows users to examine other related entities, such as diseases and chemicals.
LitVar has several known limitations. First, as a variant search system, LitVar currently only support searches by variant or variant with a gene. Second, variants in the LitVar database are currently limited to those found in the title, abstract, and full texts but not including supplementary materials. Third, LitVar endeavours to recognize a wide variety of variant formats, but a query may still yield zero results in LitVar either because we could not map it to a proper record in our database (e.g. g.28612G>A) or the variant has no associated publications in LitVar (e.g. rs115735611).
LitVar is also bound to the accuracy of the current text mining algorithms, which are known to be imperfect in both entity recognition and relation extraction. For entity tagging, our tools are mostly trained on abstracts, and their results on full text may therefore be inferior due to its structure and complexity (26). For relation extraction, LitVar currently relies on sentence co-occurrence. While it is a robust method for building real-world biological databases and web-servers such as STRING (27) and GeneView (17), its results may include false positives (e.g. when a sentence states that two entities are not related). Recently, there are a few studies showing the potential of using machine learning for extracting associations between variants and specific diseases such as cancer (28,29), but further investigation is warranted for validating and generalizing such methods across diseases and other entities (e.g. chemicals), as well as for testing their performance with full-text articles. In both cases, a large-scale human-annotated corpus would be required.
In the future, we would like to extend the current scope of LitVar by supporting queries containing other types of key entities (such as genes and diseases) and provide keyword-based queries, while continuing to improve LitVar's performance in speed and accuracy. To improve the quality of our relations, we plan to filter out sentences expressing uncertain or negative findings. We also plan to add new filters (a) to display publications that are only found in LitVar (i.e. not existing in other curated databases), as they may be of high interest to some users (e.g. curators), or (b) to show results found in specific sections of an article (e.g. Results versus Discussion section).
DATA AVAILABILITY
LitVar is publicly available at https://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/LitVar.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Shazia Dharssi and Dr Nicolas Fiorini for proofreading our manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This research is supported by the Intramural Research Programs of the National Institutes of Health, National Library of Medicine.Funding for open access charge: the Intramural Research Programs of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Khare R., Leaman R., Lu Z.. Accessing biomedical literature in the current information landscape. Methods Mol. Biol. 2014; 1159:11–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017; 45:D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pundir S., Martin M.J., O’Donovan C.. UniProt protein knowledgebase. Methods Mol. Biol. 2017; 1558:41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sherry S.T., Ward M., Sirotkin K.. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999; 9:677–679. [PubMed] [Google Scholar]
- 6. Wei C.H., Phan L., Feltz J., Maiti R., Hefferon T., Lu Z.. tmVar 2.0: integrating genomic variant information from literature with dbSNP and ClinVar for precision medicine. Bioinformatics. 2018; 34:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lapatas V., Stefanidakis M., Jimenez R.C., Via A., Schneider M.V.. Data integration in biological research: an overview. J. Biol. Res. (Thessalon). 2015; 22:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fiorini N., Lipman D.J., Lu Z.. Towards PubMed 2.0. Elife. 2017; 6:e28801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wei C.H., Kao H.Y., Lu Z.. PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013; 41:W518–W522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doughty E., Kertesz-Farkas A., Bodenreider O., Thompson G., Adadey A., Peterson T., Kann M.G.. Toward an automatic method for extracting cancer- and other disease-related point mutations from the biomedical literature. Bioinformatics. 2011; 27:408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caporaso J.G., Baumgartner W.A. Jr, Randolph D.A., Cohen K.B., Hunter L.. MutationFinder: a high-performance system for extracting point mutation mentions from text. Bioinformatics. 2007; 23:1862–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cejuela J.M., Bojchevski A., Uhlig C., Bekmukhametov R., Kumar Karn S., Mahmuti S., Baghudana A., Dubey A., Satagopam V.P., Rost B.. nala: text mining natural language mutation mentions. Bioinformatics. 2017; 33:1852–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thomas P., Rocktaschel T., Hakenberg J., Lichtblau Y., Leser U.. SETH detects and normalizes genetic variants in text. Bioinformatics. 2016; 32:2883–2885. [DOI] [PubMed] [Google Scholar]
- 14. Liu Y., Liang Y., Wishart D.. PolySearch2: a significantly improved text-mining system for discovering associations between human diseases, genes, drugs, metabolites, toxins and more. Nucleic Acids Res. 2015; 43:W535–W542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee S., Kim D., Lee K., Choi J., Kim S., Jeon M., Lim S., Choi D., Kim S., Tan A.C. et al. BEST: Next-Generation biomedical entity search tool for knowledge discovery from biomedical literature. PLoS One. 2016; 11:e0164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poon H., Quirk C., DeZiel C., Heckerman D.. Literome: PubMed-scale genomic knowledge base in the cloud. Bioinformatics. 2014; 30:2840–2842. [DOI] [PubMed] [Google Scholar]
- 17. Thomas P., Starlinger J., Vowinkel A., Arzt S., Leser U.. GeneView: a comprehensive semantic search engine for PubMed. Nucleic Acids Res. 2012; 40:W585–W591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei C.H., Harris B.R., Kao H.Y., Lu Z.. tmVar: a text mining approach for extracting sequence variants in biomedical literature. Bioinformatics. 2013; 29:1433–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schuemie M.J., Weeber M., Schijvenaars B.J., van Mulligen E.M., van der Eijk C.C., Jelier R., Mons B., Kors J.A.. Distribution of information in biomedical abstracts and full-text publications. Bioinformatics. 2004; 20:2597–2604. [DOI] [PubMed] [Google Scholar]
- 20. Westergaard D., Stærfeldt H.-H., Tønsberg C., Jensen L.J., Brunak S.. A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts. PLOS Computational Biology. 2018; 14:e1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leaman R., Lu Z.. TaggerOne: joint named entity recognition and normalization with semi-Markov Models. Bioinformatics (Oxford, England). 2016; 32:2839–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wei C.H., Kao H.Y., Lu Z.. GNormPlus: An integrative approach for tagging genes, gene families, and protein domains. Biomed. Res. Int. 2015; 2015:918710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei C.H., Kao H.Y., Lu Z.. SR4GN: a species recognition software tool for gene normalization. PLoS One. 2012; 7:e38460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Comeau D.C., Islamaj Dogan R., Ciccarese P., Cohen K.B., Krallinger M., Leitner F., Lu Z., Peng Y., Rinaldi F., Torii M. et al. BioC: a minimalist approach to interoperability for biomedical text processing. Database (Oxford). 2013; 2013:bat064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bird S., Klein E., Loper E.. Natural Language Processing with Python: Analyzing Text with the Natural Language Toolkit. 2009; Sebastopol: O’Reilly Media, Inc. [Google Scholar]
- 26. Lu Z., Kao H.Y., Wei C.H., Huang M., Liu J., Kuo C.J., Hsu C.N., Tsai R.T., Dai H.J., Okazaki N. et al. The gene normalization task in BioCreative III. BMC Bioinformatics. 2011; 12(Suppl. 8):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singhal A., Simmons M., Lu Z.. Text mining Genotype-Phenotype relationships from biomedical literature for database curation and precision medicine. PLoS Comput. Biol. 2016; 12:e1005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lever J., Mungall A.J., Jones S.J.M.. CancerMine: Knowledge Base Construction for Personalised Cancer Treatment. Proceedings of the Joint International Conference on Biological Ontology and BioCreative. 2016; 3–1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LitVar is publicly available at https://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/LitVar.