Abstract
Computational protein docking is a powerful strategy to predict structures of protein-protein interactions and provides crucial insights for the functional characterization of macromolecular cross-talks. We previously developed InterEvDock, a server for ab initio protein docking based on rigid-body sampling followed by consensus scoring using physics-based and statistical potentials, including the InterEvScore function specifically developed to incorporate co-evolutionary information in docking. InterEvDock2 is a major evolution of InterEvDock which allows users to submit input sequences – not only structures – and multimeric inputs and to specify constraints for the pairwise docking process based on previous knowledge about the interaction. For this purpose, we added modules in InterEvDock2 for automatic template search and comparative modeling of the input proteins. The InterEvDock2 pipeline was benchmarked on 812 complexes for which unbound homology models of the two partners and co-evolutionary information are available in the PPI4DOCK database. InterEvDock2 identified a correct model among the top 10 consensus in 29% of these cases (compared to 15–24% for individual scoring functions) and at least one correct interface residue among 10 predicted in 91% of these cases. InterEvDock2 is thus a unique protein docking server, designed to be useful for the experimental biology community. The InterEvDock2 web interface is available at http://bioserv.rpbs.univ-paris-diderot.fr/services/InterEvDock2/.
INTRODUCTION
Computational modeling of protein assemblies provides crucial insights for the functional characterization of macromolecular interactions occurring in the crowded cellular environment. Predictions of protein–protein interfaces can be used to design experiments to investigate the role of important interactions and possibly interfere with them, typically using mutagenesis. Models of macromolecular complexes are also useful to complement integrative structural biology (1) and to deepen our understanding of disease-associated mutations (2) and protein interaction networks (3).
A number of servers have been developed for protein–protein docking, which can be separated into template-based modeling servers, which aim to identify suitable structural templates for the protein complex, and template-free (ab initio) docking servers, which implement sampling of interface structural models followed by scoring protocols. Recent resources to find templates for interface modeling starting from the sequences of two protein partners include KBDOCK (4), focused on domain–domain interactions and PPI3D (5). Recently released servers taking protein sequences as input for homology-based interface modeling include SnapDock (6), HOMCOS (7) and SWISS-MODEL Oligo (8). Many template-free docking servers implement a rigid-body docking approach, sometimes followed by rescoring: PatchDock (9), FireDock (10), HexServer (11), ZDOCK (12), FRODOCK 2.0 (13), pyDockWEB (14), ClusPro (15), GRAMM-X (16), InterEvDock (17). A hybrid approach combining template-based and template-free docking was recently proposed in the HDOCK server (18). Some free docking servers include specific features such as symmetric docking (SymmDock (9), ZDOCK); local docking around an initial guess (RosettaDock (19), recently moved to the ROSIE server (20)); docking with more than two proteins (ClusPro, GRAMM-X); and docking including degrees of flexibility (SwarmDock (21), ATTRACT (22), HADDOCK (23)).
Attempts to address the limitations of computational docking have led to placing increasing focus on data-driven docking (24) and many servers now allow the user to specify interface residues and/or distance restraints, including ZDOCK, FRODOCK 2.0 (for refinement), pyDockWEB, ClusPro, GRAMM-X, SwarmDock, HADDOCK and ATTRACT. Some servers such as ClusPro and pyDockSAXS (25) can specifically use experimental small angle X-ray scattering (SAXS) data.
One of the main features differentiating existing docking servers is the nature of the scoring function used to discriminate correct from incorrect docking models. Most scoring strategies use either physics-based or statistical potentials. Understanding how binding partners co-evolved can provide essential clues to improve the structural prediction of protein interfaces. Following pioneering studies (26–28), the field of covariation-based contact prediction has been very active in recent years, with several servers enabling the prediction of inter-molecular contacts such as EVcomplex (29), GREMLIN (30) and I-COMS (31); however, such methods still have limited applicability due to the difficulty in building large enough joint multiple sequence alignments (MSAs) for the two protein partners. We developed the InterEvScore scoring function incorporating co-evolutionary information into the docking process, which improves predictions for as few as 10 sequences in the joint MSAs (32). We integrated this scoring function into the InterEvDock pipeline (17). InterEvDock is based on rigid-body sampling by FRODOCK (33) followed by re-scoring using the SOAP-PP atomic statistical potential (34) and InterEvScore (32) and consensus model selection. To date, the InterEvDock web server is the only free docking server allowing to directly predict the structure of protein–protein interactions using co-evolutionary information. We successfully used the InterEvDock strategy to guide our predictions in recent Critical Assessment of Predicted Interactions (CAPRI) rounds: for CAPRI rounds 28–35, our group ranked first by making correct predictions for 10 out of 18 targets (35).
Very often, the individual structures of the exact proteins involved in a complex of biological interest are not known. On the other hand, structural models can be obtained for a large fraction of proteins in interaction networks thanks to homology modeling (36), making them amenable to protein–protein docking. To date, most free docking servers, except the HDOCK server, allow users to provide only input structures but no input sequences for the protein partners.
Based on the user-oriented considerations mentioned above, here we introduce the InterEvDock2 server which represents a major evolution over the original InterEvDock. Protein sequences can now be provided as input, and not only 3D structures. To handle sequence inputs, we have added a module that performs comparative modeling prior to docking based on an automatic template search protocol. In case the user has biological input such as a position that is known to be involved in the interface between the two protein partners or a pair of residues known to be in contact, restraints with a tunable distance threshold can be specified for use in the docking procedure. This is crucial to ensure that all available biologically relevant information is used for InterEvDock2 predictions. In addition, InterEvDock2 implements the possibility to submit structures of oligomers as input to the pairwise free docking. Such an option is generally complicated in co-evolution analyses since the joint MSAs have to be generated for every chain of an oligomer. This process is now fully automatized in InterEvDock2, allowing users to submit inputs such as homodimers or more complex structures as that of the nucleosome made of ten subunits. InterEvDock2 also benefits from improved accuracy by integrating the most recent FRODOCK 2.1 algorithm for rigid-body docking and scoring (13) and implementing an improved consensus selection and from a speed-up in the generation of joint MSAs for the two protein partners. The InterEvDock2 pipeline was benchmarked on 812 complexes from the PPI4DOCK database (37) designed to ensure unbiased evaluation of the performance of free docking from unbound homology models. A total of 29% of those 812 cases have an acceptable or better solution among the top 10 consensus models returned by InterEvDock2. As InterEvDock, InterEvDock2 also outputs a list of the 10 residues most likely involved in the interface and at least one residue was correctly predicted in 91% of the 812 benchmark cases.
THE InterEvDock2 SERVER
Web interface
Users are expected to provide for each protein partner either an input sequence or an input structure (Figure 1). Input structures can be uploaded or retrieved automatically from the Protein Data Bank (PDB) by typing in the PDB code and optionally one or more chain identifier(s). More options are available through the ‘advanced options’ menu (see Figure 2 and Supplementary Figure S1). Optional breakpoints can be selected, either after template search to choose among up to 20 suggested templates prior to modeling, or after modeling for interactive visualization of the models prior to docking. When input sequences are provided, users can specify which template to use for homology modeling; as for structure inputs, the template can be uploaded or directly retrieved from the PDB. If providing a template, users can also optionally enforce the query-template alignment for modeling. It is also possible to provide only a query-template alignment obtained from a previous server run in which a template search was performed (without modifying the identifiers), in which case the input sequence and the template PDB will be automatically retrieved based on the alignment. Several options are offered to tune the modeling: by default only loops (insertions) shorter than 14 residues are rebuilt during the modeling and N-terminal and C-terminal extensions are not modeled, but maximal lengths for modeling of loops, N-terminal and C-terminal extensions can be defined by the user. Additionally, for input structures or sequences, users may define constraints that will be used to filter docking solutions; these constraints can be a single interface residue or a pair of residues in contact. Users can optionally specify the distance that will be used for each constraint. An InterEvDock2 session identifier can also be provided in order to re-use docking results from a previous run and test different constraints. As in InterEvDock, users may input the joint MSAs used for co-evolution-based scoring; otherwise the joint MSAs will be built by the server through an automated procedure. In case an oligomeric structure is submitted as one of the two docking partners, the joint MSAs will also be automatically calculated and processed by the server for every chain of the oligomer. A demonstration case using sequences as input to the docking (and optionally a constraint) is available from the InterEvDock2 submission page.
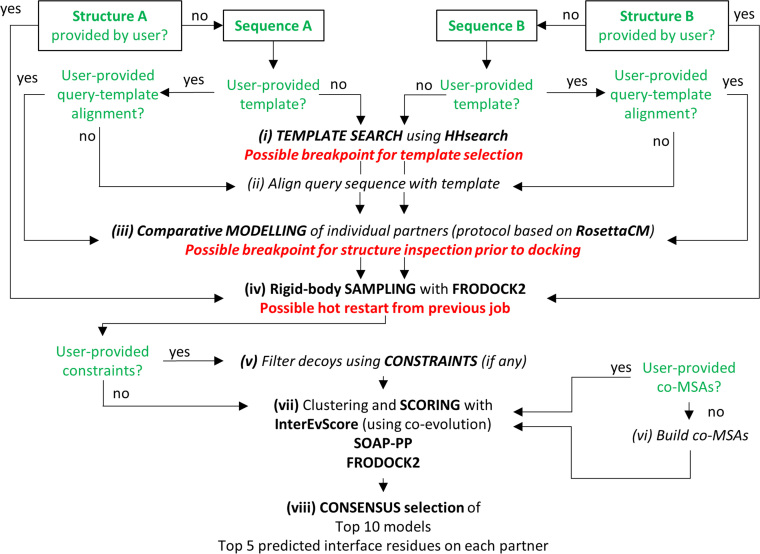
Figure 1.
Workflow of the InterEvDock2 pipeline. Eight steps can be performed in InterEvDock2 depending on the user input, out of which three mandatory steps (iv), (vii) and (viii) are always performed as they were in the original InterEvDock pipeline (17), except that the FRODOCK algorithm was updated to version 2.1 (13) and the consensus calculation was slightly modified to save time and improve results. New features are available allowing the user to provide only an input sequence for one or both partners: (i) if the user does not provide a template, search for a suitable template using HHsearch (38,39); (ii) if the user provides a template but no query-template alignment, alignment of query sequence with template sequence using MAFFT (40); (iii) once a template and a query-template alignment are available for each partner with no user-provided structure, comparative modeling using a RosettaScripts (41) protocol based on RosettaCM (42) to build a 3D model for (at least part of) the input sequence. Once a 3D structure or a structural model is available for each partner: (iv) exhaustive sampling using the rigid-body method FRODOCK 2.1; (v) (new feature) if the user provides information on residues (or pairs of residues) involved in the interface, applying constraints to filter sampled solutions; (vi) if the user does not provide a joint MSA for the two protein partners, co-MSA generation; (vii) clustering and scoring by three scores, FRODOCK 2.1, SOAP-PP (34) and InterEvScore (32); (viii) clustering and selection of the InterEvDock2 consensus top 10 decoys. Green text indicates user input. Red text indicates possible breakpoints and hot restart. Italics indicate the optional steps in the pipeline, depending on the input provided by the user. Details for each step are provided in the Supplementary Methods and Figure S1.
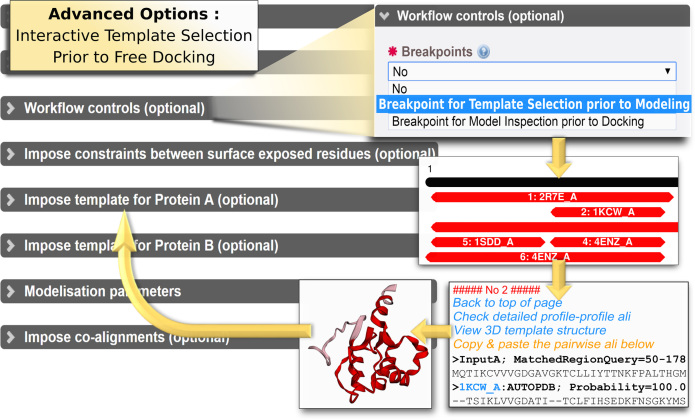
Figure 2.
Illustration of new features available in InterEvDock2. Among advanced options, interactive template selection prior to modeling and docking is now available as an advanced workflow option (breakpoint). The list of templates returned by the server can be interactively explored by the user to select a template. The provided query-template alignment can then be submitted as input to the server for modeling and docking.
The web page resulting from an InterEvDock2 submission contains information about the best ranked decoys, which can be explored interactively thanks to the PV WebGL applet (M. Biasini, https://dx.doi.org/10.5281/zenodo.12620). Detailed results are available in a downloadable archive, also containing a script for easy loading and offline visualization of the best docking solutions with PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC, New York, NY).
The InterEvDock2 server benefits from parallelized implementation in the dedicated infrastructure built at RPBS and from data privacy ensured in the Mobyle framework.
Molecular docking procedure
Figure 1 presents the InterEvDock2 pipeline which consists of eight steps; more details about each step are provided in Supplementary Figures S1 and S2 and in the Supplementary Methods. The three core docking steps – sampling with FRODOCK2.1 (iv), clustering with FRODOCK2.1 and scoring with InterEvScore and SOAP-PP (vii) and consensus calculation (viii) – are always performed. Step (vi) consists in automatically generating the joint MSAs used by InterEvScore to account for co-evolution in the scoring process, unless the joint MSAs are provided by the user. Steps (iv), (vi) and (vii) are unchanged compared to the original InterEvDock pipeline (17), except that the FRODOCK algorithm was updated to version 2.1 (13). In the final step (viii) a consensus list of 10 most likely models is calculated. Since decoys well ranked by at least two different scoring methods (out of the three methods used in InterEvDock2) have higher chances of being correct, the 3*top 10 models for each score are re-ranked according to the number of similar decoys (defined as ligand RMSD ≤ 10 Å) within the top 50 models of the other two scores (down to a minimum of two similar decoys). In case of a tie, priority is given to InterEvScore top 10 models, then SOAP-PP, then FRODOCK. If necessary, the consensus list is then filled up to 10 models by selecting the best models from each score (four from InterEvScore, three from SOAP-PP and three from FRODOCK). When building the consensus, models that are structurally redundant (i.e. ligand RMSD ≤ 10 Å) with previously selected models are excluded, so that the final list contains 10 structurally non-redundant models.
Docking from input sequences
If the user provides only an input sequence for one or both partners, steps (i) to (iii) can be applied. (i) If the user does not provide a template, the profile–profile comparison tool HHsearch is used to search for templates (38,39); only templates with HHsearch probability higher than 95% are selected. The web server returns a list of up to 20 templates selected according to HHsearch probability, query-template sequence identity and structural resolution (see details in the Supplementary Methods). By setting the breakpoint after template search, the user can choose to start modeling from any of these templates by copy-pasting the query-template alignment to the server submission form; otherwise the best template found by the automatic procedure is used. If no suitable template is identified, no modeling is performed. (ii) If the user provides a template but no query-template alignment, the query sequence is aligned with the template sequence using MAFFT (40). (iii) Once a template and a query-template alignment are available for each protein with no user-provided structure, comparative modeling using a RosettaScripts (41) protocol based on RosettaCM (42) is performed to build a 3D model for (at least part of) the input sequence. Due to runtime considerations, compared to the procedure used to build the PPI4DOCK database (37), the comparative modeling protocol implemented in the InterEvDock2 web server involves fewer optimization cycles (see protocol details in the Supplementary Methods). This protocol is quite robust for templates with relatively high homology but it can lead to loss of precision for more remote templates (typically when both templates have <50% sequence identity with the query proteins). By default, to avoid spending time reconstructing regions that are not present in the template, only loops (insertions) shorter than 14 residues are rebuilt during the modeling and N-terminal and C-terminal extensions are not modeled, but maximal lengths for modeling of loops, N-terminal and C-terminal extensions can be tuned by the user.
User-defined constraints
Step (v) applies if the user provides information on residues (or pairs of residues) involved in the interface: restraints are applied to filter sampled solutions. The distance used to enforce restraints can be modulated which offers the possibility to integrate data from various sources. The default distance was set to 8 Å for constraints on single positions and 11 Å for pair constraints (see Supplementary Results for a detailed justification of these thresholds). When constraints are provided by the user, the output returned by the server will provide information about whether or not each constraint was used during docking (e.g. constraints on residues not exposed on the surface of the protein are excluded).
Runtime
The core docking steps (iv), (vii) and (viii) take altogether around 30 min for proteins of size 200 residues and 1 h for proteins of size 400–500 residues. Template search and query-template alignment steps (i) and (ii) take only a few minutes, whatever the size of the proteins. The comparative modeling step (iii) was optimized for speed as reported above and typically takes 5–20 min depending on the size of the proteins and the query-template sequence identities. Compared to InterEvDock, InterEvDock2 benefits from a large speed-up in step (vi) for the generation of joint MSAs for two protein partners which was a key bottleneck. This step now typically lasts ∼3 min for proteins of 200 residues and ∼15 min for proteins of 400–500 residues.
RESULTS
Benchmarking on PPI4DOCK
To assess the predictive power of the InterEvDock2 server on 3D models, we have set up the most extensive benchmark to date, using unbound models as input of the docking simulations. The PPI4DOCK database (37) was designed to ensure unbiased evaluation of free docking performance and contains 1417 non-redundant heterodimeric docking targets based on unbound homology models. The InterEvDock2 pipeline was tested on the subset of 812 protein complexes from PPI4DOCK for which pairs of joint MSAs with more than 10 sequences could be obtained (excluding any antibody complex) and FRODOCK 2.1 (13) was able to generate at least one acceptable or better decoy among the top 50 000 decoys. The list of the 812 complexes used for benchmarking and detailed results are provided in http://bioserv.rpbs.univ-paris-diderot.fr/services/InterEvDock2/table.html. This benchmark dataset is roughly an order of magnitude larger than other typical docking benchmarks, among which the widely used Weng benchmark (43). For each of the 812 targets, PPI4DOCK provides unbound homology models of the two protein partners as well as the joint MSAs used for docking and scoring in the InterEvDock2 pipeline. As on the web server, the predictions for each case consist in the top 10 consensus interface models and the top 10 interface residues, which are used to assess the InterEvDock2 performance. A solution is defined as acceptable or better according to the criteria defined by the CAPRI consortium (44).
The prediction performance of InterEvDock2 is reported in Table 1. Among the 812 targets, 29% (239) have at least one model of acceptable or better quality in the top 10 consensus obtained from the InterEvDock2 pipeline, which represents a significant improvement over the top 10 success rates of the three individual scores used to build the consensus (see also Supplementary Figure S3). The 812 complexes belong to four difficulty levels (PPI4DOCK categories) based on the quality of the superimposed interface model (two unbound models superimposed on the bound structure): ‘very easy’ (174 complexes), ‘easy’ (498 complexes), ‘hard’ (118 complexes) and ‘very hard’ (22 complexes). Other ‘very hard’ and all ‘super hard’ PPI4DOCK targets do not satisfy the condition that FRODOCK 2.1 was able to generate at least one acceptable or better decoy among the top 50 000 decoys, since they may require flexibility in the docking process (37), and are therefore not included in the present benchmark. As expected, the InterEvDock2 top 10 consensus success rate decreases with increasing difficulty of the test cases, from 43% for the ‘very easy’ PPI4DOCK category to 30% for the ‘easy’ category, 11% for the ‘hard’ category and 5% for the ‘very hard’ category. Analysis of InterEvDock2 performance depending on the minimum sequence identity between the target and template shows a moderate drop in success rate for models built with remote templates (<30% sequence identity) and an increased success rate for models built with very close templates (≥95% sequence identity), compared to the overall InterEvDock2 success rate (see Supplementary Table S1).
Table 1. Prediction performance of the InterEvDock2 server on 812 complexes of the PPI4DOCK benchmark, split into four levels of difficulty: very easy, easy, hard and very hard.
| All | Very easy | Easy | Hard | Very hard | ||
|---|---|---|---|---|---|---|
| Number of cases | 812 | 174 | 498 | 118 | 22 | |
| Top 10 success rate | InterEvScore | 171 (21%) | 44 (25%) | 115 (23%) | 11 (9%) | 1 (5%) |
| SOAP_PP | 194 (24%) | 55 (32%) | 126 (25%) | 12 (10%) | 1 (5%) | |
| FRODOCK 2.1 | 164 (20%) | 55 (32%) | 102 (20%) | 5 (4%) | 2 (9%) | |
| InterEvDock2 consensus | 239 (29%) | 75 (43%) | 150 (30%) | 13 (11%) | 1 (5%) | |
| Zdock 3.0.2 | 126 (15%) | 33 (19%) | 83 (17%) | 9 (8%) | 1 (5%) | |
| Residue interface prediction (≥1 correct in top 5 receptor OR top 5 ligand) | InterEvDock2 | 735 (91%) | 160 (92%) | 450 (90%) | 103 (87%) | 22 (100%) |
| Zdock3.0.2 | 680 (84%) | 145 (83%) | 427 (86%) | 91 (77%) | 17 (79%) | |
| Residue interface prediction (≥1 correct in top 5 receptor AND top 5 ligand) | InterEvDock2 | 414 (51%) | 103 (59%) | 263 (53%) | 39 (33%) | 9 (41%) |
| Zdock3.0.2 | 345 (43%) | 76 (44%) | 228 (46%) | 33 (28%) | 8 (34%) | |
| Residue interface prediction (≥1 correct in top 1 receptor OR top 1 ligand) | InterEvDock2 | 613 (75%) | 140 (80%) | 385 (77%) | 71 (60%) | 17 (77%) |
| Zdock3.0.2 | 532 (66%) | 111 (64%) | 344 (69%) | 64 (54%) | 13 (58%) | |
| Residue interface prediction (≥1 correct in top 1 receptor AND top 1 ligand) | InterEvDock2 | 278 (34%) | 75 (43%) | 184 (37%) | 17 (14%) | 2 (9%) |
| Zdock3.0.2 | 195 (24%) | 44 (25%) | 133 (27%) | 15 (12%) | 3 (14%) |
The benchmark is made of the 812 targets of the PPI4DOCK benchmark (1417 cases) (37) for which pairs of co-evolved MSAs with more than 10 sequences could be obtained and FRODOCK 2.1 (13) was able to generate at least one acceptable or better decoy (44) among the top 50 000 decoys. In the upper part of the table, top 10 success rates are reported as the number of cases (and percentage between brackets) for which at least one model out of 10 is an acceptable or better solution. Assessed methods are InterEvScore (32), SOAP-PP (34), FRODOCK 2.1 (13), InterEvDock2 consensus (this work and (17)) and Zdock3.0.2 (45). In the lower part of the table, the number (and percentage) of cases for which at least one residue out of the top 10 or top 2 residues was correctly predicted as present in the complex interface is assessed for InterEvDock2 and Zdock3.0.2 (details are provided in the Supplementary Data). The best results for each category are highlighted in bold.
Direct comparisons with previous benchmarks are difficult because the benchmark dataset used here is much larger than other datasets typically used to assess docking and scoring performance. Comparison with previously reported success rates on the Weng benchmark (17,43) are provided in Supplementary Tables S2 and S3. An interesting feature of the Weng benchmark compared to PPI4DOCK is that it contains targets where one partner is multimeric. Out of the 85 cases from the Weng benchmark that can be used for InterEvDock2 benchmarking, 16 contain a multimeric partner. The InterEvDock2 top 10 consensus contains an acceptable or better solution for 7 out of these 16 cases (44%). This success rate is comparable to the overall success rate of InterEvDock2 on the much larger PPI4DOCK benchmark (29%) and on the 85 cases of the Weng benchmark (32%). Additionally, docking using multimeric partners has the advantage that potentially ‘sticky’ interface regions involved in multimeric interactions of one partner are buried in the multimeric interface and therefore masked for the docking process.
In Table 1, the InterEvDock2 performance is also compared to the performance of the widely used rigid-body docking program Zdock3.0.2 (45), assessed on the same 812 complexes from the PPI4DOCK benchmark. For each case, 54 000 decoys are generated and ranked by Zdock3.0.2. In 126 out of 812 cases (15%), an acceptable or better solution is found among the top 10 decoys. Altogether, these benchmarking results highlight the added value of the InterEvDock2 processing pipeline, in particular the clustering and consensus scoring steps.
Of key interest for experimental biologists, the InterEvDock2 output offers a list of 10 residues most likely involved in the complex interface (5 predicted residues on each partner) that can be targeted for mutagenesis. For these residue predictions, we reach 91% success rate, with 735 of the 812 benchmark cases having at least one of the 10 predicted residues involved in the actual interface (Table 1). As was found for the 85 cases from the Weng benchmark used to assess the original InterEvDock performance (17), this success rate is remarkably stable with increasing difficulty: from 92% for very easy cases to 90% for easy cases to 87% for hard cases. Predictions of the InterEvDock2 server can also be used as a prior to constrain more thorough docking simulations including flexibility. In that perspective, in 51% of the cases, at least one correct residue is predicted on both sides of the interface (59% for very easy targets, 53% for easy targets and 33% for hard targets). Results are also presented in Table 1 and Supplementary Figure S4 for only the top 2 predicted residues (one on each partner): at least one of the two predicted residues is correct in 75% of the cases and both are correct in 34% of the cases, highlighting the practical value of InterEvDock2 residue prediction. All those results are significantly higher than a reference interval given by random selection of residues on the surface of the protein (see Supplementary Methods and Figure S4).
Predictions of CAPRI targets
The InterEvDock2 pipeline was challenged through our participation in all CAPRI rounds since 2013. Focusing on heteromeric targets evaluated at the sixth CAPRI evaluation meeting (rounds 28–35), our group ranked first with 10 correctly predicted targets out of 18. Among those 10 targets, our best prediction among 10 submitted models was of high quality in one case, medium quality in seven cases and acceptable quality in two cases (35). In 15 of the total 18 targets, evolutionary information was available in the form of either co-evolution or conservation, providing key constraints to guide docking toward the correct solution. Although the InterEvDock2 pipeline was not specifically designed to handle homo-oligomeric docking, we were also among the highest ranking groups in the two joint CASP-CAPRI experiments involving mostly predictions of homodimers (46,47). Of note, for most CAPRI targets since 2013, only sequence information was provided to the participants. Figure 3 illustrates an InterEvDock2 run for CAPRI target T95 (round 31) involving docking between the nucleosome (a decameric structure) and the PRC1 ubiquitin ligase (a trimeric structure).
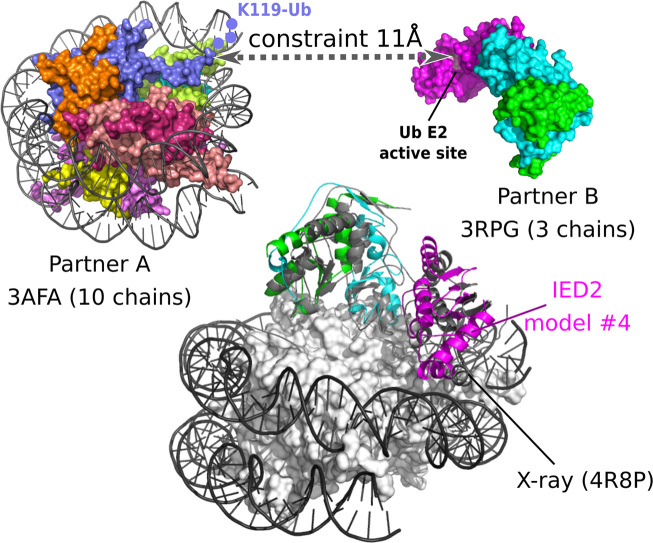
Figure 3.
Successful example of docking from multimeric inputs in a CAPRI target. Prediction for CAPRI target T95 involving docking between two multimeric inputs: the nucleosome and the PRC1 ubiquitin ligase. These multimeric inputs were directly used as inputs in the InterEvDock2 server (PDB identifier: 3afa for the nucleosome and 3rpg for the ubiquitin ligase). A constraint is additionally used between a residue close to the ubiquitinated lysine K119 and the active site of the ubiquitin ligase (constraint between residues 117C and 85A at distance 11 Å). The first acceptable solution (ranked #4 in the Top 10 InterEvDock2 consensus) is superimposed on the reference crystal structure (PDB identifier: 4r8p).
Description of docking case studies from input sequences and using constraints
To illustrate the biological relevance of InterEvDock2 predictions, we consider two docking case studies derived from the PPI4DOCK benchmark (Figure 4). The first case is a complex between the Rho-family GTPase Cdc42 and the conserved, catalytic domains of exchange factor intersectin. Details of this interaction (PDB identifier: 1ki1) and structure-based mutagenesis revealed key features of the activation of Cdc42 by intersectin (48). This case was tested on the InterEvDock2 server by providing input sequences of the interacting regions in the two partners. Unbound templates were imposed for both proteins as in the PPI4DOCK benchmark; otherwise the automatic template search might have found the bound partners belonging to PDB identifier: 1ki1 or other bound templates. The unbound templates (4f38A and 3odoA) have sequence identities of 54 and 25% with the modeled regions of Cdc42 and intersectin, respectively. Among the top 10 consensus models returned by InterEvDock2, one acceptable solution is found as top 2 (Figure 4A).
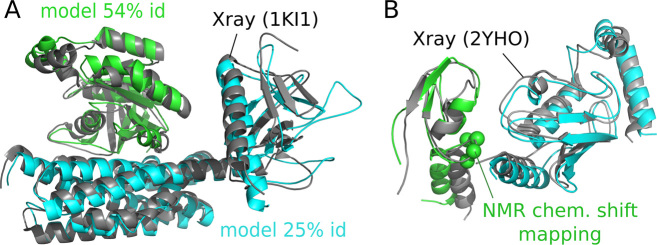
Figure 4.
Successful examples from the PPI4DOCK database. (A) Top 2 consensus model found by InterEvDock2 for docking between unbound homology models of Cdc42 (green, modeled using an unbound template at 54% sequence identity) and the conserved, catalytic domains of intersectin (cyan, modeled using an unbound template at 25% sequence identity). The model is superimposed on the reference crystal structure (PDB identifier: 1ki1) (gray). It is acceptable with interface RMSD 4.03 Å. (B) Best model found in the InterEvDock2 top 10 consensus for docking between PPI4DOCK unbound homology models of the RING domain of IDOL (green) and UBE2D (cyan) when four residues experimentally known to be important for the interaction are used as constraints (with default distance 8 Å). The model is superimposed on the reference crystal structure (PDB identifier: 2yho) (gray). The model is acceptable with interface RMSD 2.29 Å and is ranked first of the top 10 consensus. The four residues used as constraints from chemical shift mapping are shown as green spheres (M388, V389, C390 and C391).
The second case illustrates the interest of docking with user-defined restraints. We consider a complex between the RING domain of E3 ubiquitin protein ligase IDOL and ubiquitin-conjugating enzyme E2 UBE2D (PDB identifier: 2yho) (49). This interaction is involved in the regulation of cholesterol uptake. Nuclear magnetic resonance (NMR) chemical shift mapping was used to confirm the interacting region prior to crystallographic studies. This NMR analysis showed four residues (M388, V389, C390 and C391) in the RING domain of IDOL to have particularly high chemical shift variation upon binding of UBE2D. The PPI4DOCK models of the interacting regions of IDOL and UBE2D (built by homology modeling using unbound templates for the two proteins, respectively, 2yhnA and 3bzhA with sequence identities of 100 and 61%) were submitted to InterEvDock2. Two runs were performed, one without constraints and one using the four residues identified by NMR as interface constraints. Among the top 10 consensus models returned by InterEvDock2, the highest-ranked acceptable solution (medium quality according to the CAPRI criteria) was ranked number six in the run without constraints. When using the constraints derived from experimental NMR data, there were two acceptable or better solutions in the InterEvDock2 top 10 consensus: one was ranked first (Figure 4B) and the second ranked sixth.
CONCLUSION
InterEvDock2 represents a major, user-oriented evolution of InterEvDock. InterEvDock2 is still the only free docking server taking into account co-evolutionary information, relying on a combination of complementary scoring functions to identify the most likely interface models. The previous InterEvDock version was limited by its requirement of only dealing with monomeric inputs. InterEvDock2 greatly expands the range of applications to homo- and hetero-oligomers by handling multimeric chains in the two input proteins used for pairwise docking and the automated processing of their joint MSAs. Benchmarking results on PPI4DOCK emphasize the usefulness of InterEvDock2 in generating interface models of good quality in the scope of integrative structural biology. The InterEvDock2 server returns docking results within typical runtimes of 30 min (for proteins of around 100 residues) to 2 h (for proteins of around 500 residues) even when starting from input sequences, while performing well on our benchmark of 812 cases docked from unbound homology models. The server also benefits from a user-friendly submission and visualization interface, including breakpoints after template search and homology modeling, and options for offline in-depth analysis with PyMOL. InterEvDock2 is thus designed as a useful tool for biologists who can very easily submit docking runs starting from simple input sequences and specify constraints to make use of any previously acquired experimental knowledge. InterEvDock2 results can assist biologists in designing hypotheses about molecular interaction mechanisms and interface mutations to investigate the functional role of an interaction.
Supplementary Material
ACKNOWLEDGEMENTS
InterEvDock2 and Zdock3.0.2 benchmarking on PPI4DOCK was done through granted access to the HPC resources of CCRT under the allocations 2015-7078, 2016-7078 and 2017-7078 by GENCI (Grand Equipement National de Calcul Intensif). We are very grateful to Pablo Chacon (FRODOCK 2.1), Andrej Sali (SOAP-PP), Tal Pupko (Rate4Site), Marco Biasini (ProteinViewer), Johannes Söding (HHblits and HHsearch) and the RosettaCommons community for making their methods available for implementation in the InterEvDock2 server.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French Infrastructure for Integrated Structural Biology (FRISBI) [ANR-10-INSB-05-01]; Agence Nationale de la Recherche ANR-IA-2011-IFB [ANR-11-INBS-0013]; CHIPSET [ANR-15-CE11-0008-01]. Funding for open access charge: Agence Nationale de la Recherche Grant CHIPSET [ANR-15-CE11-0008-01].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ward A.B., Sali A., Wilson I.A.. Biochemistry. Integrative structural biology. Science. 2013; 339:913–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gress A., Ramensky V., Kalinina O.V.. Spatial distribution of disease-associated variants in three-dimensional structures of protein complexes. Oncogenesis. 2017; 6:e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vakser I.A. Protein-protein docking: from interaction to interactome. Biophys. J. 2014; 107:1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ghoorah A.W., Devignes M.D., Smail-Tabbone M., Ritchie D.W.. Classification and exploration of 3D protein domain interactions using Kbdock. Methods Mol. Biol. 2016; 1415:91–105. [DOI] [PubMed] [Google Scholar]
- 5. Dapkunas J., Timinskas A., Olechnovic K., Margelevicius M., Diciunas R., Venclovas C.. The PPI3D web server for searching, analyzing and modeling protein-protein interactions in the context of 3D structures. Bioinformatics. 2017; 33:935–937. [DOI] [PubMed] [Google Scholar]
- 6. Estrin M., Wolfson H.J.. SnapDock-template-based docking by geometric hashing. Bioinformatics. 2017; 33:i30–i36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawabata T. HOMCOS: an updated server to search and model complex 3D structures. J. Struct. Funct. Genomics. 2016; 17:83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bertoni M., Kiefer F., Biasini M., Bordoli L., Schwede T.. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017; 7:10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J.. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005; 33:W363–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H.J.. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008; 36:W229–W232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Macindoe G., Mavridis L., Venkatraman V., Devignes M.D., Ritchie D.W.. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010; 38:W445–W449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T., Weng Z.. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014; 30:1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramirez-Aportela E., Lopez-Blanco J.R., Chacon P.. FRODOCK 2.0: fast protein-protein docking server. Bioinformatics. 2016; 32:2386–2388. [DOI] [PubMed] [Google Scholar]
- 14. Jimenez-Garcia B., Pons C., Fernandez-Recio J.. pyDockWEB: a web server for rigid-body protein-protein docking using electrostatics and desolvation scoring. Bioinformatics. 2013; 29:1698–1699. [DOI] [PubMed] [Google Scholar]
- 15. Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S.. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017; 12:255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tovchigrechko A., Vakser I.A.. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006; 34:W310–W314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu J., Vavrusa M., Andreani J., Rey J., Tuffery P., Guerois R.. InterEvDock: a docking server to predict the structure of protein-protein interactions using evolutionary information. Nucleic Acids Res. 2016; 44:W542–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yan Y., Zhang D., Zhou P., Li B., Huang S.Y.. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017; 45:W365–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lyskov S., Gray J.J.. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008; 36:W233–W238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moretti R., Lyskov S., Das R., Meiler J., Gray J.J.. Web-accessible molecular modeling with Rosetta: the Rosetta online server that includes everyone (ROSIE). Protein Sci. 2018; 27:259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Torchala M., Moal I.H., Chaleil R.A., Fernandez-Recio J., Bates P.A.. SwarmDock: a server for flexible protein-protein docking. Bioinformatics. 2013; 29:807–809. [DOI] [PubMed] [Google Scholar]
- 22. de Vries S.J., Schindler C.E., Chauvot de Beauchene I., Zacharias M.. A web interface for easy flexible protein-protein docking with ATTRACT. Biophys. J. 2015; 108:462–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Vries S.J., van Dijk M., Bonvin A.M.. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010; 5:883–897. [DOI] [PubMed] [Google Scholar]
- 24. Rodrigues J.P., Bonvin A.M.. Integrative computational modeling of protein interactions. FEBS J. 2014; 281:1988–2003. [DOI] [PubMed] [Google Scholar]
- 25. Jimenez-Garcia B., Pons C., Svergun D.I., Bernado P., Fernandez-Recio J.. pyDockSAXS: protein-protein complex structure by SAXS and computational docking. Nucleic Acids Res. 2015; 43:W356–W361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lichtarge O., Bourne H.R., Cohen F.E.. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 1996; 257:342–358. [DOI] [PubMed] [Google Scholar]
- 27. Pazos F., Helmer-Citterich M., Ausiello G., Valencia A.. Correlated mutations contain information about protein-protein interaction. J. Mol. Biol. 1997; 271:511–523. [DOI] [PubMed] [Google Scholar]
- 28. de Juan D., Pazos F., Valencia A.. Emerging methods in protein co-evolution. Nat. Rev. Genet. 2013; 14:249–261. [DOI] [PubMed] [Google Scholar]
- 29. Hopf T.A., Scharfe C.P., Rodrigues J.P., Green A.G., Kohlbacher O., Sander C., Bonvin A.M., Marks D.S.. Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife. 2014; 3:e03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ovchinnikov S., Kamisetty H., Baker D.. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife. 2014; 3:e02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iserte J., Simonetti F.L., Zea D.J., Teppa E., Marino-Buslje C.. I-COMS: interprotein-COrrelated mutations server. Nucleic Acids Res. 2015; 43:W320–W325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andreani J., Faure G., Guerois R.. InterEvScore: a novel coarse-grained interface scoring function using a multi-body statistical potential coupled to evolution. Bioinformatics. 2013; 29:1742–1749. [DOI] [PubMed] [Google Scholar]
- 33. Garzon J.I., Lopez-Blanco J.R., Pons C., Kovacs J., Abagyan R., Fernandez-Recio J., Chacon P.. FRODOCK: a new approach for fast rotational protein-protein docking. Bioinformatics. 2009; 25:2544–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dong G.Q., Fan H., Schneidman-Duhovny D., Webb B., Sali A.. Optimized atomic statistical potentials: assessment of protein interfaces and loops. Bioinformatics. 2013; 29:3158–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu J., Andreani J., Ochsenbein F., Guerois R.. Lessons from (co-)evolution in the docking of proteins and peptides for CAPRI Rounds 28-35. Proteins. 2017; 85:378–390. [DOI] [PubMed] [Google Scholar]
- 36. Mosca R., Ceol A., Aloy P.. Interactome3D: adding structural details to protein networks. Nat. Methods. 2013; 10:47–53. [DOI] [PubMed] [Google Scholar]
- 37. Yu J., Guerois R.. PPI4DOCK: large scale assessment of the use of homology models in free docking over more than 1000 realistic targets. Bioinformatics. 2016; 32:3760–3767. [DOI] [PubMed] [Google Scholar]
- 38. Remmert M., Biegert A., Hauser A., Soding J.. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2011; 9:173–175. [DOI] [PubMed] [Google Scholar]
- 39. Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005; 21:951–960. [DOI] [PubMed] [Google Scholar]
- 40. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fleishman S.J., Leaver-Fay A., Corn J.E., Strauch E.M., Khare S.D., Koga N., Ashworth J., Murphy P., Richter F., Lemmon G. et al. . RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One. 2011; 6:e20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song Y., DiMaio F., Wang R.Y., Kim D., Miles C., Brunette T., Thompson J., Baker D.. High-resolution comparative modeling with RosettaCM. Structure. 2013; 21:1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hwang H., Vreven T., Janin J., Weng Z.. Protein-protein docking benchmark version 4.0. Proteins. 2010; 78:3111–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mendez R., Leplae R., De Maria L., Wodak S.J.. Assessment of blind predictions of protein-protein interactions: current status of docking methods. Proteins. 2003; 52:51–67. [DOI] [PubMed] [Google Scholar]
- 45. Pierce B.G., Hourai Y., Weng Z.. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One. 2011; 6:e24657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lensink M.F., Velankar S., Kryshtafovych A., Huang S.Y., Schneidman-Duhovny D., Sali A., Segura J., Fernandez-Fuentes N., Viswanath S., Elber R. et al. . Prediction of homoprotein and heteroprotein complexes by protein docking and template-based modeling: A CASP-CAPRI experiment. Proteins. 2016; 84(Suppl. 1):323–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lensink M.F., Velankar S., Baek M., Heo L., Seok C., Wodak S.J.. The challenge of modeling protein assemblies: The CASP12-CAPRI experiment. Proteins. 2018; 86:273–257. [DOI] [PubMed] [Google Scholar]
- 48. Snyder J.T., Worthylake D.K., Rossman K.L., Betts L., Pruitt W.M., Siderovski D.P., Der C.J., Sondek J.. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat. Struct. Biol. 2002; 9:468–475. [DOI] [PubMed] [Google Scholar]
- 49. Zhang L., Fairall L., Goult B.T., Calkin A.C., Hong C., Millard C.J., Tontonoz P., Schwabe J.W.. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 2011; 25:1262–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.