Abstract
Gene expression regulation is highly dependent on binding of RNA-binding proteins (RBPs) to their RNA targets. Growing evidence supports the notion that both RNA primary sequence and its local secondary structure play a role in specific Protein-RNA recognition and binding. Despite the great advance in high-throughput experimental methods for identifying sequence targets of RBPs, predicting the specific sequence and structure binding preferences of RBPs remains a major challenge. We present a novel webserver, SMARTIV, designed for discovering and visualizing combined RNA sequence and structure motifs from high-throughput RNA-binding data, generated from in-vivo experiments. The uniqueness of SMARTIV is that it predicts motifs from enriched k-mers that combine information from ranked RNA sequences and their predicted secondary structure, obtained using various folding methods. Consequently, SMARTIV generates Position Weight Matrices (PWMs) in a combined sequence and structure alphabet with assigned P-values. SMARTIV concisely represents the sequence and structure motif content as a single graphical logo, which is informative and easy for visual perception. SMARTIV was examined extensively on a variety of high-throughput binding experiments for RBPs from different families, generated from different technologies, showing consistent and accurate results. Finally, SMARTIV is a user-friendly webserver, highly efficient in run-time and freely accessible via http://smartiv.technion.ac.il/.
INTRODUCTION
RNA binding proteins (RBPs) are involved in regulating the different steps of the gene expression pathway by binding to coding and non-coding RNAs. Most regulatory RBPs bind their RNA target in a specific manner. Accumulating data support that the RNA recognition requires identification of both the sequence and the local structure attributes of the binding sites (1). In the last decade, different high-throughput RNA binding technologies have been developed to study the binding of RBPs, providing information on the binding preferences of hundreds of diverse RBPs. To determine the binding preferences of RBPs in-vitro Ray et al. introduced RNAcompete and applied it to a large cohort of RBPs from human and Drosophila melanogaster (2,3). More recently, Cook et al. introduced RNAcompete-S, which combines a single-step in-vitro selection methodology with a dedicated computational pipe-line to extract the intrinsic sequence and structural specificity of RBPs (4). A different method, named RNA Bind-n-Seq (RBNS), was developed for quantitative mapping of RNA binding specificity in-vitro and was applied to study the binding specificities of several splicing factors, such as RBFOX2, CELF1/CUGBP1 and MBNL1 (5). In parallel, high-throughput methods to identify endogenous protein–RNA interactions were developed. The first in-vivo based methods (such as RIP (6), RIP-chip (7), RIP-seq (8)) relied solely on RNA immunoprecipitation. Later, higher resolution methodologies, combining crosslinking and immunoprecipitation (CLIP), have been developed to study RBP specificities in-vivo and identify the specific interaction sites of the protein on the RNA target. Over the years, many different variants of the CLIP method have been introduced, including HITS-CLIP (9), PAR-CLIP (10), iCLIP (11), eCLIP (12), irCLIP (13) and others. For recent review on CLIP advances see Lee and Ule (14). In accordance with the growing number of experimental high-throughput RNA binding data, many different algorithms have been developed to extract the RNA preferences of RBPs. The binding preferences of RBPs can be retrieved from several databases, such as RBPDB (15), CISBP-RNA (3), ATtRACT (16).
Despite the accumulating data generated from the large variety of in-vivo and in-vitro experimental methods, deciphering the binding preferences of RBPs at both the sequence and the structural level is still a great challenge. To address this, during the last decade several different algorithms have been developed. Based on the assumption that most RBPs preferably bind in RNA accessible regions, Hiller et al. developed MEMERIS, that implements the Expectations Minimization motif discovery algorithm (MEME) to specifically search for RNA binding motifs in single-stranded RNA regions (17). A decade later the GraphProt machine learning based algorithm was introduced by the same group for learning the binding preferences of RBPs at the sequence and structural levels from experimental high-throughput binding data, without considering any prior knowledge on the structural preferences of the RBPs (18). The GraphProt model considers the generic shape of the RNA substructures, learned from RNA shape predictions (19). A different algorithm to models the sequence and structural preferences of RBPs is the RNAcontext algorithm (20). RNAcontext takes into account the probability of the sequences to be in different types of RNA secondary structure (paired, hairpin loop, bulges etc.), predicted by the RNA folding algorithm RNAplfold (21) with no prior assumptions on the RBP binding preferences. RNAcontext was implemented in the RBPmotif webserver that can be applied to both in-vitro and in-vivo binding data (22). RBPmotif webserver takes as an input a set of bound and unbound sequences and outputs the results of the RBP binding preferences at the sequence and structural levels, presenting the sequence preferences in a standard motif logo representation with additional information on the probabilities of the secondary structure of the motif as modeled for the entire binding site. The RCK k-mer based motif discovery algorithm (23) was developed for extracting the sequence and structural binding preferences specifically from RNAcompete data (2,3) and more recently implemented to predict the binding preferences of RBPs from a large set of in-vitro and in-vivo data (24). The TEISER (Tool for Eliciting Informative Structural Elements in RNA) algorithm employs a different computational approach for extracting enriched sequence and structural motifs for high-throughput data (25). TESIER uses context-free grammars (CFGs) (26) to model the RNA secondary structure preferences of the RBPs and was originally designed for discovering enriched sequence and structure motifs of RBPs associated with RNA stability, learning from whole genome expression data. TIESER was employed for extracting the sequence and structural preferences of RBPs from CLIP data (27). Recently, we have introduced the SMARTIV algorithm, which is a highly efficient algorithm for discovering combined sequence and structure motifs (28). The uniqueness of SMARTIV algorithm is that it combines the RNA sequence and secondary structure information in a single representation. The combined information is then used to extract enriched k-mers that are further clustered to generate Position Weight Matrices (PWMs), representing the joint sequence and structural preferences of the protein. By this approach SMARTIV algorithm selects in a one-step manner the overrepresented combined sequence and structure motifs as opposed to adding structural information to the enriched sequence motifs. Moreover, SMARTIV provides a variety of models for RNA secondary structure prediction, including free energy minimization, ensemble and abstract shape models. Finally, SMARTIV uses a novel approach, employing the minimum-minimum Hyper Geometric (mmHG) statistics (29,30) to extract enriched motifs in ranked data and assigns an occurrence score and P-values to PWMs based on the correspondence between the input data (sorted by the experimental information) and the output list (sorted by the assigned PWM score). Later, Heller et al. (31) developed ssHMM to identify combined sequence and structure motifs from RBP-bound sequences based on Hidden Markov Models and Gibbs sampling. Similar to SMARTIV, ssHMM uses a combined sequence a structure alphabet for extracting enriched motifs, which are visualized in a graph representation.
Here, we describe a new webserver we name SMARTIV, a Sequence and Structure Motif enrichment Analysis tool for Ranked RNA daTa generated from In-Vivo binding experiments, that employs our recently developed SMARTIV algorithm for discovering combined RNA sequence and structure motifs from in-vivo data (28). The input for SMARTIV is a list of sequences generated from a high-throughput binding experiment, specifically CLIP-based experiment. SMARTIV outputs the best combined sequence and structure motif generated from a given input dataset in a unified graphical logo representation and as a PWM with an assigned P-value. For motif representation SMARTIV uses an eight-letter alphabet that represents the sequence and structure preferences per each nucleotide. The occurrences of the enriched sequence and structure motifs in the original data are also provided in both html and text formats. SMARTIV is freely available via the website http://smartiv.technion.ac.il/.
SMARTIV METHODOLOGY
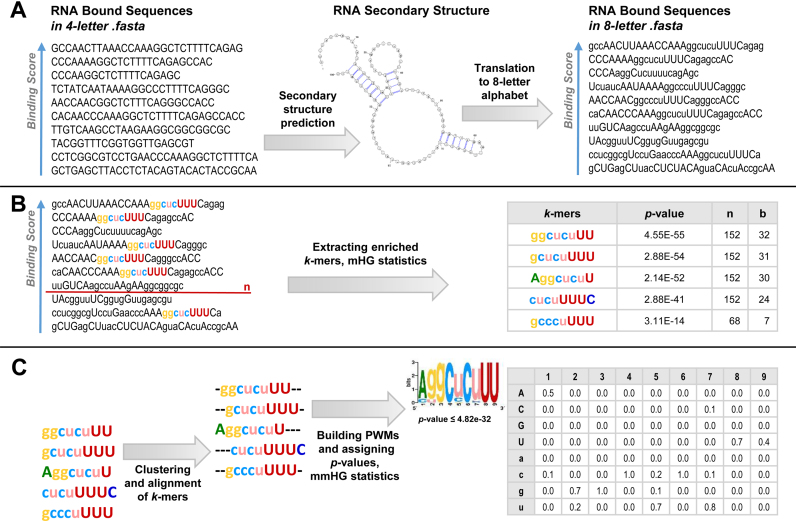
SMARTIV webserver is based on our previously developed algorithm for discovering combined sequence and structure motifs in RNA (28). A workflow summarizing the main steps of SMARTIV algorithm is given in Figure 1. Briefly, SMARTIV uses as an input a list of ranked RNA sequences from any type of CLIP experiment (PAR-CLIP, iCLIP, eCLIP etc.) that were processed by the appropriate peak calling algorithm and ranked according to a calculated sequence score/value in a descending order. As a first step, we employ RNA secondary structure predictions to the sequences, defining each nucleotide in the sequence as either paired or unpaired and integrate the sequence and structural information to a new eight-letter alphabet (A,G,C,U for unpaired nucleotides and a,g,c,u for paired nucleotides). In SMARTIV webserver, we implemented different RNA secondary structure prediction approaches: the Minimum Free Energy (MFE) (21), the Maximum Expected Accuracy (MEA) and centroid structures calculated from partition function, all three generated by the RNAfold method (21). In addition we employed a probability approach to retrieve the MFE structures from most probable shapes, using RNAshapes (32,33). Following the folding step, SMARTIV extracts k-mers, over the eight-letter alphabet, that are significantly enriched at the top of the ranked list compared to the bottom of the list, using the minimum Hyper Geometric (mHG) statistics that has been implemented in our previous DRIMUST algorithm (34). Consequently, we cluster and align the k-mers, build a Position Weight Matrix (PWM) for each cluster, select motifs (28,35), and assign P-values to PWMs (29,30). A detailed description of the algorithm is given in Polishchuk et al. (28).
Figure 1.
A visualized summary of SMARTIV methodology. (A) The input for SMARTIV is a list of sequences ranked in a descending order according to the sequence binding scores. As a first step, we employ secondary structure predictions, defining each nucleotide in the sequence as either paired or unpaired and integrate the sequence and structural information to a new eight-letter alphabet (A,G,C,U for unpaired nucleotides and a,g,c,u for paired nucleotides). (B) We extract k-mers that are significantly enriched at the top of the ranked list compared to the bottom of the list, using the mHG statistics. (C) We cluster and align the k-mers. Consequently, we build a Position Weight Matrix (PWM) for each cluster, assigning it a P-value based on its correspondence to the original ranking of the sequences, based on the experimental binding scores, using the mmHG statistics.
Input
SMARTIV server is designed for discovering combined sequence and structure binding motifs for RBPs from in-vivo high-throughput experiments. The data uploaded by the user can be generated from any type of CLIP experiment, either from an in-house experiment, data downloaded from GEO (36), ENCODE (37) or from dedicated CLIP databases, as for example, DoRiNA (38), or CLIPdb (39). An input list of the RBP-target sequences can be provided as coordinates in BED format or as sequences in FASTA format. Data in BED format must contain chromosome coordinates and strand information. The user must specify the species and a genome assembly information for the uploaded dataset (current version supports human hg18, hg19, hg38 and mouse mm9 and mm10 genomes assemblies). When the sequences are provided in FASTA format (with no coordinate information) SMARTIV employs the BLAT alignment tool (40) to map sequences to the genome and extend the sequences for the folding prediction step. A list of input sequences must contain at least 2000 sequences, each comprising of at least 20 nucleotides. Given that SMARTIV employs a ranked-based algorithm to extract enriched motifs, it is suitable only for ranked lists, sorted by any given sequence score. Sequence scores are usually generated by the dedicated peak-calling algorithms that process the raw binding data generated by the different CLIP experiments (such as Piranha (41) that can be applied to all available CLIP-based methods or PARalyser (42) that is dedicated to PAR-CLIP experiments). Input sequences should be uploaded as pre-sorted lists according to sequence score in a descending order (higher binding signal/noise ratio sequences at the top). Alternatively, for sequences provided in common formats (current version supports the two most common BED formats, BED 6-column and ENCODE narrowPeak and FASTA format) SMARTIV will sort the input data automatically.
As aforementioned in the methodology section, as a first step SMARTIV assigns the RNA secondary structure prediction (paired or unpaired) to each nucleotide in the sequence within the input list. As a default SMARTIV uses the free energy minimization approach for predicting whether the nucleotide is in a paired or unpaired conformation, using RNAfold (21). The webserver provides the option to predict RNA secondary structure using a variety of models: (a) MFE structure predicted by RNAfold (21) (default), (b) centroid structure based on partition function predicted by RNAfold (21), (c) MEA structure based on partition function predicted by RNAfold (21), (d) most probable shape structure predicted by RNAshapes (32). Additionally, users can choose to upload directly to the webserver sequences to which they have assigned a secondary structure predicted by any other method of their choice or obtained from experimental data. By default, SMARTIV searches for combined sequence-structure motif (presented in an eight-letter alphabet). However, the user can request to search for sequence motifs too (in standard four-letter alphabet). SMARTIV employs a k-mer-based algorithm to search for enriched motifs and provides the user the ability to define the k-mers length range or specific length. SMARTIV default k-mer length range is 5–7 (suitable for most of the cases). Finally, SMARTIV assigns a unique name to each job. The user is also given the option to choose a desired name for their job and to provide an e-mail address to which the result will be sent.
Output
SMARTIV outputs the most significant combined sequence and structure (or sequence only) motifs in a graphical presentation together with their corresponding P-values (See Figure 2). SMARTIV presents the most significant motif (below the defined P-value cutoff), generated per each k-mer length within the range chosen by the user. In addition, SMARTIV presents the most representative motifs grouped by similarity (35). For easier perception of SMARTIV unique eight-letter alphabet logo, the color scheme is provided in the result page at the top right corner. Each motif predicted by SMARTIV can be downloaded as a graphical logo in JPG or PDF formats or as a probability matrix (PWM) in text format. In addition, the output contains extensive information for further analysis of the results, including input parameter values, aligned list of the motif occurrences mapped to the input sequences, k-mers that were used to build the PWM along with detailed mHG statistics information. SMARTIV provides the user also with intermediate files, such as the combined sequence and structure eight-letter alphabet FASTA sequences. All data is available for viewing and downloading as text files by clicking the relevant links. For convenience, the results of all different runs per an individual session are saved on the server during the entire session and can be retrieved by the active user by clicking the ‘Results’ tab in the home page.
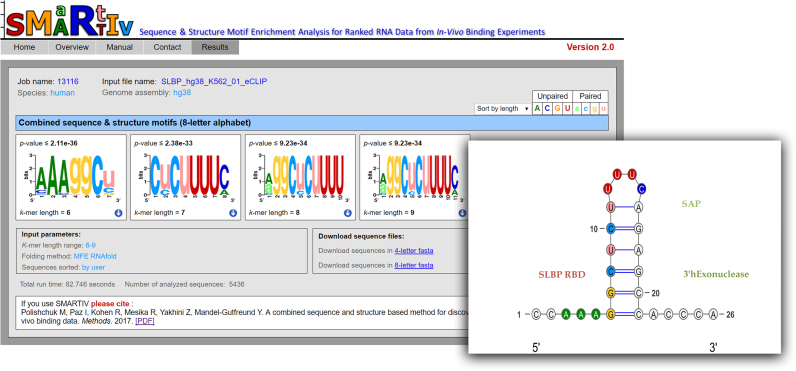
Figure 2.
SMARTIV results for extracting the combined sequence and structure motifs for SLBP. Rank list sequences from eCLIP experiment conducted for human SLBP in K562 cell were provided an input to SMARTIV webserver. Parameters were set to k-mer range: 6–9, and folding method: MFE RNAfold. Shown are the four most significant motifs in an eight-letter alphabet. On the right is a cartoon representing the secondary structure of the known SLBP binding motifs, which was solved by X-ray crystallography in complex with the SLBP protein. As shown, all four most significant motifs predicted by SMARTIV fit exactly to the known stem–loop binding site of SLBP on the histone mRNA, at both the sequence and structural level. For illustration, SMARTIV motif is mapped to the known stem–loop structure, using SMARTIV standard color-coding.
RESULTS
We have developed a new method to extract enriched sequence and structure motifs of RBPs from in-vivo high-throughput binding experiments (28), which we have implemented to a user friendly webserver named SMARTIV. We tested SMARTIV webserver on 52 sets of RNA binding data for 33 different RBPs, generated from different types of CLIP technologies, including CLIP-seq (43), HITS-CLIP (44), PAR-CLIP (10,45,46), iCLIP (47,48) and eCLIP (12). Results for the three most significant motifs (when available) from the 52 datasets are provided in Supplementary Table S1. Along with SMARTIV results we provide the known sequence motifs identified in-vitro (2,3) as well as the structural information when available from experimental data (4).
As shown, in the majority of cases the best motifs generated by SMARTIV are highly consistent between each other, e.g. hnRNPL, KHSRP, PCBP2, PTB1, QKI, SRSF1, TARDB and others (Supplementary Table S1). However, in some cases, whereas the significant motifs all show highly similar sequence preferences, some motifs differ in their secondary structure preferences, as for example in the case of EIF4G2. While the differences at the secondary structure level between the two most significant motifs of EIF4G2 could result from variability in the folding predictions, it could also suggest that the protein preferably binds to a specific RNA sequences in either single stranded or double stranded conformation, with no structural preference. To our knowledge the binding motif of EIF4G2 has not been identified by in-vitro assays and its binding motif has not been reported in the literature and thus EIF4G2 predictions could not be validated. Nevertheless, EIF4G2 is one of the few proteins for which the GUG codon serves as the exclusive translation initiation codon for its own mRNA translation (49). The motifs predicted by SMARTIV, generated from eCLIP data obtained from K562 cell, suggest that GUG stretches may be the preferred binding sites of EIF4G2 on its RNA targets, possibly in either paired or unpaired conformation.
Among the 33 RBPs we have tested, 26 had a known motif that was extracted by in-vitro assays (from RNA SELEX, RNAcompete, or EMSA experiments). Overall, for 21 RBPs (ELAV1, hnRNPA1, hnRNPC, hnRNPL, IGF2BP2, IGF2BP3, KHDRBS1, KHSRP, PCBP2, PTBP1, PUM2, QKI, RBFOX2, SLBP, SRSF1, SRSF2, SRSF9, TARDBP, TIA1, TRA2A, USAF2) the sequence preferences, revealed by the best motifs predicted by SMARTIV from different datasets (generated from different CLIP methodologies and/or different human and mouse cell lines), were generally consistent with the in-vitro identified motifs. For five RBPSs (FMR1, FUS, hnRNPK, IGF2BP1 and SRSF7) the SMARTIV predicted motifs did not fully agree with the in-vitro known motifs (see Supplementary Table S1). Note that is some cases, the motifs predicted by SMARTIV were in higher agreement with the known motifs extracted from the same in-vivo experiments by other motif finding algorithms, such as GraphProt (see for example SRSF7 in Supplementary Table S2).
Consistent with the knowledge that RBPs prefer binding to single strand regions, for the majority of the RBPs tested (27/33) the most significant motifs predicted by SMARTIV indicate a strong preference of the nucleotides to be in an unpaired conformation (presented in upper case letters). Among them, for four RBPs (ELAV1, PTBP1, QKI, SLBP and SRSF1) the combined sequence and structure motifs predicted by SMARTIV (using the default MFE-based algorithm for folding) matched exactly the sequence-structure preferences identified by RNAcompete-S(4). While in many cases it is assumed that the RBP binds to an MFE structure of the RNA, given the highly dynamic nature of the RNA it is possible that RBPs could bind the RNA in different structural contexts. To this end SMARTIV provides an option to the user for folding the input RNA sequences using different models for secondary structure prediction (as described in the input section above). As exemplified in Supplementary Table S3, when running SMARTIV on RNA sequences from CLIP datasets that were folded using four different folding algorithms in some cases the most significant motif extracted by SMARTIV gave highly consistent results, independent of the method used for folding (see results for RBFOX and SLBP in Supplementary Table S3). Nevertheless, in other cases (as for example EIF4G2, PUM2) the preferred sequence and structure motifs did differ when employing different folding prediction methods, possibly indicating the dynamic nature of the protein–RNA interactions. Among the proteins in our test set for which the structural information was available is SLBP (stem–loop Binding Protein). SLBP is a regulator of histone mRNA metabolism that binds to a highly conserved stem loop structure at the 3′ end of the histone mRNAs that is further trimmed by a 3′ hEXO endonuclease (50). The crystal structure of the human SLBP in complex with the RNA and the endonuclease was solved by Tan et al. (51). We ran SMARTIV on SLBP eCLIP data from human K562 downloaded from ENCODE. As shown in Figure 2, all four best motifs predicted by SMARTIV for human SLBP fit exactly the sequence and structure of the conserved histone mRNA stem loop bound by SLBP in the solved X-ray crystallography structure (51). As mentioned above, in the case of SLBP, the predicted sequence and structure motif was highly consistent when using all four folding prediction methods (Supplementary Table S3). Strikingly, by using a wider k-mer range (length 6–9) SMARTIV could capture the entire stem loop AAAggcucuUUUC (uppercase letters representing single stranded RNA and lowercase letters representing double strand RNA) bound by SLBP, both at the sequence and the structural levels. As mentioned above, SMARTIV results are also in high accordance to the in-vitro results of RNAcompete-S generated for the Drosophila SLBP RBP(4). It is worth noting that in RNAcompete-S, extraction of the full SLBP motif required a multi-step motif extraction process, while we identified the full stem loop motif of SLBP using SMARTIV webserver with default parameters.
Overall, running SMARTIV on different datasets for the same RBPs, generated from different CLIP methodologies and/or from different cell lines, in the majority of cases produced highly consistent results both at the sequence and the structural levels (Supplementary Table S1). As for example, the preference for the major part of the PUM2 motif to be in paired conformation was predicted by SMARTIV when extracted from either PARCLIP (HEK293 cells) or eCLIP (K562) data. These results are also consistent with predictions obtained by other sequence-structure algorithms (24). However, in a few cases, such as in the case of EWSR1, TAF15 RBPs from the FET protein family (52), SMARTIV produced different significant motifs (consistent for all k-mers tested) when running it on PARCLIP input data from HEK293 cells or with eCLIP data from K562 or HepG2 cells. The latter could likely result from differences in the binding preferences of the RBPs in different cells or from different biases related to the CLIP technology. Such differences have been previously reported using other RNA motif prediction algorithms (18). We further compared SMARTIV best predicted motifs for 16 RBPs to motifs predicted by the only available webserver for predicting combined sequence and structure motifs in RNA, RBPmotif (22), and to the state-of-the-art GraphProt algorithm (18), which we ran locally on our computers. The three predictors were tested on the same datasets generated from in-vivo CLIP data (detailed information on the datasets are given in Supplementary Table S2). Overall, as shown in Supplementary Table S2, the combined sequence and structure motifs predicted by SMARTIV are usually in agreement with the motifs predicted by RBPmotif webserver (22) and by GraphProt algorithm (18).
Finally, we compared the runtime of SMARTIV webserver to the time required to obtain motifs using RBPmotif webserver using default parameters. As shown in Supplementary Figure S1, SMARTIV run times ranges from ∼30 s to ∼2 min for different datasets. The running time for RBPmotif was between 2- and 12-fold longer for these datasets.
DISCUSSION
Accumulating data resulting from high-throughput RNA binding experiments have provided highly valuable insights on the principles of protein–RNA recognition (9–13). Taken together, the results from in-vitro and in-vivo high-throughput binding experiments have broadened our knowledge on binding preferences of a large variety of RBPs. As aforementioned, a major limitation of most high-throughput RNA binding experiments is that they do not capture the RNA structural context of the RBP binding sites. In recent years, several computational approaches have been developed to predict the binding preferences of RBPs from high-throughput binding data, considering both the sequence and the structural binding preferences of the proteins (17,18,20,23,25,53). RBPmotif (22), which implements the RNAcontext algorithm for learning the sequence and structural binding preference of RBPs (20), is currently the only webserver available for extracting sequence and structure motifs from high throughput RNA binding experiments. Here we present a new webserver, named SMARTIV, for extracting combined motifs from CLIP data. We show that SMARTIV results, generated from many different RNA binding datasets, are in good agreement with results from other tools for discovering combined sequence and structural motifs, with a great advantage of presenting the sequence and structural preferences in one unified motif representation. Nevertheless, it is important to note that since SMARTIV uses the reported sequence binding score from the CLIP peak calling algorithm as the basis for ranking the input sequences, results could be strongly influenced by the specific algorithm used to extract the binding scores. Therefore, it is recommended that users choose carefully the most appropriate peak calling algorithm for generating their input data or alternatively chose the most suitable data from CLIP databases.
Similar to all other algorithms for predicting sequence and structure motifs, SMARTIV results depend on the accuracy of the RNA folding algorithm, used for predicting the RNA secondary structure of the sequences identified in the binding experiment. To overcome this SMARTIV employs a ranked-based enrichment algorithm, selecting only the subset of k-mers, that are consistently found in the same RNA conformation, to generate the combined sequence and structure PWM. Furthermore, SMARTIV provides the user the option for folding the RNA using different methods, which are based on different secondary structure prediction models (MFE structures, Centroid structures, MEA structures and Shapes probabilities). This additional feature of SMARTIV can allow the user to both capture the dynamics in the system as well as identify motifs that are highly consistent using the different folding approaches. Moreover, users can upload sequences folded by any alternative method of interest or even obtained from known RNA secondary structures if available.
In conclusion, SMARTIV provides an easy, user friendly tool available via the web for inferring accurate sequence and structure motifs of RBPs from high-throughput in-vivo RNA binding data. To our knowledge SMARTIV is currently the most efficient algorithm of its kind, processing a standard CLIP dataset in 1–2 min on average. SMARTIV is designed to process very large datasets that are expected to be generated from current and future high-throughput RNA binding technologies. Finally, SMARTIV motif-presentation is highly intuitive, providing the combined motifs in both graphical representation and as a PWMs with an assigned P-values.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Limor Leibovich who developed the DRIMUST algorithm and provided access to the source code and to Leon Anavy for adjusting the DRIMUST code to SMARTIV. We thank Refael Kohen for his contribution to SMARTIV algorithm. We also thank Shlomi Dvir for his help in conducting statistical testing and Ilya Vorontsov for providing the source code for motif similarity identification and adapting it to the webserver.
Author Contribution: M.P. and I.P. designed and developed the website. M.P., Z.Y. and Y.M.G.. developed the method and the statistical approach. M.P. designed the algorithm, implemented the method and conducted all the validation experiments. M.P. and Y.M.G. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Lady Davis Foundation (in part) (to M.P.). Funding for open access charge: Technion internal funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Li X.C., Kazan H., Lipshitz H.D., Morris Q.D.. Finding the target sites of RNA-binding proteins. Wiley Interdiscip. Rev. RNA. 2014; 5:111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ray D., Kazan H., Chan E.T., Pena Castillo L., Chaudhry S., Talukder S., Blencowe B.J., Morris Q., Hughes T.R.. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009; 27:667–670. [DOI] [PubMed] [Google Scholar]
- 3. Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A. et al. . A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013; 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cook K.B., Vembu S., Ha K.C.H., Zheng H., Laverty K.U., Hughes T.R., Ray D., Morris Q.D.. RNAcompete-S: combined RNA sequence/structure preferences for RNA binding proteins derived from a single-step in vitro selection. Methods. 2017; 126:18–28. [DOI] [PubMed] [Google Scholar]
- 5. Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B.c.m.e. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell. 2014; 54:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Niranjanakumari S., Lasda E., Brazas R., Garcia-Blanco M.A.. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002; 26:182–190. [DOI] [PubMed] [Google Scholar]
- 7. Keene J.D., Komisarow J.M., Friedersdorf M.B.. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006; 1:302–307. [DOI] [PubMed] [Google Scholar]
- 8. Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T.. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010; 40:939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X. et al. . HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008; 456:464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.C., Munschauer M. et al. . Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J.. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010; 17:909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Nostrand E.U., Pratt G.U., Shishkin A.A., Gelboin-Burkhart C.U., Fang M.U., Sundararaman B.U., Blue S.U., Nguyen T.U., Surka C., Elkins K.U. et al. . Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zarnegar B.J., Flynn R.A., Shen Y., Do B.T., Chang H.Y., Khavari P.A.. irCLIP platform for efficient characterization of protein–RNA interactions. Nat. Methods. 2016; 13:489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee F.C.Y., Ule J.. Advances in CLIP technologies for studies of protein–RNA interactions. Mol. Cell. 2018; 69:354–369. [DOI] [PubMed] [Google Scholar]
- 15. Cook K.B., Kazan H., Zuberi K., Morris Q., Hughes T.R.. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2011; 39:D301–D308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giudice G., Sanchez-Cabo F., Torroja C., Lara-Pezzi E.. ATtRACT-a database of RNA-binding proteins and associated motifs. Database. 2016; 2016:baw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hiller M.G., Pudimat R., Busch A., Backofen R.. Using RNA secondary structures to guide sequence motif finding towards single-stranded regions. Nucleic Acids Res. 2006; 34:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maticzka D., Lange S.J., Costa F., Backofen R.. GraphProt: modeling binding preferences of RNA-binding proteins. Genome Biol. 2014; 15:R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Steffen P., Voss B., Rehmsmeier M., Reeder J., Giegerich R.. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006; 22:500–503. [DOI] [PubMed] [Google Scholar]
- 20. Kazan H., Ray D., Chan E.T., Hughes T.R., Morris Q.. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS Comput. Biol. 2010; 6:e1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kazan H., Morris Q.. RBPmotif: a web server for the discovery of sequence and structure preferences of RNA-binding proteins. Nucleic Acids Res. 2013; 41:W180–W186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Orenstein Y., Wang Y., Berger B.. RCK: accurate and efficient inference of sequence- and structure-based protein–RNA binding models from RNAcompete data. Bioinformatics. 2016; 32:i351–i359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orenstein Y., Ohler U., Berger B.. Finding RNA structure in the unstructured RBPome. BMC Genomics. 2018; 19:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goodarzi H., Najafabadi H.S., Oikonomou P., Greco T.M., Fish L., Salavati R., Cristea I.M., Tavazoie S.. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature. 2012; 485:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Searls D.B. The language of genes. Nature. 2002; 420:211–217. [DOI] [PubMed] [Google Scholar]
- 27. Goodarzi H.U., Tavazoie S.F., Tavazoie S.. TARBP2 binding structured RNA elements drives metastasis. Cell Cycle. 2014; 13:2799–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Polishchuk M.R., Paz I., Kohen R., Mesika R., Yakhini Z., Mandel-Gutfreund Y.. A combined sequence and structure based method for discovering enriched motifs in RNA from in vivo binding data. Methods. 2017; 118–119:73–81. [DOI] [PubMed] [Google Scholar]
- 29. Leibovich L., Yakhini Z.. Mutual enrichment in ranked lists and the statistical assessment of position weight matrix motifs. Algorithms Mol. Biol. 2014; 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Steinfeld I., Navon R., Ach R., Yakhini Z.. miRNA target enrichment analysis reveals directly active miRNAs in health and disease. Nucleic Acids Res. 2013; 41:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heller D., Krestel R., Ohler U., Vingron M., Marsico A.. ssHMM: extracting intuitive sequence-structure motifs from high-throughput RNA-binding protein data. Nucleic Acids Res. 2017; 45:11004–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Janssen S., Giegerich R.. The RNA shapes studio. Bioinformatics. 2015; 31:423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Steffen P., Voss B., Rehmsmeier M., Reeder J., Giegerich R.. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006; 22:500–503. [DOI] [PubMed] [Google Scholar]
- 34. Leibovich L., Paz I., Yakhini Z., Mandel-Gutfreund Y.. DRIMust: a web server for discovering rank imbalanced motifs using suffix trees. Nucleic Acids Res. 2013; 41:W174–W179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vorontsov I.E., Kulakovskiy I.V., Makeev V.J.. Jaccard index based similarity measure to compare transcription factor binding site models. Algorithms Mol. Biol. 2013; 8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. . NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blin K., Dieterich C., Wurmus R., Rajewsky N., Landthaler M., Akalin A.. DoRiNA 2.0–upgrading the doRiNA database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 2015; 43:D160–D167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang Y.C., Di C., Hu B., Zhou M., Liu Y., Song N., Li Y., Umetsu J., Lu Z.J.. CLIPdb: a CLIP-seq database for protein–RNA interactions. BMC Genomics. 2015; 16:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Uren P.J., Bahrami-Samani E., Burns S.C., Qiao M., Karginov F.V., Hodges E., Hannon G.J., Sanford J.R., Penalva L.O., Smith A.D.. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012; 28:3013–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Corcoran D.L., Georgiev S., Mukherjee N., Gottwein E., Skalsky R.L., Keene J.D., Ohler U.. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011; 12:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xue Y., Zhou Y., Wu T., Zhu T., Ji X., Kwon Y.S., Zhang C., Yeo G., Black D.L., Sun H. et al. . Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009; 36:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pandit S., Zhou Y., Shiue L., Coutinho-Mansfield G., Li H., Qiu J., Huang J., Yeo G.W., Ares M. Jr., Fu X.D.. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell. 2013; 50:223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ascano M. Jr., Mukherjee N., Bandaru P., Miller J.B., Nusbaum J.D., Corcoran D.L., Langlois C., Munschauer M., Dewell S., Hafner M. et al. . FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012; 492:382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M. Jr., Tuschl T., Ohler U., Keene J.D.. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011; 43:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., Konig J., Hortobagyi T., Nishimura A.L., Zupunski V. et al. . Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011; 14:452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z., Kayikci M., Briese M., Zarnack K., Luscombe N.M., Rot G., Zupan B., Curk T., Ule J.. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010; 8:e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takahashi K., Maruyama M., Tokuzawa Y., Murakami M., Oda Y., Yoshikane N., Makabe K.W., Ichisaka T., Yamanaka S.. Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2). Genomics. 2005; 85:360–371. [DOI] [PubMed] [Google Scholar]
- 50. Battle D.J., Doudna J.A.. The stem–loop binding protein forms a highly stable and specific complex with the 3′ stem–loop of histone mRNAs. RNA. 2001; 7:123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tan D., Marzluff W.F., Dominski Z., Tong L.. Structure of histone mRNA stem–loop, human stem–loop binding protein, and 3′hExo ternary complex. Science. 2013; 339:318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hoell J.I., Larsson E., Runge S., Nusbaum J.D., Duggimpudi S., Farazi T.A., Hafner M., Borkhardt A., Sander C., Tuschl T.. RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Biol. 2011; 18:1428–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dao P., Hoinka J., Takahashi M., Zhou J., Ho M., Wang Y., Costa F., Rossi J.J., Backofen R., Burnett J. et al. . AptaTRACE elucidates RNA sequence-structure motifs from selection trends in HT-SELEX experiments. Cell Syst. 2016; 3:62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.