Abstract
Motivation
Sequencing of human genomes is now routine, and assembly of shotgun reads is increasingly feasible. However, assemblies often fail to inform about chromosome-scale structure due to a lack of linkage information over long stretches of DNA—a shortcoming that is being addressed by new sequencing protocols, such as the GemCode and Chromium linked reads from 10 × Genomics.
Results
Here, we present ARCS, an application that utilizes the barcoding information contained in linked reads to further organize draft genomes into highly contiguous assemblies. We show how the contiguity of an ABySS H.sapiens genome assembly can be increased over six-fold, using moderate coverage (25-fold) Chromium data. We expect ARCS to have broad utility in harnessing the barcoding information contained in linked read data for connecting high-quality sequences in genome assembly drafts.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The Chromium sequencing library preparation protocol from 10 × Genomics (10 × G, Pleasanton, CA) builds on the Illumina sequencing technology (San Diego, CA) to provide indexing/barcoding information along with short reads to localize the latter on long DNA fragments, thus benefiting the economies of scale of a high-throughput platform. As sequence reads from 20 to 200 kb molecules are barcoded/linked, applications of the technology has mainly focused on phasing variant bases in human genomes (Narasimhan et al., 2016; Zheng et al., 2016).
The ability to generate linked reads with 10 × G is akin to that of Illumina TruSeq (Kuleshov et al., 2014). The latter technology provides useful complementary information to whole genome shotgun assembly projects, as the pseudo-long reads it generates may help resolve long repeats. However, to generate pseudo-long reads, TruSeq requires high coverage data of the co-localized reads for a priori fragment assemblies (by default, transparent to the user), essentially generating low fragment coverage data for its target genome. Hence, TruSeq may be relatively expensive for providing mammalian-sized genomes with adequate fragment coverage. Conversely, the Chromium platform typically provides low-coverage for each single barcoded molecule, limiting its utility for individual fragment assembly. However, it makes up for this limitation in throughput, providing higher fragment coverage.
Recently this data type has been utilized for scaffolding a draft genome assembly (Mostovoy et al., 2016), using a software designed to scaffold sequences using contiguity preserving transposition sequencing (CPT-seq) and another long-range information data source (Hi-C) (Adey et al., 2014). In their paper, Mostovoy et al. (2016) showed 12-fold improvement in contiguity of a human genome assembly draft using GemCode sequencing (precursor to Chromium, from 10 × G) at 97-fold coverage, demonstrating the potential of the technology for scaffolding draft genomes.
Here we present ARCS, the Assembly Round-up by Chromium Scaffolding algorithm, a method that leverages the rich information content of high-volume long sequencing fragments to further organize draft genome sequences into contiguous assemblies that characterize large chromosome segments. We use the recent Genome In A Bottle (GIAB) human genome sequence data (Zook et al., 2016), and compare ARCS to fragScaff, the only other technology shown in a publication to utilize 10 × G linked reads for scaffolding genome assembly drafts (Mostovoy et al., 2016). In the fragScaff scaffolding algorithm, a barcoded alignment file is parsed to determine which barcodes map to the ends of each sequence. For each possible pair of sequence ends, a shared barcode fraction metric is calculated. These values produce a distribution of shared barcode fractions for each sequence end. Edges are added to a scaffold graph based on these distributions, resulting in sequence end nodes being linked when a high fraction of barcodes are shared. For each connected component, the maximum-weight minimum spanning tree (MST) is determined, followed by iterations to incorporate any branches into the main trunk of the MST to produce the final scaffolds.
We also present similar benchmarks to Architect, a recently published scaffolder (Kuleshov et al., 2016) shown to work on Illumina TruSeq synthetic long sequences' underlying short reads (read clouds) and suggested to be adaptable to Chromium data. The Architect algorithm utilizes evidence from read clouds by first identifying barcode ‘hits’ to each scaffold, based on input barcoded read alignments. Then, a scaffold graph is constructed, where edges are created based on the number of shared barcode hits and the fraction of shared barcode hits between two scaffolds. Following pruning of potentially spurious edges, unambiguous edges are contracted to produce the final output scaffolds.
We show how our implementation yields assemblies that are more contiguous and accurate than fragScaff and Architect over a wide range of parameters, while using less time and compute resources. Using two human linked read datasets from different experiments, we demonstrate that ARCS scaffolding of pre-existing human genome drafts can yield assemblies whose contiguity and correctness are on par with or better than those assembled with the newly released 10 × G Supernova de novo assembler (Weisenfeld et al., 2017). ARCS is implemented in C ++ and runs on Unix.
2 Materials and methods
2.1 ARCS algorithm
The modular pipeline collectively referred to as ARCS first pairs sequences within a draft assembly, then lays out the pairing information for scaffolding. In the sequence pairing stage (Fig. 1), input alignments in BAM format are processed for sets of read pairs from the same barcode that align to different sequences. These form a link between the two sequences, provided that there is a sufficient number of read pairs aligned (parameter –c, set to 5 by default). Each link represents evidence that one barcode/molecule connects the sequences. To account for barcode sequencing errors, only barcodes within a specified multiplicity range (parameter –m) are considered (default 50–10 000). The multiplicity refers to the read frequency of each barcode, and the range defines a specific slice of reads considered by ARCS.
Fig. 1.
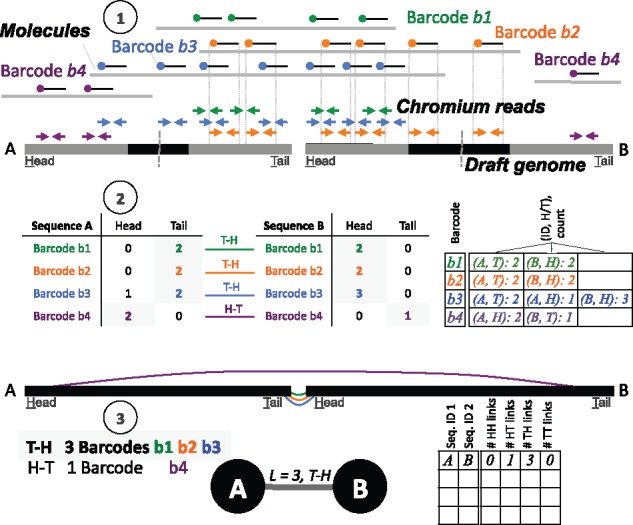
ARCS algorithm. (1) 10×G Chromium reads (blue, green, orange and purple arrows) are aligned to the draft genome. (2) Sequences are split in half by length and the ends of each are considered the head (H) or tail (T) regions (represented with grey boxes, length of the ends controlled by ARCS parameter –e). The number of read pairs derived from the same barcode and aligning to the head (H) or tail (T) regions of the sequence are tallied. These tallies are stored in memory using a map data structure, where the key is the barcode sequence. The value maps a tuple of the sequence ID and ‘H’ or ‘T’ to the count of the number of read pairs. (3) The number of barcodes supporting each link orientation (H-H, H-T, T-H, T-T) between sequence pairs is tallied. The tallies are stored in memory using an additional map data structure, where the key is a pair representing the two potentially linked sequences, and the value is a vector of integers representing the number of barcodes supporting each possible link orientation. For a given barcode to contribute linking evidence, the distribution of reads of that barcode aligning to the ‘H’ or ‘T’ regions of both sequences in the potential pair must significantly differ from a uniform distribution. A dot file is then generated which encodes the linkage evidence, where links (edges) between two sequences (nodes) are only added if the link orientation with the maximum support is predominant
As we are interested in ordering and orienting sequences, we consider reads that align near the 5′ and 3′ ends of each sequence (within a window defined by the parameter –e, default 30 000). This parameter effectively sets the maximum window length at the end of sequences, where Chromium reads align. Reads aligned outside of these windows are not considered. Thus, depending on the level of contiguity of the input assembly, adjusting –e to a lower or higher value would account for shorter contigs or focus on longer contigs. When ARCS encounters shorter sequences (less than twice the specified –e length), the length of the head and tail regions are assigned as half the total sequence length. This is important, as the selection of –e will impact how ambiguity is mitigated when creating an edge between any two sequences. In addition, as the BAM file is read, only reads that align to a sequence with at least the specified sequence identity (parameter –s, set to 98% by default), map in proper pairs and align with a non-zero mapping quality are considered. This ensures that only high-quality alignments provide evidence for the subsequent linking stages, as alignments involving reads with long repeat regions or chimeric reads will be skipped. Therefore, contigs that end in long repeats will not be linked in downstream stages due to the lack of unambiguous aligned read support.
The relative orientations of sequences are inferred through the read alignment positions. Using read alignments, we first determine subsets of reads with the same barcode that co-locate within one end of a sequence (Fig. 1, step 1); Within each sequence, the 5′ end region is arbitrarily labeled the head (H), and the 3′ end, the tail (T). The number of read pairs of the same barcode aligning to the head or the tail of a sequence (within –e bp or less of the end) is tallied as the BAM file is read. For a given barcode, we track the number of reads that map to the ‘H’ or ‘T’ of a sequence (Fig. 1, step 2).
Once the alignment file is read into memory, every possible pair of sequences that have a sufficient number of aligned reads from a given barcode (–c) are considered. For each sequence in a potential pair, a binomial test is used to calculate whether the observed distribution of reads aligning to the 5′ or 3′ end of a sequence is significantly different from a uniform distribution (threshold P = 0.05, set by parameter –r). Likewise, the number of linking barcodes that support each of the four possible link orientations (H-H, H-T, T-H, T-T) is tallied in a map data structure for each potential sequence pair (Fig. 1, step 3).
Using the link orientation tallies for each sequence pair, a graph data structure is constructed, where the nodes are sequences, and the edges represent links between them. An edge is formed only if the link orientation, defined by the order of a sequence pair‘s head and tail regions, is the most represented combination across supporting barcodes. Once pairing between sequences is complete, ARCS outputs a single file in the graph description language (gv) format.
2.2 Scaffolding
In preparation for the layout building stage, ARCS’ gv file is converted to a tab-separated value (tsv) file listing all possible oriented sequence pairs, the number of supporting barcodes with gap sizes arbitrarily set at 10 bp. This is facilitated by the supplied python script (makeTSVfile.py). Since positional information of reads within the molecule of origin is not known, estimation of gap sizes is not a straightforward problem, and would require more sophisticated approaches. The latter tsv file is read and scaffolds constructed using the algorithm implemented in LINKS, as described previously (Warren et al., 2015) (v1.7 and later).
Briefly, starting with the longest sequence as seed sequence, a layout is progressively built by considering all possible pair of sequences suggested by ARCS (Supplementary Fig. S1a), adjusting the orientation of likely pairs relative to the seed. Because linked sequence pairs may be ambiguous (a given sequence may link to multiple sequences), sequences are joined only if the number of links connecting a sequence pair is equal to or greater than a minimum (Supplementary Fig. S1b, LINKS parameter –l, default of 5) and satisfies a minimum sequence cutoff (LINKS parameter –z, default 500). Ambiguous pairings are resolved when the ratio of barcode links of the second-most to top-most supported edge is equal to or below a threshold (Supplementary Fig. S1b, LINKS parameter –a, default of 0.3; we recommend higher values such as –a 0.7 and 0.9 when running LINKS within ARCS). In the Supplementary Information, we show a section of the LINKS scaffold graph demonstrating the influence of the –a parameter on scaffolding an experimental human dataset used in the present study (Supplementary Fig. S1c). When sequence merging is exhausted in 3′, the scaffold is extended in 5′ following the same procedure.
We point out that other stand-alone scaffolding algorithms may be used within the ARCS pipeline instead of LINKS, at the user‘s discretion. For instance, we have experimented with abyss-scaffold (Jackman et al., 2017), which implements a non-greedy graph-based approach, and found the results to be comparable to that of LINKS. A modular pipeline ensures that in the future, improved scaffolders may be used within ARCS without having to alter the code base.
2.3 Data sources
We used two human Chromium datasets to illustrate the performance of ARCS on baseline sequence assemblies.
The first dataset is from an Ashkenazi female individual (NA24143) from GIAB (Zook et al., 2016), sequenced using various Illumina library protocols (accession number NIST HG004 NA24143 SRS823307; Supplementary Table S1). In preparation for whole genome de novo sequence assembly (referred herein as baseline assemblies), we downloaded Illumina whole genome shotgun (WGS) 2 × 250 bp paired-end and 6 kbp mate-pair sequencing reads. Adapter sequences from the mate-pair reads were removed using NxTrim v0.4.0 (O’Connell et al., 2015) (with parameters –norc –joinreads –preserve-mp). NxTrim also classifies reads as mate-pair, paired-end, single-end or unknown. Only reads classified as mate-pair were subsequently used for assembly. Both paired-end and mate-pair reads were corrected with BFC v181 (Li, 2015) (with the parameter –s3G). We also downloaded 10 × G linked reads from the same repository for the purpose of de novo assembly with Supernova, and scaffolding of baseline assemblies with ARCS, Architect and fragScaff (Supplementary Table S1).
The second individual‘s genome (NA12878) was sequenced and assembled by 10 × G. The corresponding raw, ∼156-fold coverage, NA12878 10 × G Chromium data was downloaded from the 10 × Genomics company website (Supplementary Table S1).
2.4 Data analysis
Whole genome shotgun paired-end and mate pair reads were assembled de novo with ABySS v2.0 (Jackman et al., 2017) with the command: abyss-pe name = hsapiens np = 64 k = 144 q = 15 v = -v l = 40 s = 1000 n = 10 S = 1000–10 000 N = 7 mp6k_de=–mean mp6k_n = 1 lib = pe400 mp = mp6k, where pe400 and mp6k are variables listing all files containing paired-end sequencing and MPET reads. The resulting contigs and scaffolds were used as baseline human genome draft assemblies for linked read scaffolding (Supplementary Table S2).
The 10 × G Chromium sequencing data was converted from a container BAM file to FASTQ format (NA24143) or processed with 10 × G longranger (Weisenfeld et al., 2017; Zheng et al., 2016) to generate barcode-containing interleaved FASTQ files (NA12878) (Supplementary Table S3). For the former dataset, the read barcodes were extracted from the RX tag in the BAM file. For both Chromium datasets the barcode was appended to the read name following an underscore. Chromium reads were then aligned to the contig and scaffold sequences using BWA mem v0.7.15 (default values, -t12) (Li and Durbin, 2010), and sorted by name. We provide instructions on how to prepare and align the Chromium reads here: ftp://ftp.bcgsc.ca/supplementary/ARCS
Based on authors’ recommendations, the input to fragScaff also included an N-base bed file. This file contained the coordinates of all undetermined base stretches when using a scaffold input, and was generated by their supplied script fasta_make_Nbase_bed.pl. A repeat bed file was also included, generated by performing a blastn v2.4.0 alignment (Altschul et al., 1990) of the input assembly to itself (with parameters –word_size 36, –perc_identity 95, –outfmt 6) and by converting the alignments using their supplied script blast_self_alignment_filter.pl.
In separate experiments, the NA24143 and NA12878 10 × G linked read data were assembled with the Supernova v1.1 de novo assembler as described (Weisenfeld et al., 2017).
The scaffolding scripts that ran on the data described above are available at ftp://ftp.bcgsc.ca/supplementary/ARCS, providing the command lines and parameters used. The corresponding assemblies are also offered through the same URL.
In separate triplicate experiments, we sub-sampled 100, 200, 300 M NA24143 and 46M, 200–1400 M NA12878 10 × G read pairs to test the effect of read coverage on the performance of ARCS, Architect and fragScaff for scaffolding the baseline scaffold assembly draft. On each file subset, we ran ARCS (–c 5 –r 0.05 –e 30000 –z 3000 –m 50–6000 for NA12878; –m 50–1000 for NA24143) and LINKS (–l 5 –a 0.9), Architect (–t 5 –rc-abs-thr 3 –rc-rel-edge-thr 0.2 –rc-rel-prun-thr 0.2) and fragScaff (–b 1 –m 3000 –E 30000 –j 1 –u 2 –C 5). For each coverage level and tool, we calculated the average contiguity and number of breakpoints (as a proxy for counting misassemblies).
Breakpoints in re-scaffolded assemblies were identified using abyss-samtobreak (-G3088269832 -l500.) (Jackman et al., 2017). Briefly, scaffolds are first broken at Ns to generate sequence ‘scaftigs’. The assembly scaftigs are aligned to the reference human genome GRCh38 with BWA mem (v0.7.15, using the –xintractg flag) (Li and Durbin, 2010). Breakpoints are identified when scaftigs do not align co-linearly to a given reference chromosome sequence. This includes cases where the order of scaftigs in the scaffold is not consistent with the respective chromosome alignments, and/or scaftigs from a given scaffolds align to two or more chromosomes. The distance between scaftigs (i.e. length of assembly gaps) is not scrutinized for length consistency between draft and reference genomes. The NG50 and NGA50 length metrics reported were calculated using a genome size of 3 088 269 832 bp. Benchmarking for computational performance was done on a DELL server with 128 Intel(R) Xeon(R) CPU E7-8867 v3, 2.50 GHz with 2.6TB RAM.
3 Results
To generate the contig and scaffold baselines for NA24143, we first assembled the paired-end and mate-pair data with ABySS-2.0 (Jackman et al., 2017). We then aligned the Chromium reads to those assemblies with BWA (Li and Durbin, 2010). Using the resulting alignments as input, we ran ARCS (v1.0.0), Architect (v0.1) and fragScaff (v140324) to further scaffold contig and scaffold baseline sequences 3 kbp and longer, as recommended (Mostovoy et al., 2016). We investigated the effects of multiple parameter combinations on scaffolding (Supplementary Table S4), reporting contiguity length metrics and number of breakpoints from sequence alignments to the reference human genome.
3.1 Scaffolding with the NA24143 GIAB Chromium data
We measured the contiguity (NG50 and NGA50 length metric) and correctness of resulting assemblies after ARCS, Architect and fragScaff scaffolding of baseline assemblies (Supplementary Table S2). During this process, we tested the effect of various parameters, including the scaffolding-specific –a, –u and –rc-rel-edge-thr (abbreviated rel) parameters in the corresponding tools, respectively (Fig. 2). Generally, these parameters affect scaffolding stringency by evaluating the validity of the linkages.
Fig. 2.
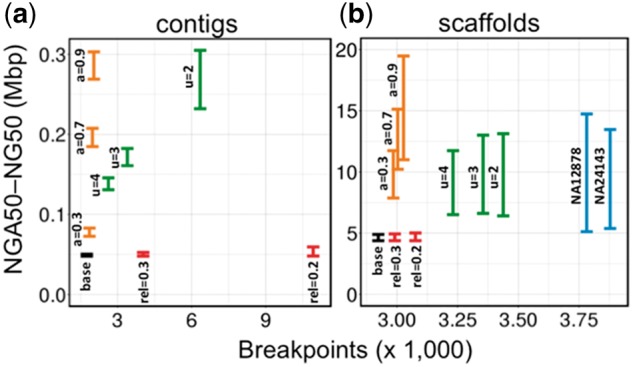
Assembly contiguity and correctness from scaffolding (a) contig or (b) scaffold baseline assemblies with 10×G Chromium reads using ARCS (orange), Architect (red) and fragScaff (green). We show the effect of the scaffolding parameters –a (ARCS), –rc-rel-edge-thr (abbreviated rel, Architect) and –u (fragScaff). The Y-axes show the range of NGA50 to NG50 lengths to indicate the uncertainty caused by real genomic variations (captured by breakpoints analysis) between individual NA24143 and the reference genome GRCh38. For comparison, we also show the same metrics for the Supernova assembly of the NA12878 and NA24143 10×G datasets (blue). The X-axes show the number of breakpoints that occur when aligning the resulting assembly to the reference
Mostovoy et al. (2016) reported their best assembly using fragScaff parameters –j 1 and –u 3, prompting us to explore similar values of –j and –u on our dataset. These parameters are described as the mean number of passing hits per node to call the P-value cutoff and modifier to the score to consider the link reciprocated in fragScaff, and are the parameters previously optimized in the study by Adey et al. (2014). In Architect, the parameter –rc-abs-thr controls the minimum number of shared barcode hits required to tentatively connect two sequence vertices in the scaffold graph. The Architect parameters –rc-rel-edge-thr and –rc-rel-prun-thr control the relative barcode support needed for creating and pruning edges in the graph, respectively.
To assess correctness, we aligned the assemblies to the primary chromosome sequences of the human reference GRCh38, and counted the number of observed breakpoints using abyss-samtobreak (Jackman et al., 2017). At the contig level (Fig. 2a), we observe that, while the ARCS and fragScaff assemblies (highest contiguity achieved at –a 0.9 and –u 2, in that order) have similar sequence contiguity (NG50 of 303 034 versus 304 926 bp, respectively), the ARCS assembly has less than one third the number of breakpoints compared to fragScaff (2030 versus 6345). In context, the corresponding ARCS and fragScaff assemblies have 16.3 and 263.4% more breakpoints than the baseline contig assembly, respectively (Supplementary Tables S5 and S6). This indicated that, while the resulting fragScaff assemblies were highly contiguous, they may harbor substantially more misassemblies. Architect scaffolding of the baseline contig assembly did not yield appreciable gains despite extensive parameter tuning (Fig. 2a and Supplementary Table S7).
At the scaffold level (Fig. 2b and Table 1), we observe that ARCS achieves a greater sequence contiguity and correctness than Architect and fragScaff (NG50 (Mbp)/breakpoints, 19.5/3027 versus 5.0/3076 versus 13.1/3438 for the three tools, in that order) when comparing amongst the most contiguous assemblies for each tool (Supplementary Tables S5–S7). We observe that, while the difference in the number of misassemblies between fragScaff and ARCS is 411 when the tools use scaffolds as input, it increases by an order of magnitude to 4315 when using contigs. The ARCS, Architect and fragScaff assemblies respectively harbor 3.6, 5.2 and 17.6% more breakpoints than the baseline scaffold assembly, which suggests that ARCS and other scaffolders for 10 × G data work best when the draft to re-scaffold is more contiguous. To see whether these 411 additional breakpoints in the fragScaff versus ARCS assemblies are large-scale misassemblies, we aligned the corresponding assemblies to the reference human genome and plotted their alignments (Fig. 3).
Table 1.
Contiguity metrics, breakpoints, total wall-clock time and peak memory usage for scaffolding 3 kbp and larger sequences from a human (NA24143) ABySS base scaffold assembly with ARCS (–c 5 –e 30000 –r 0.05 –l 5), Architect (–t 5 –rc-abs-thr 3 –rc-rel-prun-thr 0.2; –rc-rel-edge-thr abbreviated to ‘rel’ in table) and fragScaff (–C 5 –E 30000 –j 1)
| Tool | ARCS | ARCS | ARCS | fragScaff | fragScaff | fragScaff | Architect | Architect | Supernovab |
|---|---|---|---|---|---|---|---|---|---|
| Parametersa | a = 0.3 | a = 0.7 | a = 0.9 | u = 2 | u = 3 | u = 4 | rel = 0.2 | rel = 0.3 | N/A |
| n: 500 | 65 191 | 64 993 | 64 922 | 64 445 | 64 625 | 64 869 | 65 780 | 65 862 | 23 693 |
| NG50 (Mbp) | 11.74 | 15.13 | 19.48 | 13.13 | 13.01 | 11.74 | 5.01 | 4.93 | 13.47 |
| NGA50 (Mbp) | 7.78 | 10.22 | 11.00 | 6.41 | 6.62 | 6.52 | 4.38 | 4.38 | 5.38 |
| N50 (Mbp) | 12.91 | 17.98 | 21.82 | 15.80 | 15.40 | 13.07 | 5.72 | 5.62 | 15.03 |
| Largest scaffold (Mbp) | 66.18 | 97.86 | 97.86 | 93.33 | 72.78 | 68.07 | 26.41 | 26.41 | 95.16 |
| Breakpoints | 2985 | 3003 | 3027 | 3438 | 3355 | 3231 | 3076 | 2991 | 3879 |
| Wall-clock time (h:min) | 0:55 | 0:55 | 0:55 | 2:03 | 1:56 | 1:59 | 6:12 | 5:39 | 50:43 |
| Peak memory (GB) | 3.4 | 3.4 | 3.4 | 16.5 | 14.8 | 14.1 | 9.6 | 9.6 | 389.0 |
Scaffolding-specific parameters.
Supernova is a de novo assembler, and its scaffolding stage cannot be decoupled from the rest of its work flow.
Fig. 3.
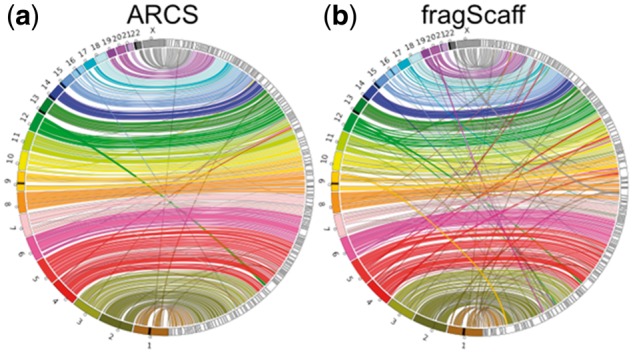
A Circos (Krzywinski et al., 2009) assembly consistency plot of conservative (a) ARCS (–c 5 –e 30000 –r 0.05 –l 5 –a 0.3) and (b) fragScaff (–C 5 –E 30000 –j 1 –u 4) scaffolding of the baseline scaffold assembly. Scaftigs from the largest 177 (ARCS) and 175 (fragScaff) scaffolds, consisting of 75% (N75) of the genome are aligned to GRCh38 with BWA mem. GRCh38 chromosomes are displayed incrementally from 1 (bottom, brown) to X (top, dark grey) on the left while scaffolds (black outlines) are displayed on the right side of the rim. Connections show aligned regions, 100 kbp and larger, between the genome and scaffolds. Large-scale misassemblies are visible as interrupting ribbons. The circles along chromosomes indicate centromeres, while the black regions on chromosomes indicate gaps in the reference
Compared to ARCS, fragScaff scaffolding of the baseline scaffold sequences yields more large-scale misassemblies, shown as inter-chromosomal translocations (Fig. 3). We note that increasing the fragScaff –j parameter (mean passing links across nodes) while relaxing –u (score cut-off multiplier) yields assemblies whose contiguity rival that of ARCS (16.9 versus 19.5 Mbp NG50, respectively), but at the cost of increased misassemblies (Supplementary Tables S5 and S6). Architect scaffolding of both contig and scaffold baseline assemblies yielded a marginal increase in contiguity figures (Fig. 2, Table 1 and Supplementary Table S7), reasons why we can only speculate on.
We also compared the resource efficiency of all three tools over the parameter range tested (See ftp://ftp.bcgsc.ca/supplementary/ARCS/benchmarks) and report its runtime and memory usage on the most contiguous assemblies of contig and baseline scaffold sequences (Table 1 and Supplementary Table S8). ARCS outperforms Architect and fragScaff for run time (average 2 fold faster than fragScaff) and memory usage on scaffolds (4 times less memory when compared to fragScaff). It should be noted that the run time of Architect and fragScaff increases quadratically with the number of input sequences, making them inefficient choices for assemblies with a large number of input sequences (more than 250 000). Running Architect on the baseline contig assemblies took roughly 7 days (187 h) for most parameter combinations when parameter –t was set to 5 (Supplementary Table S8). In contrast, equivalent runs of this tool on the scaffold baseline assemblies were faster (6 h) due to having 20 times less sequences to process (Table 1 and Supplementary Table S2). The execution speed of ARCS on the contig and scaffold baseline assemblies was consistent, both finishing in approximately 1 h (1 h 12 min and 55 min, respectively).
3.2 Scaffolding with the NA12878 Chromium data
Recently, 10 × G released their de novo assembly software called Supernova, which implements a scaffolding stage, and is developed specifically for assembling Chromium data (Weisenfeld et al., 2017). The authors presented a variety of human genome assemblies, each yielding N50 contiguity lengths 15 Mbp or higher, factoring in scaffolds 10 kbp and larger. We re-capitulated the Supernova experiment on the 156-fold Chromium sequencing data for the NA12878 individual, and corroborate their results (Supplementary Table S9). When applying a scaffold sequence length cut-off on par with that used in our study (500 bp), we report N50 length metrics corrected for genome size, NG50, in the megabase range (14.7 Mbp), which is consistent with what was maximally achievable with ARCS using the same dataset applied to the baseline scaffold assembly (NG50 = 18.3 Mbp). A Supernova assembly of 51-fold raw NA24143 chromium reads produced a similarly contiguous genome draft (NG50 = 13.5 Mbp), albeit with a higher overall number of assembly breakpoints (Fig. 2b, Supplementary Table S9). Interestingly, upon investigation of the breakpoints, we concede that fewer events ≥1 kb are observed in the largest, N75, Supernova scaffolds (data not shown).
3.3 Effect of sequence coverage on scaffolding
Despite the NA12878 Chromium read data having substantially deeper coverage than that of NA24143 (over 5 × deeper, Supplementary Table S3 datasets 5 versus 3), we observe that ARCS performs consistently across both datasets. Perhaps more interesting is the observation that there are only marginal gains in N50 length contiguity with the higher coverage Chromium dataset (21.8 versus 22.2 Mbp when using NA24143 versus NA12878 datasets with parameters –e 30 000 –r 0.05 –c 5 –l 5 –a 0.9) in spite of making 406 additional merges in the latter (64 516 versus 64 922 scaffolds, with NA12878 and NA24143 respectively, Supplementary Table S9). When we sub-sample both 10 × G data, we observe that ∼20-fold sequence read coverage of the human genome (approximately 200 M read pairs) is sufficient to achieve near-to-optimum scaffolding results with ARCS (Supplementary Fig. S2 and Table S10). This indicates that, under the conditions tested herein, with the draft assembly utilized and parameters set, the solution may work optimally even if presented with less data. We do stress the importance of characterizing the distribution of read multiplicities within barcodes to tune runtime parameters, as distributions may vary between datasets, as observed for NA24143 and NA12878 (Supplementary Fig. S3).
When comparing the impact of sequence coverage on scaffolding by ARCS as compared to fragScaff (parameters –E 30 000 –C 5 –j 1 –u 2) and Architect (parameters -t 5 –rc-abs-thr 3 –rc-rel-edge-thr 0.2 –rc-rel-prun-thr 0.2), ARCS was also found to produce assemblies with a higher NGA50 metric at all fold coverage subsets assessed (Supplementary Tables S9, S11, S12 and Fig. S2). This demonstrates that as well as being robust to the coverage of supplied linked reads, ARCS produces more contiguous assemblies than the other scaffolders evaluated in low (∼4.0-fold) to high (∼127-fold) sequence coverage conditions.
4 Discussion
Here, we have demonstrated the utility of a new algorithm, ARCS, for using 10 × Genomics read locality information to scaffold draft human genomes. We note that ARCS can perform well even when presented with relatively low coverage (25-fold) linked read data, generating accurate and megabase-range scaffolding results, starting from draft assemblies built entirely from short (250 bp) sequencing reads. We expect this performance to be generalizable to de novo assembly and scaffolding of other complex genomes.
Compared to fragScaff and Architect, ARCS produces more contiguous and correct assemblies. While running fragScaff with less stringent parameters yields assemblies with similar contiguity metrics to ARCS (16.9 versus 19.5 Mbp NG50, respectively), the fragScaff assemblies contain noticeably more misassemblies (3813 versus 3027). It is important to note that ARCS was developed specifically to utilize 10 × G data, where fragScaff was designed for scaffolding with CPT-seq data (Adey et al., 2014), which could have an impact on the resulting scaffolds. Furthermore, while fragScaff uses a more global approach of using maximum-weight minimum spanning trees based on the connected components of the scaffold graph, the ARCS graph is traversed based on local linkage support information. Although further study is required to fully understand the difference in assembly correctness between ARCS and fragScaff, it is possible that both of these factors influence the occurrence of misassemblies.
Despite thorough parameter sweeps, we found that Architect did not markedly improve the contiguity of the baseline H.sapiens assemblies. Apart from the tool design, which is intended for Illumina TruSeq read clouds (Kuleshov et al., 2016), it is possible that Architect‘s approach to ordering and orienting the sequences impacted the resulting assembly contiguity in our experiments. Whereas our scaffolding approach will take into account the ratio of barcode links between the most and second most supported edges when linking sequences, Architect requires sequence pairs to be linked by unambiguous edges in the graph. This requirement may have limited the resulting assembly contiguity for more complicated graphs, as the pruning stage may not remove all spurious edges.
We show that the contiguity of a H.sapiens contig assembly can be increased over six-fold with the use of 10 × G data with only a marginal increase in probable errors with an average ± SD of 196 ± 77 total breakpoints compared to the baseline contig assembly. However, there are limitations to the approach. As mentioned by Adey et al. (2014), when using barcodes, it is difficult to confidently place short sequence contigs into a scaffold due to a lower number of barcoded read pools aligning to the sequence. In addition, as the barcoded molecules may be over 100 kbp in length (Goodwin et al., 2016), they may span several entire short input sequences, preventing ARCS from extracting orientation information from read alignment positions, as they do not preferentially align to one end. This is exacerbated when scaffolding fragmented assemblies (N50 length < 10 kbp). To alleviate this problem, the minimum number of aligned reads required per barcode may be lowered to prevent possible links from being disregarded. Barcode reuse across molecules or incorrect alignment of linked reads due to repeats can also introduce false linkages at the sequence pairing stage, potentially resulting in incorrect merges during scaffolding. Although, with their Chromium technology, 10 × Genomics improved upon their GemCode protocol by increasing the number of fragment partitions and curbing barcode reuse—a trend we expect to continue as the technology matures further. While the positional information of linked reads within a given fragment is not known, making it challenging for estimating gap or overlap sizes in genome assemblies, it remains an attractive technology for scaffolding draft genomes. This is especially true when the technique is applied to later stages of scaffolding, when the contiguity of the draft sequence assembly is high.
As is the case with other bioinformatics software, fine-tuning parameters for best results necessitates ample testing. For ARCS, we generally find that requiring five or more aligned read pairs per sequence edge and setting the read alignment window to 30 kbp (ARCS –c and –e parameters), when possible, provides the best trade-off between assembly contiguity and accuracy. For LINKS, the ability to control the minimum number of supporting barcodes required to make a merge is most critical and increasing it decreases the chance of spurious joins. The evaluations presented in the paper, which serve as guidelines, indicate that requiring as low as five or more barcodes per sequence edge produces contiguous assemblies with accuracy similar to that of the starting, baseline assembly. We recommend testing ARCS using a range of parameters, and re-iterate that best scaffolding outcomes in terms of contiguity and accuracy track with both the contiguity and quality of the starting assembly draft.
Recently, 10 × G released their de novo assembly algorithm, Supernova (Weisenfeld et al., 2017), designed specifically for the Chromium sequencing technology. It uses read locality information early in the assembly process, as opposed to exclusively at the scaffolding stage, as ARCS and the other tools tested in this work do. This could help prevent misassemblies at the earlier stages of an assembly project, alleviating issues around the propagation of errors when scaffolding genome drafts. Stand-alone tools, on the other hand, make possible retrospective scaffolding of pre-existing drafts, and are beneficial to genome-finishing efforts (Hunt et al., 2014). However, as we have seen here, the degree at which existing scaffolding technologies perform varies, and the need of specialized bioinformatics solutions for this task is paramount.
To our knowledge, ARCS is the first publicly available stand-alone application for scaffolding draft genomes that is designed specifically for using 10 × Genomics linked reads. ARCS is freely available in open source for public use.
Funding
This work has been partly supported by the National Human Genome Research Institute of the National Institutes of Health (under award number R01HG007182). Additional funds were received by IB through Genome Canada, Genome Quebec, Genome British Columbia and Genome Alberta for the Spruce-Up (243FOR) project (www.spruce-up.ca). The content reported here is solely the responsibility of the authors, and does not necessarily represent the official views of the National Institutes of Health or other funding organizations.
Conflict of Interest: none declared.
Supplementary Material
References
- Adey A. et al. (2014) In vitro, long-range sequence information for de novo genome assembly via transposase contiguity. Genome Res., 24, 2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Goodwin S. et al. (2016) Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet., 17, 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M. et al. (2014) A comprehensive evaluation of assembly scaffolding tools. Genome Biol., 15, R42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman S.D. et al. (2017) ABySS 2.0: resource-efficient assembly of large genomes using a bloom filter. Genome Res., doi: 10.1101/gr.214346.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov V. et al. (2014) Whole-genome haplotyping using long reads and statistical methods. Nat. Biotechnol., 32, 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov V. et al. (2016) Genome assembly from synthetic long read clouds. Bioinformatics, 32, i216–i224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. (2009) Circos: an information aesthetic for comparative genomics. Genome Res., 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2015) BFC: correcting Illumina sequencing errors. Bioinformatics, 31, 2885–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics, 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostovoy Y. et al. (2016) A hybrid approach for de novo human genome sequence assembly and phasing. Nat. Methods, 13, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan V.M. et al. (2016) Health and population effects of rare gene knockouts in adult humans with related parents. Science, 352, 474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’connell J. et al. (2015) NxTrim: optimized trimming of Illumina mate pair reads. Bioinformatics, 31, 2035–2037. [DOI] [PubMed] [Google Scholar]
- Warren R.L. et al. (2015) LINKS: Scalable, alignment-free scaffolding of draft genomes with long reads. Gigascience, 4, 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld N.I. et al. (2017) Direct determination of diploid genome sequences. Genome Res, 27, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X.Y. et al. (2016) Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol., 34, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook J.M. et al. (2016) Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci. Data, 3, 160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


