Abstract
SV-plaudit is a framework for rapidly curating structural variant (SV) predictions. For each SV, we generate an image that visualizes the coverage and alignment signals from a set of samples. Images are uploaded to our cloud framework where users assess the quality of each image using a client-side web application. Reports can then be generated as a tab-delimited file or annotated Variant Call Format (VCF) file. As a proof of principle, nine researchers collaborated for 1 hour to evaluate 1,350 SVs each. We anticipate that SV-plaudit will become a standard step in variant calling pipelines and the crowd-sourced curation of other biological results.
Code available at https://github.com/jbelyeu/SV-plaudit
Demonstration video available at https://www.youtube.com/watch?v=ono8kHMKxDs
Keywords: structural variants, visualization, manual curation
Background
Large genomic rearrangements, or structural variants (SVs), are an abundant form of genetic variation within the human genome [1,2], and they play an important role in both species evolution [3,4] and human disease phenotypes [5–9]. While many methods have been developed to identify SVs from whole-genome sequencing (WGS) data [10–14], the accuracy of SV prediction remains far below that of single-nucleotide and insertion-deletion variants [1]. Improvements to SV detection algorithms have, in part, been limited by the availability and applicability of high-quality truth sets. While the Genome in a Bottle [15] consortium has made considerable progress toward a gold-standard variant truth set, the incredibly high quality of the data underlying this project (300x and PCR free) calls into question the generality of the accuracy obtained in typical quality WGS datasets (30x with PCR amplification).
Given the high false positive rate of SV calls from genome and exome sequencing, manual inspection is a critical quality control step, especially in clinical cases. Scrutiny of the evidence supporting an SV is considered to be a reliable “dry bench” validation technique, as the human eye can rapidly distinguish a true SV signal from alignment artifacts. In principle, we could improve the accuracy of SV call sets by visually validating every variant. In practice, however, current genomic data visualization methods [16–21] were designed primarily for spot checking a small number of variants and are difficult to scale to the thousands of SVs in typical call sets. Therefore, a curated set of SVs requires a new framework that scales to thousands of SVs, minimizes the time needed to adjudicate individual variants, and manages the collective judgment of large and often geographically dispersed teams.
Here we present SV-plaudit, a fast, highly scalable framework enabling teams of any size to collaborate on the rapid, web-based curation of thousands of SVs. In the web interface, users consider a curation question for a series of pre-computed images (Fig. 1, Supplementary Fig. S1) that contain the coverage, paired-end alignments, and split-read alignments for the region surrounding a candidate SV for a set of relevant samples (e.g., tumor and matched normal samples). The curation question is defined by the researcher to match the larger experimental design (e.g., a cancer study may ask if the variant is a somatic variant, a germline variant, or a false positive). Responses are collected and returned as a report that can be used to identify high-quality variants.
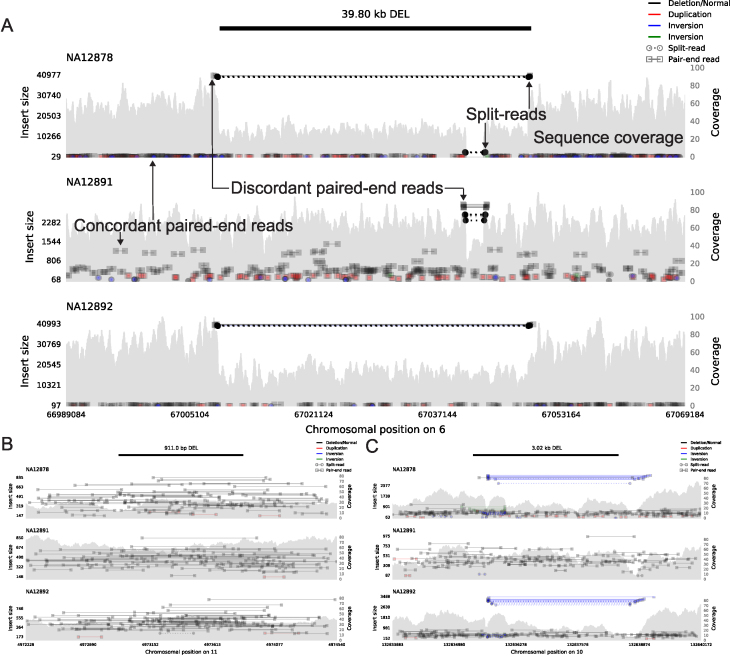
Figure 1:
Example samplot images of putative deletion calls that were scored as (A) unanimously GOOD, (B) unanimously BAD, and (C) ambiguous with a mix of GOOD and BAD scores with respect to the top sample (NA12878) in each plot. The black bar at the top of the figure indicates the genomic position of the predicted SV, and the following subfigures visualize the alignments and sequence coverage of each sample. Subplots report paired-end (square ends connected by a solid line, annotated as concordant and discordant paired-end reads in A) and split-read (circle ends connected by a dashed line, annotated in A) alignments by their genomic position (x-axis) and the distance between mapped ends (insert size, left y-axis). Colors indicate the type of event the alignment supports (black for deletion, red for duplication, and blue and green for inversion) and intensity indicates the concentration of alignments. The grey filled shapes report the sequence coverage distribution in the locus for each sample (right y-axis, annotated in A). The samples in each panel are a trio of father (NA12891), mother (NA12892), and daughter (NA12878).
While a team of curators is not required, collecting multiple opinions for each SV allows SV-plaudit to report the consensus view (i.e., a “curation score”) of each variant. This consensus is less susceptible to human error and does not require expert users to score variants. With SV-plaudit, it is practical to inspect and score every variant in a call set, thereby improving the accuracy of SV predictions in individual genomes and allowing curation of high quality-truth sets for SV method tuning.
Results
To assess SV-plaudit's utility for curating SVs, nine researchers in the Quinlan laboratory at the University of Utah manually inspected and scored the 1,350 SVs (1,310 deletions, 8 duplications, 4 insertions, and 28 inversions) that the 1000 Genomes Project [1] identified in the NA12878 genome (Supplemental File 1). Since we expect trio analysis to be a common use case of SV-plaudit, we included alignments from NA12878 and her parents (NA12891 and NA12892), and participants considered the curation question “The SV in the top sample (NA12878) is:” and answers “GOOD,” “BAD,” or “DE NOVO.” In total, the full experiment took less than 2 hours with Amazon costs totaling less than $0.05. The images (Supplemental File 2) were generated in 3 minutes (20 threads, 2.7 seconds per image) and uploading to S3 required 5 minutes (full command list in Supplemental File 3). The mean time to score all images was 60.1 minutes (2.67 seconds per image) (Fig. 2A, reports in Supplemental Files 4, 5). In the scoring process, no de novo variants were identified. Forty images did not render correctly due to issues in the alignment files (e.g., coverage gaps) and were removed from the subsequent analysis (Supplemental File 6).
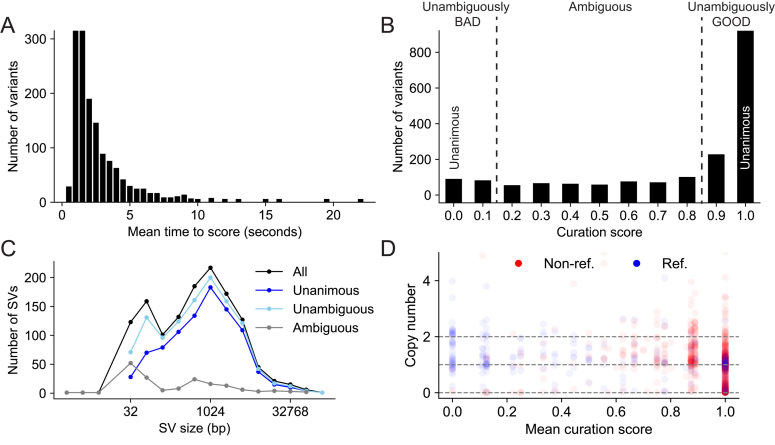
Figure 2:
(A) The distribution of the time elapsed from when an image was presented to when it was scored. (B) The distribution of curation scores. (C) The SV size distribution for all, unanimous (score 0 or 1), unambiguous (score <0.2 or >0.8), and ambiguous (score >= 0.2 and <= 0.8) variants. (D) A comparison of predictions for deletions between CNVNATOR copy number calls (y-axis), SVTYPER genotypes (color, “Ref.” is homozygous reference and “Non-ref.” is heterozygous or homozygous alternate), and curation scores (x-axis). This demonstrates a general agreement between all methods with a concentration of reference genotypes and copy number 2 (no evidence for a deletion) at curation score <0.2, and non-reference and copy number one or zero events (evidence for a deletion) at curation score >0.8. There are also false positives for CNVNATOR (copy number <2 at score = 0) and false negatives for SVTYPER (reference genotype at score = 1).
For this experiment, we used a curation score that mapped “GOOD” and “DE NOVO” to the value one, “BAD” to the value zero, and the mean as the aggregation function (Fig. 2B). Most (70.5%) variants were scored unanimously, with 67.1% being unanimously “GOOD” (score = 1.0, e.g., Fig. 1A) and 3.4% being unanimously “BAD” (score = 0.0, e.g., Fig. 1B). Since we had nine scores for each variant, we expanded our definition of “unambiguous” variants to be those with at most one dissenting vote (score <0.2 or >0.8), which accounted for 87.1% of the variants. The 12.9% of SVs that were “ambiguous” (more than one dissenting vote, 0.2 <= score <= 0.8) were generally small (median size of 310.5 bp vs 899.5 bp for all variants, Fig. 2C) or contained conflicting evidence (e.g., paired-end and split-read evidence indicated an inversion and the read-depth evidence indicated a deletion, e.g., Fig. 1C.).
Other methods, such as SVTYPER [22] and CNVNATOR [23], can independently assess the validity of SV calls. SVTYPER genotypes SVs for a given sample by comparing the number of discordant paired-end alignments and split-read alignments that support the SV to the number of pairs and reads that support the reference allele. CNVNATOR uses sequence coverage to estimate copy number for the region affected by the SV. Both of these methods confirm the voting results (Fig. 2D). Considering the set of “unambiguous” deletions, SVTYPER and CNVNATOR agree with the SV-plaudit curation score in 92.3% and 81.7% of cases, respectively. Here, agreement means that unambiguous false SVs (curation score <0.2) have a CNVNATOR copy number near 2 (between 1.4 and 2.4) or an SYTYPER genotype of homozygous reference. Unambiguous true SVs (curation score >0.8) have a CNVNATOR copy number near 1 or 0 (<1.4), or an SYTYPER genotype of nonreference (heterozygous or homozygous alternate).
Despite this consistency, using either SVTYPER or CNVNATOR to validate SVs can lead to false positives or false negatives. For example, CNVNATOR reported a copy number loss for 44.2% of the deletions that were scored as unanimously BAD, and SVTYPER called 30.7% of the deletions that were unanimously GOOD as homozygous reference. Conversely, CNVNATOR had few false negatives (2.4% of unanimously GOOD deletions were called as copy neutral), and SVTYPER had few false positives (0.2% of nonreference variants were unanimously BAD). This comparison is meant to demonstrate that different methods have distinct strengths and weaknesses and should not be taken as a direct comparison between SVTYPER and CNVNATOR, since CNVNATOR was one of nine methods used by the 1000 Genomes project while SVYTPER was not.
These results demonstrate that, with SV-plaudit, manual curation can be a cost-effective and robust part of the SV detection process. While we anticipate that automated SV detection methods will continue to improve, due in part to the improved truth sets that SV-plaudit will provide, directly viewing SVs will remain an essential validation technique. By extending this validation to full call sets, SV-plaudit not only improves specificity but can also enhance sensitivity by allowing users to relax quality filters and rapidly screen large sets of calls. Beyond demonstrating SV-plaudit's utility, our curation of SVs for NA12878 is useful as a high-quality truth set for method development and tuning. A Variant Call Format (VCF) file of these variants annotated with their curation score is available in Supplementary File 5.
Discussion
SV-plaudit is an efficient, scalable, and flexible framework for the manual curation of large-scale SV call sets. Backed by Amazon S3 and DynamoDB, SV-plaudit is easy to deploy and scales to teams of any size. Each instantiation of SV-plaudit is completely independent and can be deployed locally for private or sensitive datasets or be distributed publicly to maximize participation. By rapidly providing a direct view of the raw data underlying candidate SVs, SV-plaudit delivers the infrastructure to manually inspect full SV call sets. SV-plaudit also allows researchers to specify the questions and answers that users consider to ensure that the curation outcome supports the larger experimental design. This functionality is vital to a wide range of WGS experiments, from method development to the interpretation of disease genomes. We are actively working on machine learning methods that will leverage the curation scores for thousands of SV predictions as training data.
Conclusions
SV-plaudit was designed to judge how well the data in an alignment file corroborate a candidate SV. The question of whether a particular SV is a false positive due to artifacts from sequencing or alignment is a broader issue that must be answered in the context of other data sources such as mappability and repeat annotations. While this second level of analysis is crucial, it is beyond the scope of this paper, and we argue this analysis be performed only for those SVs that are fully supported by the alignment data. While SV-plaudit combines samplot and PlotCritic to enable the curation of structural variant images, we emphasize that the PlotCritic framework can be used to score images of any type. Therefore, we anticipate that this framework will facilitate “crowd-sourced” curation of many other biological images.
Methods
Overview
SV-plaudit (Fig. 3) is based on two software packages: samplot for SV image generation and PlotCritic for staging the Amazon cloud environment and managing user input. Once the environment is staged, users log into the system and are presented with a series of SV images in either a random or predetermined order. For each image, the user answers the curation question and responses are logged. Reports on the progress of a project can be quickly generated at any point in the process.
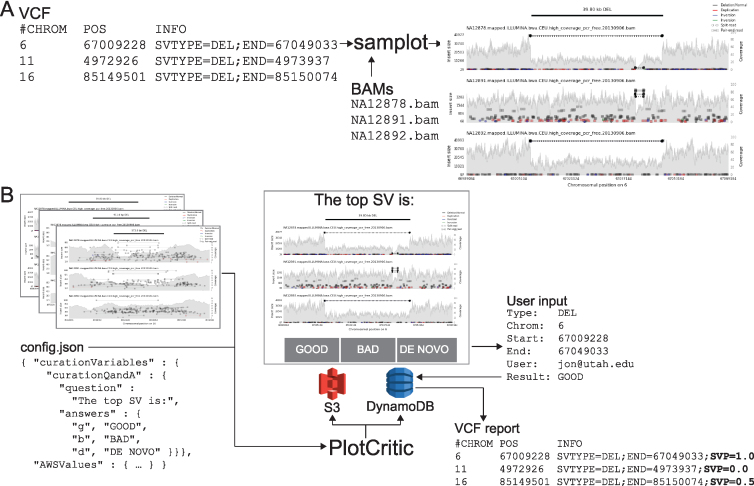
Figure 3:
The SV-plaudit process. (A) Samplot generates an image for each SV from VCF considering a set of alignment (BAM or CRAM) files. (B) PlotCritic uploads the images to an Amazon S3 bucket and prepares DynamoDB tables. Users select a curation answer (“GOOD,” “BAD,” or “DE NOVO”) for each SV image. DynamoDB logs user responses and generates reports. Within a report, a curation score function can be specified by mapping answer options to values and selecting an aggregation function. Here “GOOD” and “DE NOVO” were mapped to 1, “BAD” to 0, and the mean was used. An especially useful output option is a VCF annotated with the curation scores (shown here in bold as a SVP).
Samplot
Samplot is a Python program that uses pysam [24] to extract alignment data from a set of BAM or CRAM files and matplotlib [25] to visualize the raw data for the genomic region surrounding a candidate SV (Fig. 3A). For each alignment file, samplot renders the depth of sequencing coverage, paired-end alignments, and split-read alignments where paired-end and split-read alignments are color-coded based by the type of SV they support (e.g., black for deletion, red for a duplication, etc.) (Fig. 1, Supplementary Fig. S2, which considers variants at different sequencing coverages, and Supplementary Fig. S3, which depicts variants supported by long-read sequencing) [26,27]. Alignments are positioned along the x-axis by genomic location and along the left y-axis by the distance between the ends (insert size), which helps users to differentiate normal alignments from discordant alignments that support an SV. Depth of sequencing coverage is also displayed on the right y-axis to allow users to inspect whether putative copy number changes are supported by the expected changes in coverage. To improve performance for large events, we downsample “normal” paired-end alignments (a +/- orientation and an insert size range that is within Z standard deviations from the mean; by default Z = 4). Plots for each alignment file are stacked and share a common x-axis that reports the chromosomal position. By convention, the sample of interest (e.g., proband or tumor) is displayed as the top track, followed by the set of related reference genomes tracks (e.g., parents and siblings, matched normal sample). Users may specify the exact order by using command line parameters to samplot. A visualization of genome annotations and genes and exons within the locus is displayed below the alignment plots to provide context for assessing the SV's relevance to phenotypes. Rendering time depends on the number of samples, sequence coverage, and the size of the SV, but most images will require less than 5 seconds, and samplot rendering can be parallelized by SV call.
PlotCritic
PlotCritic (Fig. 3B) provides a simple web interface for scoring images and viewing reports that summarize the results from multiple users and SV images. PlotCritic is both highly scalable and easy to deploy. Images are stored on Amazon Web Services (AWS) S3 and DynamoDB tables store project configuration metadata and user responses. These AWS services allow PlotCritic to dynamically scale to any number of users. It also precludes the need for hosting a dedicated server, thereby facilitating deployment.
After samplot generates the SV images, PlotCritic manages their transfer to S3 and configures tables in DynamoDB based on a JSON configuration file (config.json file in Fig. 3B). In this configuration file, one defines the curation questions posed to reviewers as well as the allowed answers and associated keyboard bindings to allow faster responses (curationQandA field in Fig. 3B). In turn, these dictate the text and buttons that appear on the resulting web interface. As such, it allows the interface to be easily customized to support a wide variety of curation scenarios. For example, a cancer experiment may display a tumor sample and matched normal sample and ask users if the SV appears in both samples (i.e., a germline variant) or just in the tumor sample (i.e., a somatic variant). To accomplish this, the curation question (question field in Fig. 3B) could be “In which samples does the SV appear?”, and the answer options (answers field in Fig. 3B) could be “TUMOR,” “BOTH,” “NORMAL,” or “NEITHER.” Alternatively, in the case of a rare disease, the interface could display a proband and parents and ask if the SV is only in the proband (i.e., de novo) or if it is also in a parent (i.e., inherited). Since there is no limit to the length of a question or number of answer options, PlotCritic can support more complex experimental scenarios.
Once results are collected, PlotCritic can generate a tab-delimited report or annotated VCF that, for each SV image, details the number of times the image was scored and the full set of answers it received. Additionally, a curation score can be calculated for each image by providing a value for each answer option and an aggregation function (e.g., mean, median, mode, standard deviation, min, max). For example, consider the cancer example from above where the values 3, 2, 1, and 0 mapped to the answers “TUMOR,” “BOTH,” “NORMAL,” and “NEITHER,” respectively. If “mode” were selected as the curation function, then the curation score would reflect the opinion of a plurality of users. The mean would reflect the consensus among all users, and the standard deviation would capture the level of disagreement about each image. While we expect mean, median, mode, standard deviation, min, and max to satisfy most use cases, users can implement custom scores by operating on the tab-delimited report.
Each PlotCritic project is protected by AWS Cognito user authentication, which securely restricts access to the project website to authenticated users. A project manager is the only authorized user at startup and can authenticate other users using Cognito's secure services. The website can be further secured using HTTPS, and additional controls, such as IP restrictions, can be put in place by configuring AWS IAM access controls directly for S3 and DynamoDB.
Availability of source code and requirements
Project name: SV-plaudit
Project home page: https://github.com/jbelyeu/SV-plaudit
Operating systems: Mac OS and Linux
Programing language: Python, bash
License: MIT
Research Resource Initiative Identification ID: SCR_01 6285
Availability of supporting data and material
The datasets generated and/or analyzed during the current study are available in the 1000 Genomes Project repository, ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/phase3/data/
All test data used or generated during this study, and a snapshot of the code, are available in the GigaScience GigaDB repository [28].
Additional files
Supplemental Figure 1. Plots for different structural variant types shown in sample NA12878. (A) A region is shown where a duplication event was called. (B) A region is shown where an inversion event was called.
Supplemental Figure 2. A deletion call for sample NA12878 using different sequencing data to compare variant plots from high, medium, and low coverage levels. Mean sequencing depth of the BAM files used was (A) 58x (1000 Genomes Project, high coverage), (B) 33x (Genome in a Bottle Consortium), (C) and 5x (1000 Genomes Project, low coverage).
Supplemental Figure 3. A selection of structural variant visualizations from the Genome in a (A and B), “LongReadHomRef” in (C), and “NoConsensusGT” in (D).
Supplemental_File_1.vcf
Supplemental_File_3.sh
Supplemental_File_4.csv
Supplemental_File_5.vcf
Supplemental_File_6.txt
Abbreviation
SV: structural variant; VCF: Variant Call Format; WGS: Whole Genome Sequencing.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Supplementary Material
4/9/2018 Reviewed
4/9/2018 Reviewed
Funding
This research was supported by US National Human Genome Research Institute awards to R.M.L. (NIH K99HG009532) and A.R.Q. (NIH R01HG006693 and NIH R01GM124355) as well as a US National Cancer Institute award to A.R.Q. (NIH U24CA209999).
Authors' contributions
J.R.B. and R.M.L. developed the software. J.R.B., T.J.N., B.S.P., T.A.S., J.M.H., S.N.K., M.E.C., B.K.L., and R.M.L. scored variants for the experiment. J.R.B., A.R.Q., and R.M.L. wrote the manuscript. A.R.Q. and R.M.L. conceived the study.
References
- 1. Sudmant PH, Rausch T, Gardner EJ et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Redon R, Ishikawa S, Fitch KR et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Newman TL, Tuzun E, Morrison VA et al. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 2005;15:1344–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–64. [DOI] [PubMed] [Google Scholar]
- 5. Payer LM, Steranka JP, Yang WR et al. Structural variants caused by Alu insertions are associated with risks for many human diseases. Proc Natl Acad Sci U S A. 2017;114:E3984–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schubert C. The genomic basis of the Williams-Beuren syndrome. Cell Mol Life Sci. 2009;66:1178–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pleasance ED, Cheetham RK, Stephens PJ et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–82. [DOI] [PubMed] [Google Scholar]
- 9. Zhang F, Gu W, Hurles ME et al. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ye K, Schulz MH, Long Q et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rausch T, Zichner T, Schlattl A et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Handsaker RE, Korn JM, Nemesh J et al. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet. 2011;43:269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kronenberg ZN, Osborne EJ, Cone KR et al. Wham: Identifying structural variants of biological consequence. PLoS Comput Biol. 2015;11:e1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Layer RM, Chiang C, Quinlan AR et al. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zook JM, Chapman B, Wang J et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32:246–51. [DOI] [PubMed] [Google Scholar]
- 16. Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fiume M, Williams V, Brook A et al. Savant: genome browser for high-throughput sequencing data. Bioinformatics. 2010;26:1938–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Munro JE, Dunwoodie SL, Giannoulatou E. SVPV: a structural variant prediction viewer for paired-end sequencing datasets. Bioinformatics. 2017;33:2032–3. [DOI] [PubMed] [Google Scholar]
- 19. O'Brien TM, Ritz AM, Raphael BJ et al. Gremlin: an interactive visualization model for analyzing genomic rearrangements. IEEE Trans Vis Comput Graph. 2010;16:918–26. [DOI] [PubMed] [Google Scholar]
- 20. Wyczalkowski MA, Wylie KM, Cao S et al. BreakPoint Surveyor: a pipeline for structural variant visualization. Bioinformatics. 2017;33:3121–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spies N, Zook JM, Salit M et al. svviz: a read viewer for validating structural variants. Bioinformatics. 2015;31:3994–6. [DOI] [PubMed] [Google Scholar]
- 22. Chiang C, Layer RM, Faust GG et al. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods. 2015;12:966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abyzov A, Urban AE, Snyder M et al. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. https://github.com/pysam-developers/pysam [PDF]pysam documentation - Read the Docs.
- 25. Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science Engineering. 2007;9:90–95. [Google Scholar]
- 26. Zook JM, Catoe D, McDaniel J et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data. 2016;3:160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Daniel Kortschak R, S Pedersen B, L Adelson D. bíogo/hts: high throughput sequence handling for the Go language. JOSS. 2017;2:168. [Google Scholar]
- 28. Belyeu JR, Nicholas TJ, Pedersen BS et al. Supporting data for “SV-plaudit: A cloud-based framework for manually curating thousands of structural variants”. GigaScience Database. 2018. http://dx.doi.org/10.5524/100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
4/9/2018 Reviewed
4/9/2018 Reviewed