Abstract
Classical simulations of protein flexibility remain computationally expensive, especially for large proteins. A few years ago, we developed a fast method for predicting protein structure fluctuations that uses a single protein model as the input. The method has been made available as the CABS-flex web server and applied in numerous studies of protein structure-function relationships. Here, we present a major update of the CABS-flex web server to version 2.0. The new features include: extension of the method to significantly larger and multimeric proteins, customizable distance restraints and simulation parameters, contact maps and a new, enhanced web server interface. CABS-flex 2.0 is freely available at http://biocomp.chem.uw.edu.pl/CABSflex2.
INTRODUCTION
The dynamics of protein structures defines their biological functions. Because the experimental investigation of protein flexibility is often difficult or impossible, computational approaches play a significant role in this field. Simulations of biologically relevant protein fluctuations remain computationally demanding (using classical modeling tools of atomistic resolution) and often require supercomputer power. An inexpensive alternative is using coarse-grained simulation models combined with the reconstruction of predicted structures to all-atom representation (1). In 2013, we developed the CABS-flex web server for fast simulations of near-native dynamics of globular proteins (2). The CABS-flex method was shown to be a computationally efficient alternative to the classical, all-atom molecular dynamics (3). We also demonstrated that fluctuations of protein residues obtained from CABS-flex are well correlated to those of NMR ensembles (4). The CABS-flex method is also used as part of the AGGRESCAN3D method for the prediction of protein aggregation propensities (5) (AGGRESCAN3D employs CABS-flex to include the influence of dynamic protein structure fluctuations on aggregation propensity). Moreover, the CABS-flex methodology is a component of the CABS-dock method for protein-peptide docking (6–8), which enables significant flexibility of a peptide and a protein receptor during explicit simulation of peptide binding.
In CABS-flex, protein dynamics is simulated using a CABS coarse-grained protein model (1). The CABS model employs the Monte Carlo dynamics and asymmetric Metropolis scheme which satisfies requirements of microscopic reversibility and Boltzmann distribution of generated ensembles (1,9). Several works demonstrating the agreement between CABS long-time dynamics and experimental data have been summarized in a recent review (1). CABS-flex provides an alternative to other efficient methods for generating protein residue fluctuation profiles, such as sequence-based predictors of protein disordered regions or other coarse-grained approaches. In comparison to sequence-based predictors (10), CABS-flex is better adapted to detecting non-obvious dynamic fluctuations, for example within the well-defined secondary structural elements that could be biologically relevant. Other computational tools use normal mode analysis (NMA) (11–15) or various kinds of coarse-grained models (16,17). Their limitations depend on their particular design, especially on the simplifications assumed, and on the modeled system (for example NMA is well suited only for certain systems (18)).
In this work, we present a major update of the original CABS-flex, which significantly extends its capabilities. CABS-flex 2.0 has three major feature upgrades:
the limitations of protein structure input have been extended from 400 to 2000 amino acids, and from single-chain proteins only to proteins consisting of up to 10 chains;
a panel of customizable simulation parameters and options that enable deeper control of the simulation process, including user-defined modifications of distance restraints;
protein contact maps for simulation trajectory (presenting frequency of residue-residue contacts during simulation) and for 10 representative protein models.
According to users’ feedback, the major drawbacks of the original CABS-flex server were protein size limitations (restricted only to single-chain proteins shorter than 400 amino acids). The significant extension of protein size limits (described above) was possible thanks to dedicating significantly larger computational resources to web server jobs and rewriting the CABS-flex code.
The original CABS-flex server did not allow modifications of simulation settings, while CABS-flex 2.0 features customizable simulation parameters and options that enable users to tailor the simulation to their requirements. The customizable simulation parameters include (among others): temperature, simulation length and distance restraints. The introduced versatility makes the CABS-flex server suitable for performing complex simulations of proteins with disordered regions of significant length, flexible loops or simulations with user-defined distance restraints.
Additionally, the newly designed interface of the CABS-flex 2.0 server provides new and intuitive input forms, as well as extended output panels for interactive result analysis. A new feature of contact maps facilitates results analysis and provides a direct insight into intramolecular interactions of the modeled protein.
MATERIALS AND METHODS
CABS-flex modeling protocol
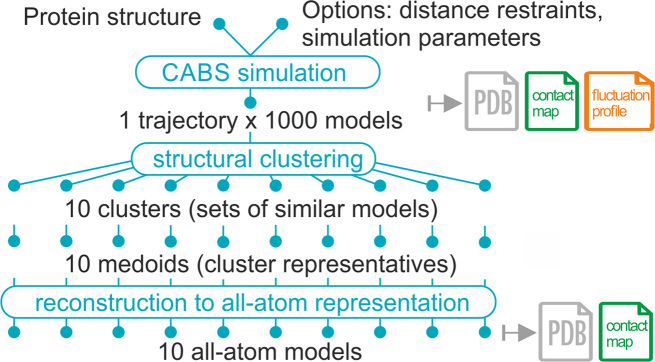
The original CABS-flex server was described in detail elsewhere (2). CABS-flex modeling results were validated against all-atom MD data (3) and NMR experimental structures (4). The method was also extensively validated as a component of modeling tools for predicting protein solubility (5) and flexible peptide docking (6–8). The key principles of CABS-flex 2.0 remain similar to these of the original web server. The overview of the method pipeline is presented in Figure 1.
Figure 1.
CABS-flex 2.0 pipeline. In the first modeling step, CABS-flex uses the CABS coarse-grained protein model as an efficient simulation engine (the CABS model design and applications have been recently reviewed (1)). Subsequently, a protein dynamics trajectory is clustered into 10 cluster medoids (a medoid is a representative model of a cluster whose average dissimilarity to all the models in the cluster is minimal), which are automatically reconstructed to all-atom representation.
DESCRIPTION OF THE WEB SERVER
Input data
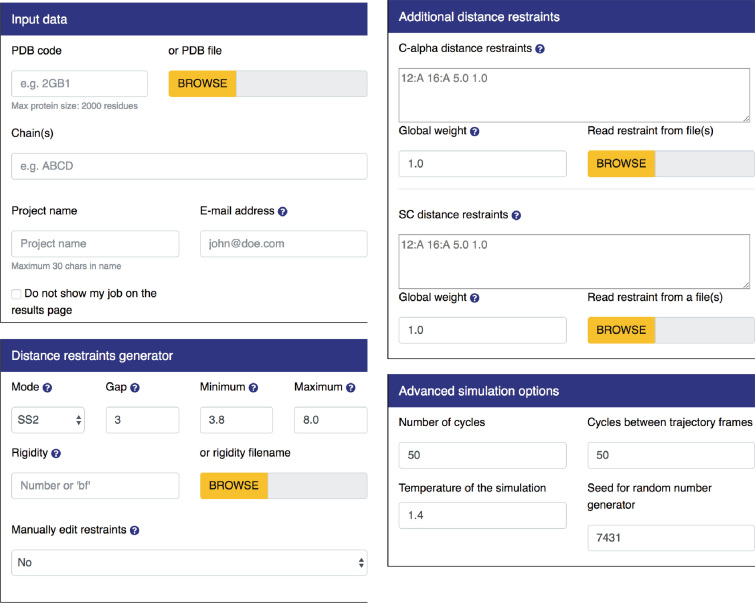
The only required input is the protein structure. It may be provided either as a PDB code or as an uploaded PDB format file (through ‘Input data’ panel, see Figure 2). In both of these cases, all the residues must comprise a complete set of backbone atoms (i.e. N, Cα, C and O). The input protein structure may contain multiple chains. However, each chain must be no longer than 2000 residues, due to limited computational resources. Additionally, CABS-flex 2.0 allows optional input information:
Chain name(s) provided as a multi-letter code used to select specific chains from the uploaded PDB file; for example, ‘ABCD’ enables selecting A, B, C and D chains from the PDB file;
Project name, which will appear in the queue list, unless the option ‘Do not show my job on the results page’ is used. If not provided, the name will be replaced with a random hashcode;
Email address, which will be used by the server to send an email notification about job completion
Figure 2.
Input panels. By default, all panels except for ‘Input data’ are hidden and can be displayed by clicking on the selected tab.
Advanced input options
By default, CABS-flex uses a set of distance restraints and simulation parameters discussed by Jamroz et al. (3). These settings were derived to provide the best possible convergence between CABS-flex simulations and the consensus picture of protein fluctuations in aqueous solution derived by all-atom Molecular Dynamics (MD) simulations (of 10 nanosecond length, with different force fields) for globular proteins. Therefore, the default set of restraints and parameters is dedicated to the short timescale dynamics of folded (globular) proteins. For other protein systems and timescales, the CABS-flex settings may need to be properly adjusted.
The advanced options enable modification of the default settings according to the user's needs and information about the modeled system. The advanced options are organized under three dropdown panels: distance restraints generator, additional distance restraints, advanced simulation options, which are described below and presented in Figure 2.
Distance restraints generator
The ‘distance restraints generator’ panel allows the user to customize Cα–Cα restraints imposed on the protein residues. The use of distance restraints enables the user to restrict the conformational search (for selected protein fragments) according to knowledge about the modeled system. For example, it is possible to prevent selected protein fragments from moving or treat them as moderately or fully flexible. By default, CABS-flex uses a set of distance restraints which have a weak stabilizing effect on secondary structure elements, in comparison to CABS-flex simulations without any restraints (3).
An individual distance restraint between two Cα atoms is defined by the following syntax:
“residue1_id residue2_id distance strength”, for example: “123:A 73:B 14.3 1” defines a single restraint between the 123th residue of chain A and the 73rd residue of chain B to be at a distance of 14.3 Angstroms with a restraint strength of 1. A restraint strength value of ‘1’ (or larger) means rigid and ‘0’ makes a restraint non-existent.
A set of restraints is automatically generated according to the scheme defined by the ‘Mode’, ‘Gap’, ‘Minimum’, ‘Maximum’ and ‘Rigidity’ fields:
‘Mode’ tells the algorithm to only generate restraints for which at least one (‘Mode’ = ‘SS1’) or both (‘Mode’ = ’SS2’) residues are assigned a regular secondary structure (helix or sheet). ‘Mode’ = ‘All’ generates restraints for all residues and ‘None’ generates no restraints. By default ‘Mode’ = ‘SS2’
‘Gap’ sets the minimum distance along the protein chain for two residues to be bound with a restraint. By default ‘Gap’ = 3, which means for example that residue number 15 cannot be restrained with residues numbered from 12th to 18th.
‘Minimum’ and ‘Maximum’ fields define the minimum and maximum length of the restraint in Angstroms. In other words, restraints will be automatically generated only for residues within these distances (default values are 3.8 and 8.0, for minimum and maximum, respectively).
‘Rigidity’ is for designing restraint strengths in three different ways. First, by providing a single number. Then, the number is set as a restraint strength in all generated restraints (by default, ‘Rigidity’ = 1). Second, by providing the ‘bf’ keyword. This allows creating restraint strengths according to numbers provided in the beta factor column (for C-alpha atoms) in the input PDB file. The smaller rigidity value of two restrained residues is taken as restraint strength. Third, by providing a custom file with one rigidity number per residue (using the format: ‘residue_id rigidity’, for example: ‘1:A 0.75’). The smaller rigidity value of two restrained residues is taken as restraint strength.
Note that the generated set of protein restraints is accessible for inspection and download after running the job from the ‘Project information’ tab.
The option ‘Manually edit protein restraints’ provides an additional web-based tool for manual restraints edition. When ‘Yes’ is selected, the option redirects the user to a new web page before the job is submitted. CABS-flex 2.0 generates a set of restraints to be used in simulation based on selected options. Each of the generated restraints can be manually edited (including its strength and distance) or deleted. Additionally, new restraints between any of the protein's residues can be created and further modified.
Additional distance restraints
This form enables inserting additional (to those automatically generated) restraints imposed on the modeled protein structure. The top panel is for the Cα–Cα restraints and the bottom panel is for the side chain–side chain restraints (SC–SC). Restraints can be added either through the text box or from an uploaded text file (for the definition of restraints, see the section above). Additionally, the global restraints weight for both Cα–Cα and SC-SC restraints can be set in respective fields.
Advanced simulation options
This panel enables modification of the parameters that control the simulation. The ‘Number of cycles’(Ncycle) field sets the total number of models saved in the trajectory to be equal to 20 x Ncycle, i.e. setting Ncycle = 50 results in 20 × 50 = 1000 models in the trajectory. It is worth noting that not all of the models generated during the CABS-flex simulation are written to the trajectory. The next field, ‘Cycles between trajectory frames’ (Nskipped), sets the number of models skipped on saving models, in other words for Nskipped = 100 every hundredth model will be saved. This field also indirectly sets the total number of models generated, i.e. for Ncycle = 50 and Nskipped = 100, the total number of generated models equals 20 × 50 × 100 = 100 000, 1000 of which will be written to the trajectory. For more details refer to the wiki pages of the standalone CABS-flex package, available at https://bitbucket.org/lcbio/cabsflex
The dimensionless reduced temperature in the CABS model cannot be straightforwardly linked to the real temperature. Its role, however, is similar: it serves as a parameter controlling the total energy of the modeled system. The higher the temperature, the more mobile are the atoms, which results in larger fluctuations. In the CABS model, T = 1.0 is usually close to the temperature of the crystal (native state), T = 2.0 typically enables complete unfolding of unrestrained small protein chains. In other words, the reduced temperature T has the same meaning as the product of Boltzmann constant and real temperature.
Output files and data
Once the job is completed, three additional tabs are added to its web page: ‘Models’, ‘Contact maps’ and ‘Fluctuation plot’.
Models tab
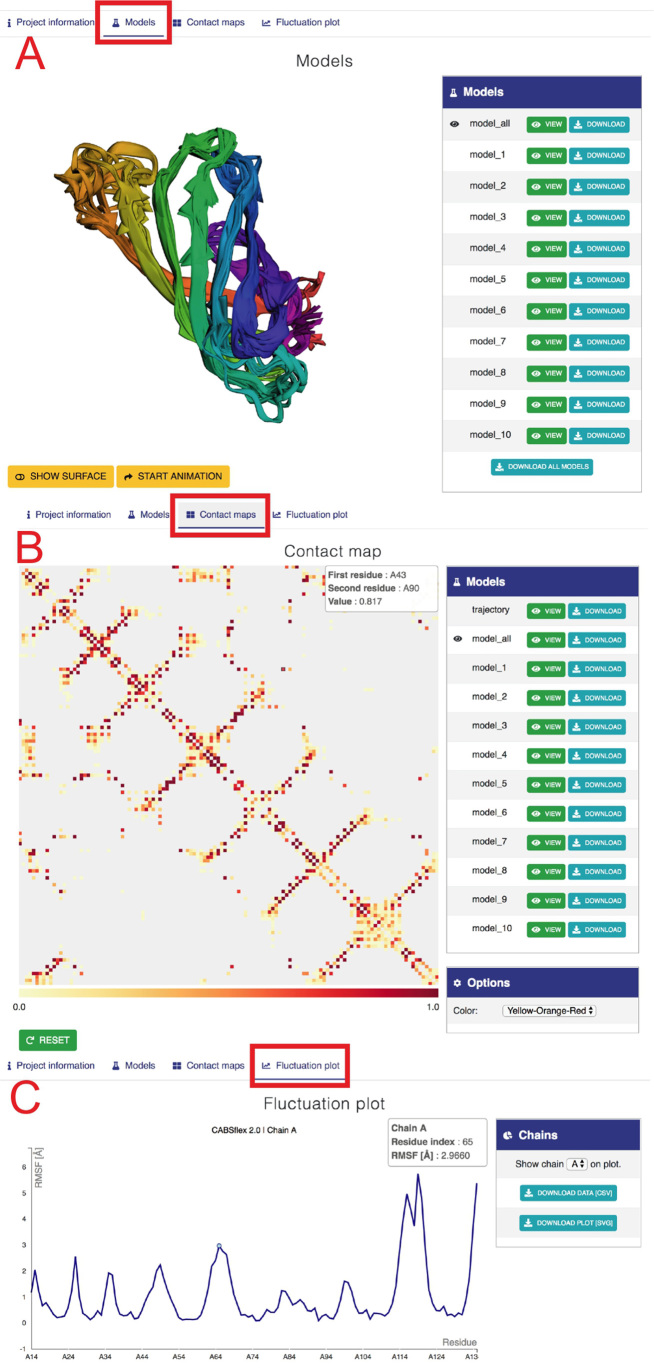
This section is used to display three-dimensional structures of 10 final models (see Figure 3A) and to provide the structure data. An interactive molecular viewer enables visual analysis of the obtained results. The ‘model_all’ set consists of all 10 final models, and its visualization shows the structure heterogeneity present in the final models. Additional viewing options are available, such as toggling between surface and cartoon representation and rotating the protein molecule about the vertical axis. Users may change what is being displayed in the viewer by selecting from the side menu one of the ten final models or the trajectory (all final models in superposition). Each final model as well as the trajectory may be downloaded as PDB files.
Figure 3.
Example view of output tabs for streptavidin protein (with default CABS-flex settings, 2RTM PDB was used as the input). (A) ‘Models’ tab. The left panel shows three-dimensional visualization of 10 models. The right panel provides buttons to display and download the selected model or all 10 models (model_all). Below: buttons to modify the surface and rotate the model. (B) ‘Contact maps’ tab. The tab shows a matrix with contacts with the color scale below (note that pointing the cursor at the contact between residue 39 and 104 in chain A invoked an information balloon with contact information). Right panels provide buttons to display and download maps (in a graphic svg format and text files, both as zip files under Download buttons), and the ‘Options’ section that allows changing the color range of contacts. (C) The ‘Fluctuation plot’ tab shows the residue fluctuation profile (RMSF) for a selected protein chain. Right panels allow selecting a protein chain and downloading the displayed data.
Contact maps tab
The ‘Contact maps’ tab provides a detailed view of the protein's residue-residue interaction pattern (see Figure 3B). The central panel displays an interactive contact map between residues. Each dot in the map represents an interaction within a pair of residues. Its color depends on the frequency of occurrence of this particular interaction in the models set that was used to generate data for the map. The user may select in the side tab which set of structures is used to populate the map, by clicking on the ‘View’ button, next to one of the twelve preset sets. Sets named model_1-10 are singletons containing just one of the final models, and by selecting one of them a binary (contact frequency is either 0 or 1) contact pattern for this model is displayed. The ‘Trajectory’ set contains all models saved in the trajectory during the simulation (1000 models by default); the ‘trajectory’ contact map displays which contacts were most frequent throughout the simulation. The ‘model_all’ set consists of all ten final models, and its map shows the most conserved structural features present in the final models. Maps are available for download as graphic (svg format) and text files (both as zip files under Download buttons). Hovering on any of the squares displays a rectangle in the upper-left corner with more detailed information about the selected contact. The color range of the contact map is adjustable in the ‘Options’ panel. Any transformation of the contact map (translation, scaling) can be reversed by clicking on the ‘Reset’ button below it.
Fluctuation plot tab
The ‘Fluctuation plot’ tab provides an interactive 2D plot presenting residue-wise fluctuations recorded throughout the simulation. Fluctuations are calculated as RMSF after global superposition. Both graphics (svg) and numerical data (csv) are available for download. For multichain proteins a separate plot is generated for each chain. They can be displayed by selecting it in the ‘Chains’ panel.
SERVER ARCHITECTURE AND DOCUMENTATION
The CABS-flex 2.0 server is equipped with an HTML web interface dynamically generated with a Flask framework and a jinja2 templating engine. Validated user-provided data are added to the MySQL database. The job is either started, if there are free server resources available, or put in the queue to wait for execution. The server notifies the user on the computation progress, reporting the job status (‘pending’, ‘in queue’, ‘running’ and ‘done’). Molecular visualization is executed using a 3Dmol library (HTML5/Javascript). The interactive contact maps and plots are created with a D3.js library (HTML5/Javascript). Information about proteins is downloaded from the PDB using RESTful services. The CABS-flex 2.0 website runs on the Apache2 server and MySQL database for user queue storage. As the simulation engine, the server uses the newly developed CABS-flex standalone package, which can be downloaded from http://bitbucket.org/lcbio/cabsflex. The CABS-flex 2.0 server is free, open to all users and there is no login requirement.
The documentation of CABS-flex 2.0 is available online under the ‘How to’ subpage.
SUMMARY
In this work, we developed an easy-to-use CABS-flex 2.0 web server interface for efficient simulations of large-scale (large in the context of protein size and timescales) structure fluctuations of proteins and protein complexes. CABS-flex 2.0 is based on the coarse-grained simulations of protein motion that have been successfully used in the CABS-flex 1.0 server (2,4) and other modeling tools (5–8). CABS-flex 2.0 server allows simulations of large protein systems and elaborate (when required) control of protein flexibility. The output visualizations and fluctuation trajectories can be used in studies of biomolecular processes. In particular, CABS-flex 2.0 may be used to identify the most mobile structural fragments, generate structures of protein receptors for molecular docking (as an alternative to available experimental structures), study allosteric equilibria and perform many other tasks that require large-timescale dynamics data.
FUNDING
National Science Center (NCN, Poland) Grant [MAESTRO2014/14/A/ST6/00088]. Funding for open access charge: National Science Center (NCN, Poland) Grant [MAESTRO2014/14/A/ST6/00088].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kmiecik S., Gront D., Kolinski M., Wieteska L., Dawid A.E., Kolinski A.. Coarse-Grained protein models and their applications. Chem. Rev. 2016; 116:7898–7936. [DOI] [PubMed] [Google Scholar]
- 2. Jamroz M., Kolinski A., Kmiecik S.. CABS-flex: Server for fast simulation of protein structure fluctuations. Nucleic Acids Res. 2013; 41:W427–W431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jamroz M., Orozco M., Kolinski A., Kmiecik S.. Consistent view of protein fluctuations from All-Atom molecular dynamics and Coarse-Grained dynamics with Knowledge-Based Force-Field. J. Chem. Theory Comput. 2013; 9:119–125. [DOI] [PubMed] [Google Scholar]
- 4. Jamroz M., Kolinski A., Kmiecik S.. CABS-flex predictions of protein flexibility compared with NMR ensembles. Bioinformatics. 2014; 30:2150–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zambrano R., Jamroz M., Szczasiuk A., Pujols J., Kmiecik S., Ventura S.. AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures. Nucleic Acids Res. 2015; 43:W306–W313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kurcinski M., Jamroz M., Blaszczyk M., Kolinski A., Kmiecik S.. CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 2015; 43:W419–W424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blaszczyk M., Kurcinski M., Kouza M., Wieteska L., Debinski A., Kolinski A., Kmiecik S.. Modeling of protein-peptide interactions using the CABS-dock web server for binding site search and flexible docking. Methods. 2016; 93:72–83. [DOI] [PubMed] [Google Scholar]
- 8. Ciemny M.P., Debinski A., Paczkowska M., Kolinski A., Kurcinski M., Kmiecik S.. Protein-peptide molecular docking with large-scale conformational changes: the p53-MDM2 interaction. Sci. Rep. 2016; 6:37532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kolinski A. Protein modeling and structure prediction with a reduced representation. Acta Biochim. Polon. 2004; 51:349–371. [PubMed] [Google Scholar]
- 10. Dosztanyi Z., Meszaros B., Simon I.. Bioinformatical approaches to characterize intrinsically disordered/unstructured proteins. Brief. Bioinform. 2010; 11:225–243. [DOI] [PubMed] [Google Scholar]
- 11. Kruger D.M., Ahmed A., Gohlke H.. NMSim web server: integrated approach for normal mode-based geometric simulations of biologically relevant conformational transitions in proteins. Nucleic Acids Res. 2012; 40:W310–W316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hollup S.M., Salensminde G., Reuter N.. WEBnm@: a web application for normal mode analyses of proteins. BMC Bioinformatics. 2005; 6:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lopez-Blanco J.R., Aliaga J.I., Quintana-Orti E.S., Chacon P.. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014; 42:W271–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lindahl E., Azuara C., Koehl P., Delarue M.. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 2006; 34:W52–W56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suhre K., Sanejouand Y.H.. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004; 32:W610–W614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Camps J., Carrillo O., Emperador A., Orellana L., Hospital A., Rueda M., Cicin-Sain D., D’Abramo M., Gelpi J.L., Orozco M.. FlexServ: an integrated tool for the analysis of protein flexibility. Bioinformatics. 2009; 25:1709–1710. [DOI] [PubMed] [Google Scholar]
- 17. Lauck F., Smith C.A., Friedland G.F., Humphris E.L., Kortemme T.. RosettaBackrub–a web server for flexible backbone protein structure modeling and design. Nucleic Acids Res. 2010; 38:W569–W575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma J. Usefulness and limitations of normal mode analysis in modeling dynamics of biomolecular complexes. Structure. 2005; 13:373–380. [DOI] [PubMed] [Google Scholar]