The transformics method was developed to support the integrated testing strategy for carcinogenesis. Results showed the main role of the immune system in the transformation by 3-MCA, with initiating events related to non-genotoxic mechanisms, suggesting the involvement of the AhR receptor.
Abstract
The development of alternative methods to animal testing is a priority in the context of regulatory toxicology. Carcinogenesis is a field where the demand for alternative methods is particularly high. The standard rodent carcinogenicity bioassay requires a large use of animals, high costs, prolonged duration and shows several limitations, which can affect the comprehension of the human relevance of animal carcinogenesis. The cell transformation assay (CTA) has long been debated as a possible in vitro test to study carcinogenesis. This assay provides an easily detectable endpoint of oncotransformation, which can be used to anchor the exposure to the acquisition of the malignant phenotype. However, the current protocols do not provide information on either molecular key events supporting the carcinogenesis process, nor the mechanism of action of the test chemicals. In order to improve the use of this assay in the integrated testing strategy for carcinogenesis, we developed the transformics method, which combines the CTA and transcriptomics, to highlight the molecular steps leading to in vitro malignant transformation. We studied 3-methylcholanthrene (3-MCA), a genotoxic chemical able to induce in vitro cell transformation, at both transforming and subtransforming concentrations in BALB/c 3T3 cells and evaluated the gene modulation at critical steps of the experimental protocol. The results gave evidence for the potential key role of the immune system and the possible involvement of the aryl hydrocarbon receptor (AhR) pathway as the initial steps of the in vitro transformation process induced by 3-MCA, suggesting that the initiating events are related to non-genotoxic mechanisms.
Introduction
In recent years, the European Union has promoted the development of alternative methods, with the aim to achieve the replacement, reduction and refinement of animal experiments, according to the 3Rs principles (1). Similarly, in the United States, there is a strong drive to move to more mechanistic-based in vitro and high-throughput chemical testing as part of the 21st century vision of toxicology (2). Despite the prodigious efforts made so far, the complete replacement of animal experimentation by validated alternative assays remains a challenging issue (3–5).
Carcinogenesis is a field where the demand for alternative methods is particularly high (1). Moreover, carcinogenicity testing has been recognized as the area with the most relevant needs for harmonization among the different regulatory approaches (1). The standard rodent carcinogenicity bioassay (RCB) requires an extensive use of animals. Apart from animal welfare considerations, the RCB shows several limitations, particularly due to the high costs, the prolonged duration (2 years) and the scarce mechanistic information, which can make it difficult to completely understand the human relevance. Furthermore, international work is ongoing with the aim to review the uncertainty and complexity of the RCB-based assessments. This shall contribute to revisiting RCB reference data evaluation and to improve the definition of acceptable performance of in vitro approaches (4).
In addition, the complexity of the tumor process cannot completely be exploited in a single in vitro test, especially for the evaluation of non-genotoxic carcinogens (NGTXCs), which are characterized by different mechanisms of action, playing a role in the initiating events and early steps of the tumor process, as well as in sustaining the tumor growth and the acquisition of the malignant phenotype (6). Indeed, the current regulatory approaches are mainly based on genotoxicity assays, which are barely able to recognize compounds acting without a direct DNA damage. As a consequence, many NGTXCs might remain unidentified (6).
Due to the limitations of the current approaches to properly identify the NGTXCs chemicals, it has been proposed to develop an integrated approach to testing and assessment (IATA) for non-genotoxic carcinogenesis. An IATA is an approach based on multiple information sources used for hazard identification, hazard characterization and/or safety assessment of chemicals. An IATA integrates and weights all relevant existing evidence and guides the targeted generation of new data, where required, to inform regulatory decision-making regarding potential hazard and/or risk (7).
Among in vitro tests, the cell transformation assay (CTA) has been proposed as a possible alternative to animal models based on some experimental evidence that cellular and molecular processes involved in in vitro cell transformation seem to resemble those sustaining in vivo carcinogenesis and occur as a result of comprehensive cellular response to direct and indirect damage to DNA (8,9).
In particular, the in vitro transformation assay on BALB/c 3T3 cells is one of the CTA models, that benefits from a convenient protocol and an easily detectable endpoint, represented by malignant foci of transformed cells (10).
BALB/c 3T3 are embryonic mouse fibroblasts, which undergo transformation, following chemical treatment, with cells escaping the contact inhibition and piling up randomly (11). The transformed cells from malignant foci are tumorigenic and metastatic when injected into suitable host animals, and they acquire invasive properties in vitro (12–14). Whilst some recent studies have concluded that there is a satisfactory predictive capability of this CTA model, the validation performance was not considered to be sufficiently robust for regulatory test guideline purposes at the Organisation for Economic Co-operation and Development (OECD) (1,15,16).
Furthermore, the use of CTAs to predict carcinogenesis cannot be fully contemplated in the regulatory context because the current protocols do not provide any information on either molecular key events supporting the carcinogenesis process, nor the mechanism of action of the test chemicals. In 2014, the OECD considered that a number of issues made the development of a test guideline for CTA models impossible at that time and recommended the development of guidance documents to allow the use of these methods as part of a weight of evidence approach in the testing of substances for carcinogenic potential (17,18).
However, it is the application of a high-throughput microarray approach to CTAs that could overcome the lack of mechanistic understanding, by providing the key information on molecular mechanisms related to the final adverse outcome, as represented by the foci formation.
To explore this potential solution to the problem of poor mechanistic understanding of the CTA, the application of advanced genomics technologies to the BALB/c 3T3 CTA, for the identification of toxicity pathways and gene signatures predictive of carcinogenicity, is presented.
Few studies are available on the use of microarray technology applied to CTA, and they essentially focus on the identification of potential biomarkers as fingerprints of the tested chemicals. Rohrbeck et al. (9) applied a toxicogenomics approach to the BALB/c 3T3 CTA by using Affymetrix GeneChips, covering about 36 000 mouse gene transcripts, in order to identify a gene signature potentially predictive for carcinogenic agents. The transcriptional response to the treatment with benzo(a)pyrene, 3-methylcholanthrene (3-MCA) and 2,4-diaminotoluene was studied by evaluating the effects of these chemicals at different time points and concentrations, trying to identify a common gene profile (9).
In order to utilize the CTA within an IATA for carcinogenesis, we developed the transformics method, which utilized transcriptomics to highlight the molecular steps leading to in vitro malignant transformation. The reference chemical 3-MCA was tested at both in vitro transforming and subtransforming concentrations, and transcripts were analyzed at different time points along the process of in vitro transformation. Gene signatures, indicative of concentration-related events, were identified and anchored to the phenotypic endpoint. The results gave evidence for the potential key role of the immune system and for the possible involvement of the aryl hydrocarbon receptor (AhR) pathway as the initial step of the in vitro transformation process induced by 3-MCA. Overall, our results showed the strength of the transformics method to highlight the carcinogenic mechanisms that are not purely connected to genotoxicity.
Materials and methods
Experimental design
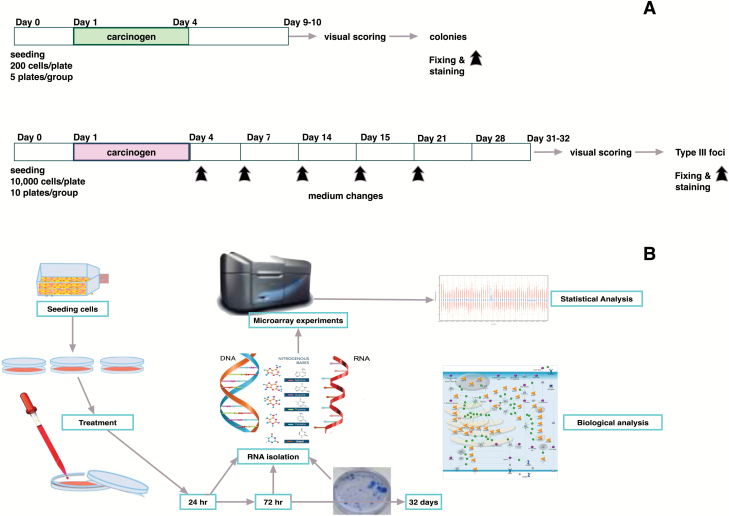
The experimental design is shown in Figure 1.
Figure 1.
(A) Experimental protocol of the CTA on BALB/c 3T3. Cells were seeded at 1 × 104 cells/plate and treated 24 h after the seeding. After 3 days, the medium was changed with culture medium. After 31–32 days, the cells were fixed and stained. A concurrent cytotoxicity assay was performed to evaluate the cell survival after the chemical treatment. (B) Integrated experimental approach. Cells were seeded and treated. At the selected time points, RNA was extracted, and microarray experiments were performed. A set of plates for each treatment were maintained in culture for the CTA. The foci formation served as the phenotypic anchoring of the microarray results. GeneSpring GX software (Agilent Technologies) was employed for the statistical analysis of microarray experiments, whereas MetaCore (Thomson Reuters, https://portal.genego.com/) was used for the biological analysis.
Cells
The original stock of BALB/c 3T3 cells, clone A31-1-1, was obtained from the Health Science Research Resource Bank (Osaka, Japan). Cells were tested, characterized and authenticated by the cell bank for the species origin. Working cultures were expanded from the original cryopreserved stock. Cells were grown in M10F, minimum essential medium supplemented with 10% fetal bovine serum (FBS, Gibco BRL). Only subconfluent cells were used, and the target cells were not maintained beyond the third passage after thawing. Cultures were maintained in a humidified incubator with an atmosphere of 5% CO2 in air at 37°C.
Chemicals and solutions
3-MCA and dimethyl sulfoxide (DMSO) were obtained by Sigma–Aldrich. DMSO was used at a 0.1% concentration as the vehicle of 3-MCA and as the negative control of the experiments. Two stock solutions of the reference chemical were prepared. The first one was obtained by diluting 3-MCA in DMSO at the final concentration of 4 mg/ml. This solution was then diluted at 1:100 in DMSO to obtain the second stock solution. Treatments were carried out by diluting 1:1000 the two stock solutions in the culture medium, to reach the final working concentration of 4 and 0.04 μg/ml.
Cytotoxicity test and CTA
The tests were performed according to the ECVAM validated protocol (15,16,19). Details are reported in Supplementary Information, available at Carcinogenesis Online.
The reference chemical 3-MCA was tested at two different concentrations: the transforming concentration of 4 µg/ml and the potentially subtransforming concentration of 0.04 µg/ml.
Total RNA extraction
Total RNA was isolated at 24 and 72 h of the exposure period and after 32 days, by using TRIzol Reagent (Invitrogen, San Diego, CA) followed by purification with Rneasy affinity column (Qiagen, Valencia, CA), according to the manufacturer’s instructions. RNA quality was assessed by Agilent Bioanalyzer 2100 (RNA 6000 Nano LabChip, Agilent Technologies, Palo Alto, CA). Four biological replicates were performed.
Total RNA labeling and hybridization
cRNA was labeled, purified and hybridized on oligonucleotide slides (SurePrint G3 Mouse Gene Expression 8x60K Microarray Kit) according to the Agilent 60-mer oligo microarray processing protocol (G4140-90040 One-Color Version 6.6 Revision B2, available online at www.chem.agilent.com).
Statistical analysis of microarray data
Raw data were filtered for intensity and quality signal by using GeneSpring GX (Agilent Technologies). All samples were then firstly analyzed with common statistical approach. One-way analysis of variance (One-way ANOVA), corrected by Benjamini–Hochberg, was used to select differentially expressed genes among the treatments and the controls with a P ≤ 0.01. Then, the selected list was analyzed by principal component analysis (PCA). The same approach was applied to samples belonging to each time point. Furthermore, a t-test analysis was carried out to underline the gene modulation induced by each treatment in comparison with DMSO (P ≤ 0.05 Benjamini–Hochberg). Venn diagrams for 24, 72 h and 32 days were obtained by crossing the resulting gene lists.
Tools of biological interpretation
The lists of differentially expressed genes in 0.04 and 4 µg/ml 3-MCA treatments were evaluated by using MetaCore, which is an integrated ‘knowledge-based’ platform with a manually annotated database of protein interactions verified by ‘small experiment’ data and gene–disease associations (https://portal.genego.com/). In particular, an enrichment analysis based on comparative analysis was performed to identify Pathways and Process Networks altered after 24 and 72 h of chemical exposure, whereas a single approach was used for the evaluation of the data at 32 days.
Results
Effects of 3-MCA on cytotoxicity and cell transformation
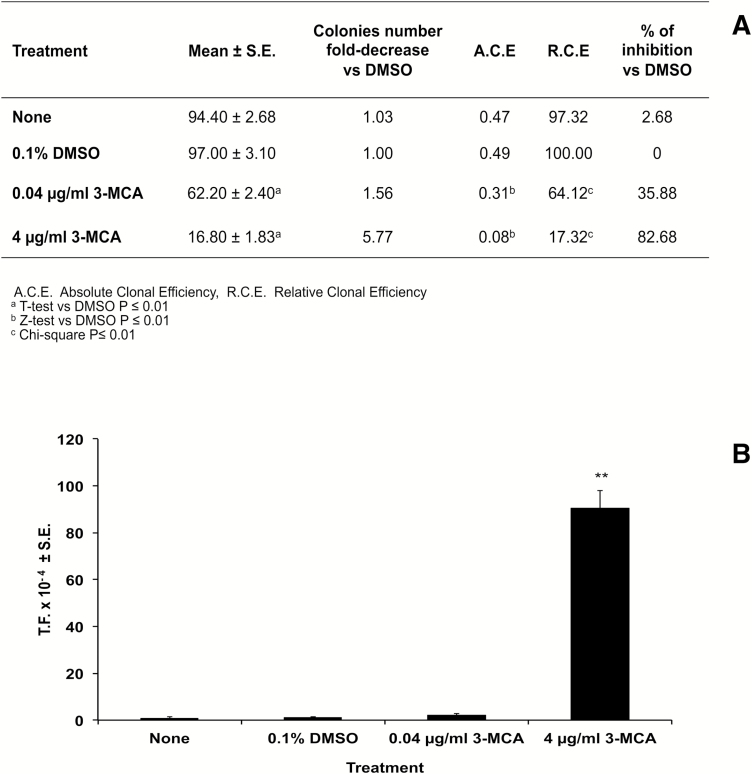
The effects on the cell survival were evaluated on the basis of the cellular capability to form colonies. A statistically significant reduction of cell growth was observed at both concentrations of 3-MCA. The transformation frequency (TF), i.e. the fraction of transformed cell, showed the absence of cell transformation at 0.04 µg/ml 3-MCA and a high and statistically significant transformation cell at 4 µg/ml treatment. Moreover, at the highest concentration, all the treated plates contained transformed foci, whose occurrence was about 16-fold greater than the negative control (Figure 2).
Figure 2.
(A) The effect on the cell survival was evaluated on the basis of the cellular capability to form colonies. BALB/c 3T3 cells exposed to 3-MCA showed, at both concentrations, a statistically significant reduction of cell growth, expressed as absolute clonal efficiency (ACE) or as relative clonal efficiency (RCE) compared with the solvent control (DMSO). Cell growth was inhibited by 35.88 and 82.68%, respectively at 0.04 and 4 µg/ml 3-MCA concentrations. (B) The transformation frequency was calculated on the basis of the foci number and the ACE values. It showed the absence of cell transformation at 0.04 µg/ml 3-MCA and a high and statistically significant TF at 4 µg/ml treatment. At the top concentration, all the treated plates contained transformed foci, whose occurrence was about 16-fold greater than the negative control.
Statistical analysis of microarray experiments
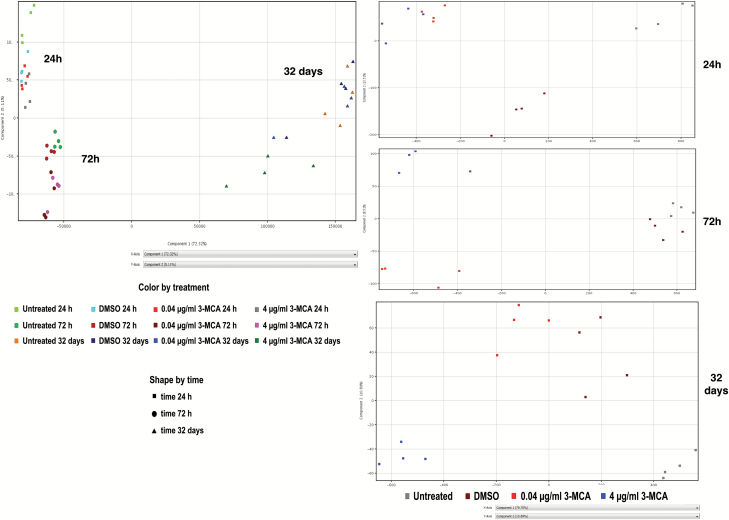
The PCA derived from the complete One-way ANOVA samples list highlighted three specific groups corresponding to the different duration of exposure, i.e. the ‘time’ point, the chemical concentration and the type of cell treatment.
For the ‘time’ point, 24 and 72 h clusters appear to be very similar to each other but differ to the 32 days cluster. A concentration-related arrangement was also observed in each group. The effect of 4 μg/ml 3-MCA concentration at 32 days was particularly evident (Figure 3A). When analyzing profiles at the single time points, the impact of each treatment was even more evident. The transcriptional profiles induced at the two tested concentrations, which were similar during the first 24 h of treatment, became progressively dissimilar as time progressed. At 32 days, the transcriptional profile following 0.04 μg/ml 3-MCA exposure was similar to that observed in the solvent control (Figure 3B). The Venn diagrams, obtained by crossing the resulting gene lists, showed the progressive decrease of the number of modulated genes in common between the two treatments, e.g. those genes that differentiate the cells exposed to the chemical from DMSO-treated cells (Supplementary Figure 1, available at Carcinogenesis Online). It is noticeable that, at 32 days, the transcriptional response due to the exposure at 4 µg/ml affected 789 genes, and this appeared to be significantly modulated (P < 0.05), while no gene modulation was detected in cells treated with 0.04 µg/ml 3-MCA. Overall, these results indicate a behavior that was also seen in the PCA, and thus, these two statistical analytical approaches mutually confirm each other.
Figure 3.
(A) PCA obtained by the list of differentially expressed genes (One-way ANOVA, P ≤ 0.01 Benjamini–Hochberg correction). The effect due to the ‘time’ parameter appears evident. (B) PCA of differentially expressed genes for each time point, 24, 72 h, 32 days. The dose effect becomes progressively more evident.
Biological interpretation of microarray experiments
Pathways and processes analysis
The MetaCore Pathway Maps and Process Networks that identified the 3-MCA treatments at the selected time points and resulted in statistically significant false discovery rate (FDR) (0.05) were considered. In the comparative analysis, the statistically significant pathways at 24 h of treatment were mainly ascribable to the highest concentration (4 µg/ml) and mostly suggestive of the immune response involvement (Figure 4A). Specifically, the Pathway Maps ‘Immune response_Classical complement pathway’, ‘Immune response_Lectin-induced complement pathway,’ ‘Immune response_Alternative complement pathway’ and ‘Complement pathway disruption in thrombotic microangiopathy’ were found to be perturbed following 4 µg/ml 3-MCA treatment. Furthermore, the highest concentration was able to significantly perturb the pathway that referred to the cytoskeleton remodeling, the ‘Cytoskeleton remodeling_RalA regulation pathway,’ while the involvement of cell adhesion mechanisms (‘Cell adhesion_ECM remodeling’) appears to be related mainly to the lowest concentration (0.04 µg/ml). The loss of cell attachment to the extracellular matrix is related to the early cell response to stress, as a cellular homeostatic mechanism that prevents cells from detachment-induced cell death (anoikis) (20). This provides key evidence of the likely pivotal event sustaining the adaptive response in cells treated with the lowest concentration. The Process Networks evaluation also confirmed these results, highlighting the involvement of inflammation processes (‘Inflammation_Complement system’) linked to the complement and cell adhesion events (‘Cell adhesion_Cell-matrix interaction’) associated, respectively, to 4 or 0.04 µg/ml 3-MCA treatment. Among the most significant Process Networks were ‘Cell cycle_G1-S Growth factor regulation,’ ‘Apoptosis_Apoptosis stimulation by external signals,’ ‘Signal transduction_ESR1-nuclear pathway’ and, also significant but to a slightly lesser degree (FDR = 0.047), the ‘Retinol metabolism’ Pathway Map, were all affected by both concentrations. At 72 h, the highest concentration treatment still showed the involvement of the immune response (Figure 4A). In addition, the involvement of the ‘Signal transduction_AKT pathway signaling’ and also the ‘Cell cycle G1-S Growth factor regulation process’ indicate that genes related to the cell cycle are influenced by the treatment. The apoptotic process identified at 24 h was no longer observed at 72 h. While the ‘Apoptosis_Apoptosis stimulation by external signals process’ was perturbed by both concentrations at 24 h, only the ‘Apoptosis_Death Domain receptors and caspases in apoptosis process’ was altered at 72 h at the lowest tested concentration. A different gene modulation was induced by the 0.04 µg/ml treatment at 24 h compared with 72 h. The immune response at the lowest concentration became evident only at 72 h, while the processes related to apoptosis persisted after 72 h. From the comparative analysis at 72 h, it is remarkable that the involvement of the ‘Estradiol metabolism’ pathway was ascribable to the lowest concentration (FDR < 0.01), rather than to the highest concentration (FDR > 0.05). Further exploration of this phenomenon could elucidate the event(s) discriminating the adaptive response from the committed step leading to the transformation. At 32 days, the pathways and the processes with FDR < 0.05 resulted exclusively related to immunity and inflammation, as shown in Figure 4B. An overall visualization of results is presented in Figure 5.
Figure 4.
Statistically significant Pathway maps (FDR < 0.05) obtained from the Metacore Enrichment Analysis. (A) Results of the Compared Enrichment Analysis performed on the lists of genes obtained by the t-test between each concentration and the solvent at 24 and 72 h of exposure. A comparative analysis was performed in Metacore for the two data sets, named ‘common’ and ‘unique’ sets. Gene IDs of possible targets in each data set (‘common’ or ‘unique’) were matched separately with gene IDs in functional ontologies in Metacore. Separate lists were obtained including genes that have been modulated exclusively by one of the two treatments and genes that have been modulated by both treatments. Solid orange line ( ) = genes exclusively modulated by 0.04 µg/ml 3-MCA treatment (0.04 µg/ml 3-MCA ‘unique’ set). Solid blue line ( ) = genes exclusively modulated by 4 µg/ml 3-MCA treatment (4 µg/ml 3-MCA ‘unique’ set). Dashed blue line ( ) = genes modulated by both 3-MCA treatments (‘common’ set). The first four pathways that are modulated at both 24 and 72 h by both treatments, as indicated by the colored arrows, are encircled in the blue box. (B) Results of the Single Enrichment Analysis performed on the list of genes obtained by the t-test between 4 µg/ml 3-MCA and the solvent at 32 days from cells seeding.
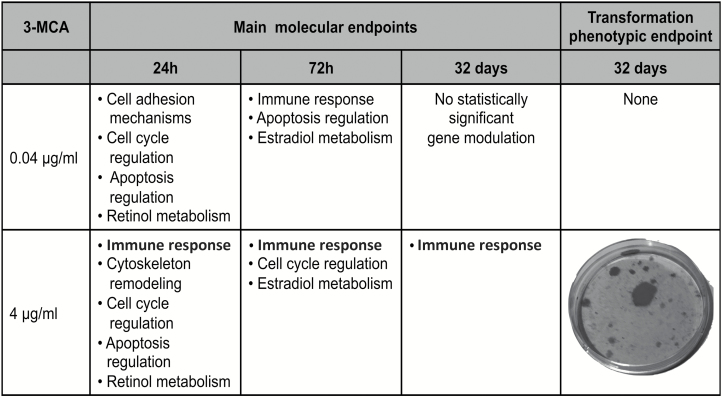
Figure 5.
Summary table of molecular and transformation phenotypic endpoints. For each treatment and time, the main biological targets are reported. At 24 h, both treatments can modulate cell cycle, apoptosis and retinol metabolism regulation. While the cell adhesion mechanism modulation is associated only with the 0.04 µg/ml 3-MCA treatment, as a cell adaptive response (see text), the cytoskeleton remodeling observed at 4 µg/ml 3-MCA treatment is the early key event toward the cell transformation. At 72 h, the immune response becomes the distinctive trait of the gene modulation by both treatments, albeit it is associated with the transcriptional modulation of the apoptotic processes at 0.04 µg/ml 3-MCA treatment and with the alteration of the cell-cycle regulation at 4 µg/ml 3-MCA treatment, marking a different fate for the two cell populations. At 32 days, no significant modulation is seen anymore at 0.04 µg/ml 3-MCA treatment, while the phenotypic outcome of the cell transformation (malignant foci) is clearly visible in the plates treated with 4 µg/ml 3-MCA treatment, still sustained by the immune response.
Genes involved in the metabolism
Cyp1a1 and Cyp1b1 were the only genes transcriptionally activated by both concentrations, at both 24 and 72 h (Table 1 and Supplementary Table 1, available at Carcinogenesis Online). Cyp1a1 and 1b1 are considered prototypical enzyme induction targets of AhR (21). The expression of Cyp1a1 is almost totally dependent on AhR activity (21). Both Cyp1a1 and 1b1 are involved in metabolism of procarcinogens, such as polycyclic aromatic hydrocarbons and dioxins. Indeed, 3-MCA is commonly and preferentially used as an inducer of these CYP enzymes for S9 mix used in metabolism studies (22). In addition to Cyp1a1 and Cyp1b1 modulation, the ‘Retinol metabolism’ pathway was also characterized by the upregulation of xanthine oxidase, which is involved in oxidative purine metabolism. The Ugt1a10 upregulation observable in the ‘Estradiol metabolism’ pathway was exclusively induced by 0.04 µg/ml 3-MCA treatment at 72 h. UDP-glucuronosyltransferases (UGTs) are phase II metabolic enzymes necessary to transform non-polar endogenous and exogenous compounds, such as environmental pollutants and carcinogenic molecules, into their hydrophilic derivatives. Besides the deactivation and detoxification of the exogenous substrates, UGTs represent key elements in the maintenance of homeostasis of several endogenous molecules. The homeostatic maintenance of adequate level of UGTs in the organism avoids adverse physiological disturbance (Supplementary Figure 2, available at Carcinogenesis Online).
Table 1.
Biological traits possibly affected by 3-MCA treatments, based on the modulated genes over time and related Pathway Maps. Gene modulation (up, ↑; down, ↓; no modulation, —)
| Biological trait / hallmark | Pathway maps | Gene | Up/down regulation | ||||
|---|---|---|---|---|---|---|---|
| 24h | 72h | 32 days | |||||
| 0.04 µg/ml 3-MCA |
4 µg/ml 3-MCA |
0.04 µg/ml 3-MCA |
4 µg/ml 3-MCA |
4 µg/ml 3-MCA |
|||
| Cellular metabolism | Retinol metabolism | Cyp1a1 | ↑ | ↑ | -- | -- | -- |
| Cyp1b1 | ↑ | ↑ | -- | -- | -- | ||
| Xo | ↑ | ↑ | -- | -- | -- | ||
| Estradiol metabolism | Cyp1a1 | -- | -- | ↑ | ↑ | -- | |
| Cyp1b1 | -- | -- | ↑ | ↑ | -- | ||
| Ugt1a10 | -- | -- | ↑ | -- | -- | ||
| Microenvironment / cell adhesion/ cytoskeleton | Cell adhesion_ Chemokines and adhesion |
Acta1 | -- | ↓ | -- | -- | -- |
| Lama4 | ↓ | -- | -- | -- | -- | ||
| Mmp 13 | ↓ | -- | -- | -- | -- | ||
| Cell adhesion_ ECM remodeling |
Lama4 | ↓ | -- | -- | -- | -- | |
| Cytoskeleton remodeling_ RalA regulation pathway |
Arrb1 | -- | ↑ | -- | -- | -- | |
| Acta1 | -- | ↓ | -- | -- | -- | ||
| Neoangiogenesis | Complement pathway disruption in thrombotic micro-angiopathy | C3 | -- | ↑ | ↑ | ↑ | -- |
| Selp | -- | ↑ | -- | -- | -- | ||
|
Ccl5
(Ccl8, Ccl2 Ccl7) |
↓ | ↓ | ↓ | ↑ | ↑ | ||
| Cell death | Apoptosis and survival_ Role of PKR in stress-induced apoptosis | P21 | -- | -- | -- | ↑ | -- |
| Nfkbia | -- | -- | -- | ↑ | -- | ||
| Regulation of GSK3 beta in bipolar disorder |
Irs-1
|
-- | -- | -- | ↓ | -- | |
| Fgfr1 | -- | -- | -- | ↑ | -- | ||
| Nxn | -- | -- | -- | ↓ | -- | ||
| Genomic instability | Development Role of HDAC and calcium/ calmodulin- dependent kinase (CaMK) in control of skeletal myogenesis | P21 | -- | -- | -- | ↑ | -- |
| Hdac5 | -- | -- | -- | ↑ | -- | ||
|
Irs-1
|
-- | -- | -- | ↓ | -- | ||
| Muscle contraction_ Relaxin signaling pathway |
Nfkbia | -- | -- | -- | ↑ | -- | |
| Signal transduction_ AKT signaling | P21 | -- | -- | -- | ↑ | -- | |
| Nfkbia | -- | -- | -- | ↑ | -- | ||
|
Irs-1
|
-- | -- | -- | ↓ | |||
| Immune response | Immune response_Classical complement pathway | C1s2 | -- | ↑ | -- | -- | -- |
| C3 | -- | ↑ | ↑ | ↑ | -- | ||
| Immune response_-Lectin induced complement pathway | C3 | -- | -- | ↑ | ↑ | -- | |
| Immune response_ Alternative complement pathway | C3 | -- | -- | ↑ | ↑ | -- | |
| Ptx3 | ↑ | ↑ | -- | -- | -- | ||
| Immune response_IL-4- responsive genes in type 2 immunity |
Ccl5
(Ccl8, Ccl2 Ccl7) |
-- | -- | ↓ | ↑ | ↑ | |
| Immune response_ Platelet activating factor/ PTAFR pathway signaling |
Arrb1 | -- | ↑ | -- | -- | -- | |
| Nfatc2 | -- | ↓ | -- | -- | -- | ||
| Adcy7 | -- | ↑ | -- | -- | -- | ||
| Nociception_ Expression and role of Nociceptin in immune system |
Ccl5 | ↓ | ↓ | -- | -- | -- | |
| Immune response_IL-4- responsive genes in type 2 immunity | Ccl17 | -- | -- | ↓ | -- | -- | |
| Tff3 | -- | -- | ↓ | -- | -- | ||
| Immune response_TNF-R2 signaling pathways | Traf1 | -- | -- | ↑ | -- | -- | |
| Birc3 | -- | -- | ↑ | -- | -- | ||
| Bmf | -- | -- | ↑ | -- | -- | ||
| Nfkbia | -- | -- | -- | ↑ | -- | ||
| Immune response_MIF- mediated glucocorticoid regulation | Nfkbia | -- | -- | -- | ↑ | -- | |
| Il-6 | -- | -- | -- | -- | ↑ | ||
| Mmp1 | -- | -- | -- | -- | ↑ | ||
| Immune response_Histamine H1 receptor signaling in immune response | Nfkbia | -- | -- | -- | -- | ↑ | |
| Immune response_ IL-4 signaling pathway |
Irs-1
|
-- | -- | -- | ↓ | -- | |
| Immune response_IL-4- induced regulators of cell growth,survival, differentiation and metabolism | Egr2 | -- | -- | -- | -- | ↑ | |
| Mmp13 | -- | -- | -- | -- | ↑ | ||
| Ccne | -- | -- | -- | -- | ↑ | ||
| Immune response_IFN alpha/beta signaling pathway | Stat2 | -- | -- | -- | -- | ↑ | |
| Pmrt1 | -- | -- | -- | -- | ↑ | ||
| Ifn-beta | -- | -- | -- | -- | ↑ | ||
| Usp18 | -- | -- | -- | -- | ↑ | ||
| Isg54 | -- | -- | -- | -- | ↑ | ||
| Immune response_Antigen presentation by MHC class I | Psme2 | -- | -- | -- | -- | ↑ | |
| Psmb8 | -- | -- | -- | -- | ↑ | ||
| Hsp90 | -- | -- | -- | -- | ↑ | ||
|
Tap1,
Tap2 |
-- | -- | -- | -- | ↑ | ||
| Immune response_Oncostatin M signaling via MAPK in human cells | Rac1 | -- | -- | -- | -- | ↑ | |
| Mmp1 | -- | -- | -- | -- | ↑ | ||
| Immune response_Histamine H1 receptor signaling in immune response | Il-6 | -- | -- | -- | -- | ↑ | |
| Mmp1 | -- | -- | -- | -- | ↑ | ||
| Mmp9 | -- | -- | -- | -- | ↓ | ||
| Mmp 13 | -- | -- | -- | -- | ↓ | ||
Full details of the roles of all genes mentioned in this article can be conveniently obtained via the hyperlinks to NCBI Entrez in the Gene Section NCBI Entrez http://www.ncbi.nlm.nih.gov/Entrez/index.html.
As shown, at 32 days, the gene modulation was exclusively ascribable to the highest concentration. At this time point, several sequences associated with tumor progression and metastasis were identified. Table includes genes that have not been discussed in the manuscript. These genes are reported in brackets.
Genes modulated at 24 h
While the most relevant pathways and processes that arose from the MetaCore analysis were all in relation to the immune response, this was not triggered at the low dose of 0.04 µg/ml. Indeed, genes responsible for the immune response, including C1s2, C3 and Selp, were overexpressed only in relation to 4 µg/ml 3-MCA treatment (see Figure 5). C1s2 encodes a serine protease constituent of the human complement subcomponent C1. C3 exerts a central role in the activation of both classical and alternative complement system, which are both identified in our study. C3 is implicated in the immune surveillance response against cancer; nevertheless, its pro-inflammatory activity plays a role in chronic inflammation associated with carcinogenesis. Indeed, the complement seems to play a role as a pro-carcinogen agent through specific mechanisms, which include increasing the level of cytokines and tumorigenic growth factors, sustaining angiogenesis and preventing apoptosis (23). Selp encodes for P-selectin, a membrane calcium-dependent receptor. This molecule appears to contribute to carcinogenic and metastatic processes, although its mechanism has not been completely elucidated yet (24). In addition, Ptx3 and Ccl5 genes were modulated respectively up and down by the two concentrations of 0.04 and 4 µg/ml with statistical significance (P ≤ 0.05). Ptx3 encodes for a member of the pentraxin protein family, involved in the regulation of inflammation and in the complement activation, besides playing a role in angiogenesis and tissue remodeling. Ccl5 belongs to a superfamily of chemokines, secreted proteins involved in immunoregulatory and inflammatory processes. Ccl5 downregulation is associated with antiangiogenic and antimetastatic effects (25). MetaCore results also highlighted the contribution of genes involved in cytoskeleton remodeling and cellular adhesion, such as Acta1 and Arrb1, which were exclusively modulated by the highest concentration, and Mmp13 and Lama4 specifically downregulated after 0.04 µg/ml 3-MCA treatment. Mmp13 gene-encoded protein belongs to the family of matrix metalloproteinases characterized by the capability to break down extracellular matrix in normal or pathological processes. The Mmp13 increase is associated with cancer (26). The alpha chain isoform laminin, alpha 4, product of Lama4 gene, is a member of laminins, a family of extracellular matrix glycoproteins. These proteins represent the major non-collagenous constituent of basement membranes. Increased Lama4 expression seems to induce clonal expansion, metastasis and poor prognosis (27). Arrb1 together with Nfatc2 and Adcy7 was also found among the genes included in the ‘Immune response_Platelet activating factor/PTAFR pathway signaling,’ exclusively regulated by the highest concentration. In our model, Arrb1 and Adcy7 were overexpressed, while Nfatc2, whose product is a DNA-binding protein, was downregulated. This nuclear factor seems to act as a tumor suppressor (28). The proteins encoded by Arrb1, β-arrestin, a cytosolic protein G-protein-coupled receptor adaptor, and by Adcy7, adenylate cyclase, are believed to have a role in carcinogenesis, through the promotion of cell proliferation and by affecting the apoptosis and/or cell survival (29,30). Moreover, β-arrestin seems to promote angiogenesis (29).
Genes modulated at 72 h
In contrast to the results determined at 24 h, the perturbation of the immune-related pathways was ascribable to both concentrations (FDR < 0.05), with the common modulation of C3 family genes. Interestingly, Ccl5 gene downregulation persisted only for the 0.04 µg/ml treatment. In addition, the ‘Immune response_IL-4-responsive genes in type 2 immunity’ and the ‘Immune response_TNF-R2 signaling pathways’ were specifically altered by the lowest concentration, including genes Ccl17, Tff3, and genes specifically related to apoptosis. Ccl17 and Tff3, whose expression generally increases in several tumor types and whose product is a stable secretory protein, are generally positively associated to poor prognosis of mammary carcinoma (31). These genes, however, were downregulated in cells exposed to the lowest concentration, confirming the absence of neoplastic transformation. The genes related to apoptosis exhibited an ambiguous behavior. Indeed, the antiapoptotic genes, Traf1 and Birc3, as well as the proapoptotic gene Bmf were all upregulated in the 0.04 µg/ml treatment, while at the highest concentration, the apoptotic machinery seems to be supported by the upregulation of the proapoptotic genes p21 and Ikb, and by the downregulation of Irs-1, whose expression confers protection against cell death (32). Other relevant genes belonging to the statistically significant pathways altered by the 4 µg/ml treatment were the upregulated genes Hdac5, Fgfr1 and the downregulated Nxn. The Hdac5 product is the hystone deacetylase 5 able to alter chromosome structure and to affect transcription factor access to DNA. The histone deacetylase (HDAC) family proteins are commonly dysregulated in several tumors (33). Nxn gene encodes for a thioredoxin-related oxidoreductase whose decrease confers more resistance to the oxidative stress (34). The fibroblast growth factor receptor, Fgfr1, is a member of the family of tyrosine kinase receptors involved in regulation of cell survival, proliferation, differentiation and its overexpression is associated with carcinogenesis (35).
Genes modulated at 32 days
Among the genes modulated by 4 µg/ml 3-MCA concentration, Stat2 and Prmt1 were upregulated. The Prmt1 product is a member of the protein arginine N-methyltransferase family responsible for the majority of cellular arginine methylation activity. Its increased expression is recognized to play a role in several types of cancers. Stat2 encodes a member of the Stat protein family of transcription factors, without the capability to directly bind the DNA. It is reported that Stat2, specifically activated by IFN α/β, seems to promote the carcinogenic process, through the activation of pro-inflammatory mediators such as Il-6. Interestingly, Il-6 and Ifn-β were both upregulated as shown in the ‘Immune response_IFN alpha/beta signaling pathway’, confirming the activation of AhR, which can induce pro-inflammatory factors, such as Il-6 (21). Besides, in this Pathway Map, Usp18 and Isg54, whose expression is connected respectively to the NF-kB inhibition and to proapoptotic events, were overexpressed (36,37). Also, Nfkbia, a NF-kB inhibitor, was overexpressed. Other cancer-related genes overexpressed in the BALB/c 3T3 cells were Psme2, Psmb8, Hsp90, Egr2, Ccne and Rac1. Psme2 and Psmb8 products belong to the immunoproteasome, a multi-catalytic proteinase complex with the function of processing class I MHC peptides. Pmse2 is hypothesized to participate in processes of carcinogenesis and tumor progression. Pmsb8 is associated with poor gastric cancer prognosis (38). Tap1 and Tap2, which were upregulated, encode proteins involved in the antigen presentation. Their deficiency is associated with immune escape mechanisms (39), but they may also be implicated in multidrug resistance (40). Egr2 is a zinc finger transcription factor modulating various physiological processes. For instance, Egr2, which is recognized to activate NF-kB in brain cancer, injury and inflammation, is associated with renal cell carcinoma and with Ewing’s sarcoma (41–43). The Ccne product belongs to the highly conserved cyclin family and functions as a regulatory subunit of Cdk2 during cell cycle G1/S transition. Its deregulation leads to chromosome instability, contributing to tumorigenesis (44). Finally, Rac1 encodes a GTPase member of the RAS superfamily of small GTP-binding proteins with the function of regulating several cellular events, including the control of cell growth, cytoskeletal reorganization and the activation of protein kinases. The overexpression of this gene appears to be implicated in carcinogenesis, cell migration and cancer invasion processes (45). Among genes differentially expressed at 32 days, Mmp1, 9 and 13 were observed with an opposite trend. Indeed, Mmp1 was upregulated, whereas Mmp9 and 13 were downregulated. The product of all these genes is metalloproteinases involved in cellular invasion, as described previously for Mmp13.
The 3-MCA-induced gene modulations and the related Pathway Maps and affected biological traits are reported in Table 1. Supplementary Table 2, available at Carcinogenesis, Online offers a rapid visualization of the modulated genes.
Discussion
According to the genotoxicity paradigm, the cancer process is conceived as a sequence of mutations affecting key genes, with chemicals acting via non-genotoxic mechanisms, supporting the proliferation of the initiated cells. Based on this conventional representation of the carcinogenesis process, three main steps can be recognized: initiation, which is irreversible and sustained by genotoxic chemicals, promotion, which is considered a reversible stage and sustained by non-genotoxic chemicals, referred as promoters, and progression. Initiating agents are thought to induce DNA damage with no threshold. Promoters are considered unable to transform cells in the absence of initiation. An experimental threshold may be identified for promoting agents, according to their mechanism and mode of action. In the experimental reproduction of the initiation/promotion process, a subtransforming concentration of a genotoxic chemical, usually a polycyclic aromatic hydrocarbon, is used to initiate cells, followed by the chronic administration of a non-genotoxic chemical to promote the proliferation of initiated cells.
The results from our study provide a new perspective to that of previous CTA reports, elucidating the mechanistic understanding that will enable the CTA to be appropriately incorporated into an IATA for non-genotoxic carcinogenesis.
We tested 3-MCA, using the foci occurrence as the phenotypic anchoring of the specific gene signature. 3-MCA is one of the reference chemicals, used as the positive control in the standard CTA. 3-MCA is much less toxic than other polycyclic aromatic hydrocarbons used as CTA reference controls, such as benzo(a)pyrene (46). We have extensively used this chemical at transforming concentrations as the positive control in CTA experiments and at subtransforming concentrations in CTA initiation/promotion protocols, collecting data on a large number of historical controls (46). 3-MCA was first described to be able to activate the estrogen receptor, either directly or indirectly, by acting as an agonist of AhR. The latter mechanism seems to play the main role, leading to the inhibition of the estrogenic response, through the inhibitory AhR-ERα crosstalk (47). The experimental design included two concentrations of 3-MCA. The lowest tested concentration was chosen on the basis of our historical controls and on the results from the EURL ECVAM CTA validation study, as the subtransforming concentration that cannot induce cell transformation when administered as a single treatment. The highest concentration of 4 µg/ml was an effective transforming concentration (16,46).
The treatment with 3-MCA triggers the modulation of common pathways, such as the ‘Retinol metabolism’ and the ‘Estradiol metabolism’ pathways, marked by the activation of Cyp1a1 and Cyp1b1, at 24 h, persisting at 72 h. However, only the subtransforming concentration induces the Ugt1a10 upregulation at 72 h.
Several literature reports have shown decreased UGTs activity in tumor tissues compared with normal counterparts. The loss of UGT expression has been proposed to be associated with the early stages of neoplastic transformation and to promote the malignant progression, as the consequence of the reduced ability to detoxify carcinogens (48).
The activation of Ugt1a10, only in the cells treated with the lowest concentration, supports the hypothesis that this concentration is still compatible with the defense mechanisms of the cell that may induce an adaptive response and, eventually, detoxification. Indeed, at 32 days, the cells treated with the lowest concentration do not show any gene modulation.
On the contrary, the cells treated with the effective transforming concentration showed a specific transcriptional profile, sustained by the phenotypic endpoint of the transformation assay. The gene profile includes Pathways Maps and Process Networks, which are mainly related to the immune system, with concurrent induction of pro-inflammatory and immune escape processes, supported by several inflammatory genes related to carcinogenesis and tumor progression.
We can speculate that the high-level stimulation of an AhR-mediated response by the top 3-MCA concentration would represent a key element in triggering and sustaining the aberrant immune system activation.
Recent studies have addressed the modulatory effect of AhR in the innate immune system. Besides its well-known role as the signal mediator of dioxin toxicity, AhR is recognized, in fact, to be involved in both innate and adaptive immune responses, with consequent implications in transplantation immunity and in the development of autoimmune diseases (49,50).
The involvement of AhR in sustaining the process of cell transformation through the activation of the immune response is also confirmed by the modulation of C3 and Ccl5, which are also described as AhR target genes (50). In particular, the overexpression of Ccl5 has been reported to enhance tumor cell proliferation and to facilitate invasion and metastasis, through the stimulation of angiogenesis (25). In our experimental model, C3 is activated, while Ccl5 is inhibited. In particular, Ccl5 is downregulated in the first 24 h of the treatment at both concentrations. After 72 h, Ccl5 downmodulation persists exclusively at the lowest concentration. At this time, it is possible that the cells treated with the highest concentration overcome the AhR-mediated control and activate a persistent inflammatory response signal.
Accumulating evidence has shown the key role of inflammation and immune evasion to sustain the early steps of cancer process and the importance of genomic changes that do not directly affect the genetic code, thus suggesting a different non-genotoxic initiation of carcinogenesis (6). However, the immune system is recognized to exert a complex role in the development and progression of cancer. Innate immune cells seem to be able to produce an inflammatory microenvironment that can promote the formation of premalignant lesions and accelerate the tumor growth. Conversely, they can also prevent tumor progression and invasion (51).
In the first 24 h of treatment at the highest concentration, it is already evident that there is a considerable involvement of the innate immune response, through the upregulated expression of genes, such as C1s2 and C3, which are connected to the complement, and whose activation is physiologically involved in the immune surveillance response against cancer. The unnecessary and prolonged stimulation of these genes has been linked to several diseases associated with inflammation and involved in the carcinogenesis process. In our model, the perturbation of ‘Immune response_Classical Complement pathway’, and the related upregulation of C3 persists after 72 h, which suggests that the complement activation represents one of the mechanisms leading to foci formation.
The upregulation of Ptx3, in the context of the modulation of the ‘Immune response_Alternative Complement pathway,’ underpins the role of the complement in the carcinogenesis process. Indeed, Ptx3 is involved in complement activation, and it is recognized to have an active role in tumor-associated inflammation. Because of its overexpression in different kind of tumors, Ptx3 has been acknowledged for its diagnostic and prognostic value. Tumor cells can induce activated Ptx3 expression in stromal fibroblasts (52). In our transformics model, 3-MCA, at the top concentration, can induce the modulation of this gene by itself, further supporting the inflammation triggered by the complement activation, as the mechanism fostering the carcinogenesis process. Ptx3 upregulation was also observed in the cells treated with the lowest concentration of 3-MCA, without being linked to the cell transformation. We can suppose that, in the absence of C1s2 and C3 regulation, Ptx3 modulation alone is not able to activate the complement genes and initiate the transformation process. At the highest concentration, the perturbation of genes implicated in chromosome instability has been also revealed, as confirmed by the overexpression of Hdac5 at 72 h and the overexpression of Ccne and Pmrt1 at 32 days (33,44,53).
These results support the hypothesis that the cell transformation is initiated by non-genotoxic events, including the stimulation of AhR, by the activation of an aberrant immune response, sustaining the modulation of inflammation markers and the production of Cyp1a1 and Cyp1b1, involved in the activation of 3-MCA to reactive metabolites, which eventually induce genotoxic damage. Thus, the transforming ability of 3-MCA associated with the highest concentration seems to be related to the persistence of the immune response signal, whereas at the lowest concentration, the detoxification pathways turnoff the abnormal immune response and restore cell homeostasis.
From the non-genotoxic carcinogenesis point of view, cancer may be the consequence of non-genotoxic mechanisms, supported by the induction of tissue inflammation and by the stimulation of the immune response, fostering the biological conditions for the occurrence of DNA damage in tissues injured by a prolonged exposure to stressors (6). Transformics is able to describe the entire process, anchoring the molecular machinery to the oncotransformation biological endpoint and as such can be regarded as a useful tool in the integrated testing strategy for carcinogenesis chemical hazard assessment. Gene modulation data can be used to calculate thresholds for events at the molecular level (54). In this context, transformics has the potential to improve the knowledge of dose–response relationships including providing the biological understanding to underpin statistical methods of analyzing dose–response data, such the benchmark dose approach, promoted by national and international Governmental organizations (55–57). Thus, the approach used here has great potential to better advance quantitative risk assessment and the statistical tools used therein. With respect to genotoxicity tests, which only highlight the DNA damage, transformics offers the possibility to understand the biological significance of the genotoxic events along the multistep carcinogenesis process. Indeed, toxicogenomics has been described as the tool to bridge both genotoxicity and non-genotoxicity events to carcinogenesis (58). In conclusion, the transformics approach has the potential to be a valuable asset to determine an easily recognizable phenotypic endpoint of oncotransformation, as provided by the CTA example shown here, with the corresponding gene signature, in chemical carcinogenicity hazard assessment. This approach is promising also for other relevant in vitro assays addressing transformation. By providing such mechanistic understanding to underpin the use of such assays within the design of an IATA, the regulatory confidence in the assays and their application will improve. Assessment of non-genotoxic carcinogenicity of chemicals has been sorely lacking in the regulatory arena, in the face of increasing cancer incidence (59). Practical attempts to take greater front end preventative action to protect public health is a high priority regulatory goal internationally.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Funding
This work has been entirely supported by Regione Emilia Romagna – Supersite Project (Act 428/08.02.2010).
Conflict of Interest Statement: None declared.
Abbreviations
- 3-MCA
3-methylcholanthrene
- AhR
aryl hydrocarbon receptor
- CTA
cell transformation assay
- DMSO
dimethyl sulfoxide
- IATA
integrated approach to testing and assessment
- NGTxCs
non-genotoxic carcinogens
- PCA
principal component analysis
- RCB
rodent carcinogenicity bioassay
- TF
transformation frequency
- UGTs
UDP-glucuronosyltransferase
References
- 1. Annys E., et al. (2014)Advancing the 3Rs in regulatory toxicology—carcinogenicity testing: scope for harmonisation and advancing the 3Rs in regulated sectors of the European Union. Regul. Toxicol. Pharmacol., 69, 234–242. [DOI] [PubMed] [Google Scholar]
- 2. National Research Council.(2007)Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: The National Academies Press; https://doi.org/10.17226/11970. [Google Scholar]
- 3. Ashton R., et al. (2014)State of the art on alternative methods to animal testing from an industrial point of view: ready for regulation?ALTEX, 31, 357–363. [DOI] [PubMed] [Google Scholar]
- 4. Paparella M., et al. (2017)Uncertainties of testing methods: what do we (want to) know about carcinogenicity?ALTEX, 34, 235–252. [DOI] [PubMed] [Google Scholar]
- 5. Madia F., et al. (2016)Analysis of Carcinogenicity Testing for Regulatory Purposes in the European Union. JRC Technical Report. Luxembourg: European Commission. pp.1.–. doi:10.2788/547846 [Google Scholar]
- 6. Jacobs M.N., et al. (2016)International regulatory needs for development of an IATA for non-genotoxic carcinogenic chemical substances. ALTEX, 33, 359–392. [DOI] [PubMed] [Google Scholar]
- 7. OECD. (2016)Guidance Document on the Reporting of Defined Approaches to be used within Integrated Approaches to Testing and Assessment. Vol. 255 Organization for Economic Co-operation and Development, Paris. [Google Scholar]
- 8. Combes R., et al. (2007)Proposed integrated decision-tree testing strategies for mutagenicity and carcinogenicity in relation to the EU REACH legislation. Altern. Lab. Anim., 35, 267–287. [DOI] [PubMed] [Google Scholar]
- 9. Rohrbeck A., et al. (2010)Toxicogenomics applied to in vitro carcinogenicity testing with Balb/c 3T3 cells revealed a gene signature predictive of chemical carcinogens. Toxicol. Sci., 118, 31–41. [DOI] [PubMed] [Google Scholar]
- 10. Mascolo M.G., et al. (2010)BALB/c 3T3 cell transformation assay for the prediction of carcinogenic potential of chemicals and environmental mixtures. Toxicol. In Vitro., 24, 1292–1300. [DOI] [PubMed] [Google Scholar]
- 11. Sakai A. (2007)BALB/c 3T3 cell transformation assays for the assessment of chemical carcinogenicity. AATEX, 14, 367–373. [Google Scholar]
- 12. Melchiori A., et al. (1992)Induction of invasive and experimental metastasis potential in BALB/c 3T3 cells by benzo(a)pyrene transformation. Invasion Metastasis, 12, 1–11. [PubMed] [Google Scholar]
- 13. Colacci A., et al. (1993)Induction of a malignant phenotype in BALB/c 3t3 cells by 1,1,2,2-tetrachloroethane. Int. J. Oncol., 2, 937–945. [DOI] [PubMed] [Google Scholar]
- 14. Adatia R., et al. (1993)Induction of chemotactic and invasive phenotype in BALB/c 3T3 cells by 1,2-dibromoethane transformation. Invasion Metastasis, 13, 234–243. [PubMed] [Google Scholar]
- 15. Corvi R., et al. (2012)ECVAM prevalidation study on in vitro cell transformation assays: general outline and conclusions of the study. Mutat. Res., 744, 12–19. [DOI] [PubMed] [Google Scholar]
- 16. Tanaka N., et al. (2012)Prevalidation study of the BALB/c 3T3 cell transformation assay for assessment of carcinogenic potential of chemicals. Mutat. Res., 744, 20–29. [DOI] [PubMed] [Google Scholar]
- 17. OECD. (2015)Guidance Document on the in Vitro Syrian Hamster Embryo (SHE) Cell Transformation Assay. Vol. 214 Organization for Economic Co-operation and Development, Paris. [Google Scholar]
- 18. OECD. (2017)Guidance Document on the in Vitro Bhas 42 Cell Transformation Assay. Vol. 231 Organization for Economic Co-operation and Development, Paris. [Google Scholar]
- 19. Sasaki K., et al. (2012)Photo catalogue for the classification of foci in the BALB/c 3T3 cell transformation assay. Mutat. Res., 744, 42–53. [DOI] [PubMed] [Google Scholar]
- 20. Vlahakis A., et al. (2017)The interconnections between autophagy and integrin-mediated cell adhesion. J. Mol. Biol., 429, 515–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murray I.A., et al. (2014)Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat. Rev. Cancer, 14, 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang W., et al. (2009)Persistent induction of cytochrome P450 (CYP)1A enzymes by 3-methylcholanthrene in vivo in mice is mediated by sustained transcriptional activation of the corresponding promoters. Biochem. Biophys. Res. Commun., 390, 1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ning C., et al. (2015)Complement activation promotes colitis-associated carcinogenesis through activating intestinal IL-1β/IL-17A axis. Mucosal Immunol., 8, 1275–1284. [DOI] [PubMed] [Google Scholar]
- 24. Reyes-Reyes M.E., et al. (2006)P-selectin activates integrin-mediated colon carcinoma cell adhesion to fibronectin. Exp. Cell Res., 312, 4056–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang S.W., et al. (2015)CCL5/CCR5 axis induces vascular endothelial growth factor-mediated tumor angiogenesis in human osteosarcoma microenvironment. Carcinogenesis, 36, 104–114. [DOI] [PubMed] [Google Scholar]
- 26. Xia J., et al. (2015)P2X7 receptor stimulates breast cancer cell invasion and migration via the AKT pathway. Oncol. Rep., 34, 103–110. [DOI] [PubMed] [Google Scholar]
- 27. Ross J.B., et al. (2015)Identification of molecular determinants of primary and metastatic tumour re-initiation in breast cancer. Nat. Cell Biol., 17, 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robbs B.K., et al. (2008)Dual roles for NFAT transcription factor genes as oncogenes and tumor suppressors. Mol. Cell. Biol., 28, 7168–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang Y., et al. (2015)β-Arrestin1 enhances hepatocellular carcinogenesis through inflammation-mediated Akt signalling. Nat. Commun., 6, 1–14. [DOI] [PubMed] [Google Scholar]
- 30. Li C., et al. (2015)ADCY7 supports development of acute myeloid leukemia. Biochem. Biophys. Res. Commun., 465, 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lau W.H., et al. (2015)Trefoil factor-3 (TFF3) stimulates de novo angiogenesis in mammary carcinoma both directly and indirectly via IL-8/CXCR2. PLoS One, 10, e0141947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takatani T., et al. (2016)IRS1 deficiency protects β-cells against ER stress-induced apoptosis by modulating sXBP-1 stability and protein translation. Sci. Rep., 6, 28177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stypula-Cyrus Y., et al. (2013)HDAC up-regulation in early colon field carcinogenesis is involved in cell tumorigenicity through regulation of chromatin structure. PLoS One, 8, e64600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Funato Y., et al. (2013)Nucleoredoxin regulates glucose metabolism via phosphofructokinase 1. Biochem. Biophys. Res. Commun., 440, 737–742. [DOI] [PubMed] [Google Scholar]
- 35. Katoh M. (2016)FGFR inhibitors: effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review). Int. J. Mol. Med., 38, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang Z., et al. (2015)USP18 negatively regulates NF-κB signaling by targeting TAK1 and NEMO for deubiquitination through distinct mechanisms. Sci. Rep., 5, 12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stawowczyk M., et al. (2011)The interferon stimulated gene 54 promotes apoptosis. J. Biol. Chem., 286, 7257–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kwon C.H., et al. (2016)PSMB8 and PBK as potential gastric cancer subtype-specific biomarkers associated with prognosis. Oncotarget, 7, 21454–21468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Doorduijn E.M., et al. (2016)TAP-independent self-peptides enhance T cell recognition of immune-escaped tumors. J. Clin. Invest., 126, 784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matassa D.S., et al. (2016)Oxidative metabolism drives inflammation-induced platinum resistance in human ovarian cancer. Cell Death Differ., 23, 1542–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nafez S., et al. (2015)Early growth response 2 (Egr-2) expression is triggered by NF-κB activation. Mol. Cell. Neurosci., 64, 95–103. [DOI] [PubMed] [Google Scholar]
- 42. Yao T., et al. (2016)Identification of genes associated with renal cell carcinoma using gene expression profiling analysis. Oncol. Lett., 12, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grünewald T.G., et al. (2015)Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat. Genet., 47, 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teixeira L.K., et al. (2015)Cyclin E deregulation promotes loss of specific genomic regions. Curr. Biol., 25, 1327–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu H., et al. (2005)Carcinogenic effect of nickel compounds Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Mol. Cell Biochem., 279, 45–67.16283514 [Google Scholar]
- 46. Colacci A., et al. (2011)Different sensitivity of BALB/c 3T3 cell clones in the response to carcinogens. Toxicol. In Vitro, 25, 1183–1190. [DOI] [PubMed] [Google Scholar]
- 47. Liu S., et al. (2006)Aryl hydrocarbon receptor agonists directly activate estrogen receptor alpha in MCF-7 breast cancer cells. Biol. Chem., 387, 1209–1213. [DOI] [PubMed] [Google Scholar]
- 48. Mazerska Z., et al. (2016)The role of glucuronidation in drug resistance. Pharmacol. Ther., 159, 35–55. [DOI] [PubMed] [Google Scholar]
- 49. Gargaro M., et al. (2016)Aryl hydrocarbon receptor-dependent pathways in immune regulation. Am. J. Transplant, 16, 2270–2276. [DOI] [PubMed] [Google Scholar]
- 50. Quintana F.J. (2013)The aryl hydrocarbon receptor: a molecular pathway for the environmental control of the immune response. Immunology, 138, 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hagerling C., et al. (2015)Balancing the innate immune system in tumor development. Trends Cell Biol., 25, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hakelius M., et al. (2016)Normal oral keratinocytes and head and neck squamous carcinoma cells induce an innate response of fibroblasts. Anticancer Res., 36, 2131–2137. [PubMed] [Google Scholar]
- 53. Yu Z., et al. (2009)A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol. Cell. Biol., 29, 2982–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Quercioli A., et al. (2017)The use of omics-based approaches in regulatory toxicology: an alternative approach to assess the no observed transcriptional effect level. Microchem. J., 136, 143–148. [Google Scholar]
- 55. EFSA. (2012)Scientific Opinion on Exploring options for providing advice about possible human health risks based on the concept of Threshold of Toxicological Concern (TTC). EFSA J., 10, pp. 103 Wiley On-line http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2012.2750/full. [Google Scholar]
- 56. US-EPA. (2012)Benchmark dose technical guidance. Risk Assessment Forum. Washington, DC: U.S. Environmental Protection Agency. https://www.epa.gov/sites/production/files/2015-01/documents/benchmark_dose_guidance.pdf. [Google Scholar]
- 57. WHO. (2009)Principles for Modeling Dose-response for the Risk Assessment of Chemicals. Environmental health criteria. World Health Organization, Geneva. [Google Scholar]
- 58. Mahadevan B., et al. (2011)Genetic toxicology in the 21st century: reflections and future directions. Environ. Mol. Mutagen., 52, 339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ferlay J., et al. (2015)Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer, 136, E359–E386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.