Abstract
Advancement in the field of computational research has made it possible for the in silico methods to offer significant benefits to both regulatory needs and requirements for risk assessments, and pharmaceutical industry to assess the safety profile of a chemical. Here, we present ProTox-II that incorporates molecular similarity, pharmacophores, fragment propensities and machine-learning models for the prediction of various toxicity endpoints; such as acute toxicity, hepatotoxicity, cytotoxicity, carcinogenicity, mutagenicity, immunotoxicity, adverse outcomes pathways (Tox21) and toxicity targets. The predictive models are built on data from both in vitro assays (e.g. Tox21 assays, Ames bacterial mutation assays, hepG2 cytotoxicity assays, Immunotoxicity assays) and in vivo cases (e.g. carcinogenicity, hepatotoxicity). The models have been validated on independent external sets and have shown strong performance. ProTox-II provides a freely available webserver for in silico toxicity prediction for toxicologists, regulatory agencies, computational and medicinal chemists, and all users without login at http://tox.charite.de/protox_II. The webserver takes a two-dimensional chemical structure as an input and reports the possible toxicity profile of the chemical for 33 models with confidence scores, and an overall toxicity radar chart along with three most similar compounds with known acute toxicity.
INTRODUCTION
An early assessment of the toxic properties for a chemical structure is not only important in the field of drug discovery but also for the regulatory decision making bodies such as European Medicines Agency (EMA), U.S. Food and Drug Administration (FDA) and environmental health protection agencies like U.S. Environmental Protection Agency (EPA), and European Environment Agency (EEA) (1). With the ever-rising number of chemicals and an exponential number of their combinations as mixtures, our exposure to chemicals also increases. Interaction with chemicals is an integral part of our everyday life as we are living in a highly active chemical environment that includes everything from the food that we eat, the medicines we are prescribed, the cosmetics we use as to the air we breathe. However, such exposure can be both harmful and beneficial depending on the amount and duration of the chemical exposure. Thus, it is important to validate the toxic potential of the chemicals and their combination experimentally (2). However, due to several challenges such as time, cost and ethical concerns with respect to animal trials, it is impossible to test all these chemicals on experimental platforms. Thus, in silico toxicity is highly evolving as an integral platform for the prediction of toxicity of chemicals that could be harmful to humans, animals, plants, and environments (3). The aim of in silico toxicity models is to complement the existing in vitro toxicity methods to predict toxicity effects of chemicals, thereby minimizing the time, the need of animal testing and cost associated with it. In silico toxicity model incorporates the knowledge from various fields such as toxicology, biostatistics, systems biology, computer science and many other relevant disciplines (1).Toxicity of a chemical can be measured in terms of toxicity endpoints, such as mutagenicity, carcinogenicity and many other endpoints. It can be further measured both quantitatively such as LD50 (lethal dose) values, and qualitatively, such as binary (active or inactive) for certain cell types and assays or indication area such as cytotoxicity, immunotoxicity and hepatotoxicity (3). One of the most important fodders for in silico models is the information-rich data. The data sources like DSSTox (Distributed Structure-Searchable Toxicity) (4), CEBS: a comprehensive annotated database of toxicological data (5), LiverTox: a database of clinical and research information on DILI (Drug Induced Liver Injury) (6), and Tox21 datasets; have largely supported the interpretation of information from large-scale high-throughput assays with hundreds to thousands of biological endpoints, such as identification of toxicity pathways (7,8). The toxicology in the 21st Century (Tox21) (an initiative by the federal collaboration of National Institute of Environmental Health Sciences, the EPA, and the FDA), has greatly supported the development of computational methods for the assessment of toxicity of chemicals, fostering the vision of transforming toxicology into a predictive science (8,9). Several computational models have been developed to predict the toxicity of chemicals (8,10,11). Initially, to make rodent acute toxicity prediction platform available to a larger community, both to experimental researchers as well as computational toxicologists, ProTox: webserver for the prediction of rodent oral toxicity was published in the year 2014 (12). ProTox methods performed comparatively better than commercial software like Discovery Studio's TOPKAT (Toxicity Prediction by Komputer Assisted Technology; Accelrys, Inc., USA) (http://accelrys.com/) as well as freely accessible tool like Toxicity Estimation Software Tools (T.E.S.T.) developed by the U.S. Environmental Protection Agency (https://www.epa.gov/chemical-research/toxicity-estimation-software-tool-test). Additionally, webservers partially freely available for the community such as the AdmetSAR which includes different types of prediction models based on QSAR methods has also been beneficial for the community (13).
The ProTox-II webserver provides several advantages over existing computational models. ProTox webserver, includes both chemical and molecular target knowledge. A novelty of the ProTox-II webserver is that the prediction scheme is classified into different levels of toxicity such oral toxicity, organ toxicity (hepatotoxicity), toxicological endpoints (such as mutagenicity, carcinotoxicity, cytotoxicity and immunotoxicity), toxicological pathways (AOPs) and toxicity targets thereby providing insights into the possible molecular mechanism behind such toxic response. The new version, ProTox-II incorporates molecular similarity, pharmacophore based, fragment propensities, most common features and machine learning models for prediction of various toxicity endpoints. ProTox-II consisting of 33 models is a freely available computational toxicity prediction webserver enabling the prediction of the largest number of toxicological endpoints to date.
ProTox-II PLATFORM
Input parameter
The user interface of the ProTox-II is easy-to-use and self-explanatory. To predict potential toxicities associated with a chemical structure, the user can type the name of the compound or insert the SMILES (Simplified Molecular-Input Line-Entry System) string of the compound. Additionally, the user has the possibility to draw the chemical structure with the help of the chemical editor (https://www.chemdoodle.com/). Furthermore, the integrated PubChem search (https:/pubchem.ncbi.nlm.nih.gov/) allows the user to search for chemical structures using the compound name. Optionally, the user may select additional models or all models for prediction. If the user does not specify any additional models, the webserver computes the prediction for acute toxicity and toxicity targets by default.
Output information
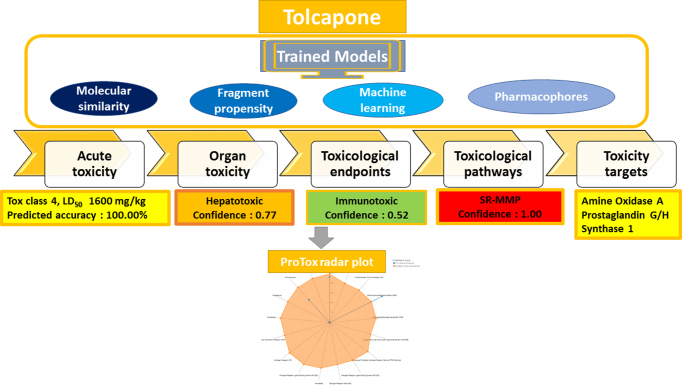
The prediction results for the acute toxicity and toxicity targets are generated instantly. The result page will show the predicted median lethal dose (LD50) in mg/kg weight, toxicity class, and prediction accuracy as well as average similarity along with three most similar toxic compounds from the dataset with the known rodent oral toxicity value. The predicted toxicity targets information, if available will be shown with the name of the target as well as the average fit and similarity of the input compound with the pharmacophore and known ligands of the respective targets. Furthermore, if the user selects additional models, the result page will show the prediction outcomes with confidence score for each model in a table. A web link to access the results will be provided to the user, in case the prediction results cannot be shown immediately. These prediction results are also displayed as a toxicity radar plot comparing the average confidence score of the active compounds in the training set of each model, to that of the input compound (Figure 1). This plot can be assessed using the ‘Open Toxicity Radar Chart’ link that will appear on the result page, once the computation is complete. The same chart can be opened by using the thumbnail below the Toxicity Models Report. More detailed information with an example compound output is made available on the ProTox-II webserver.
Figure 1.
Application case: Tolcapone (a withdrawn drug) is considered as an input structure, predicted using 33 models with respective confidence scores, and prediction results are provided as an overall toxicity radar chart. Tolcapone is predicted to be active for seven endpoints, connecting different layers of the ProTox-II classification scheme.
METHODS
The ProTox-II platform is divided into a five different classification steps: (i) acute toxicity (oral toxicity model with six different toxicity classes); (2) organ toxicity (one model); (3) toxicological endpoints (four models); (4) toxicological pathways (12 models) and (5) toxicity targets (15 models). Here, we provide short description for each of the models available on the ProTox-II server. A detailed information with references, performance scores and frequency distribution of most common features present in the training set (both for active and inactive) molecule are available under model info on the ProTox-II webserver. A complete description on the number of data sets used in this study is provided as Supplementary Table S1.
Acute toxicity
Oral toxicity
The acute toxicity models are developed based on chemical similarities between compounds with known toxic effects and the presence of toxic fragments as explained in our previous paper (12). The acute toxicity data are extracted from the updated version of the in-house database SuperToxic (14).
Toxicity targets
The prediction of toxicity targets is based on 15 different targets from the Novartis in vitro safety panels of the protein targets linked to adverse drug-reactions (15) as reported in our previous work (12).
Organ toxicity
Hepatotoxicity
Drug-induced hepatotoxicity is a significant cause of acute liver failure and one of the major reasons for the withdrawal of drugs from the market (16). Drug-induced liver injury (DILI) is either a chronic process or a rare event. However, prediction of DILI is important and one of the safety concerns for the drug developers, regulators and clinicians (17). The data for the prediction of DILI are taken from DILIrank (18) and the NIH LiverTox database (6). The ProTox-II hepatotoxicity prediction model has a balanced accuracy of 82.00% on cross-validation and 86.00% on external validation. The AUC–ROC scores of cross-validation and external validation are 0.86 and 0.91 respectively (Tables 1 and 2). The kappa value is 0.69 for the model (Tables 1 and 2).
Table 1. Cross-validation results for the newly included models in the ProTox-II platform in terms of balanced accuracy, AUC–ROC and kappa value.
| Models | Balanced accuracy (%) | AUC–ROC | Kappa | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| Organ toxicity | |||||
| DILI | 82.00 | 0.86 | 0.69 | 75.00 | 89.00 |
| Toxicity endpoints | |||||
| Mutagenicity | 84.00 | 0.90 | 0.70 | 83.00 | 85.00 |
| Carcinogenicity | 81.24 | 0.85 | 0.69 | 80.00 | 81.00 |
| Cytotoxicity | 85.00 | 0.89 | 0.65 | 92.00 | 78.00 |
| Immunotoxicity | 75.00 | 0.76 | 0.35 | 69.50 | 79.50 |
| Toxicological pathways | |||||
| nr-ahr | 91.00 | 0.89 | 0.80 | 87.00 | 94.00 |
| nr-ar | 93.00 | 0.84 | 0.75 | 89.00 | 97.00 |
| nr-ar-lbd | 89.00 | 0.87 | 0.76 | 79.50 | 97.00 |
| nr-aromatase | 92.00 | 0.86 | 0.79 | 78.00 | 96.00 |
| nr-er | 90.00 | 0.75 | 0.71 | 85.00 | 95.00 |
| nr-er-lbd | 89.00 | 0.85 | 0.73 | 83.00 | 95.00 |
| nr-ppar-gamma | 92.00 | 0.81 | 0.71 | 86.00 | 97.00 |
| sr-are | 91.00 | 0.84 | 0.69 | 85.00 | 97.00 |
| sr-hse | 90.00 | 0.79 | 0.73 | 89.00 | 91.00 |
| sr-mmp | 91.00 | 0.90 | 0.74 | 82.50 | 96.50 |
| sr-p53 | 89.00 | 0.84 | 0.74 | 83.00 | 95.00 |
| sr-atad5 | 89.00 | 0.84 | 0.71 | 81.50 | 96.50 |
Table 2. External validation results for the newly included models in the ProTox-II platform in terms of balanced accuracy, AUC–ROC and kappa value.
| Models | Balanced accuracy (%) | AUC–ROC | Kappa | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| Organ toxicity | |||||
| DILI | 86.00 | 0.91 | 0.66 | 81.00 | 90.00 |
| Toxicity endpoints | |||||
| Mutagenicity | 85.00 | 0.91 | 0.71 | 83.00 | 87.00 |
| Carcinogenicity | 83.30 | 0.87 | 0.65 | 79.00 | 78.00 |
| Cytotoxicity | 83.60 | 0.90 | 0.60 | 93.00 | 74.00 |
| Immunotoxicity | 70.00 | 0.74 | 0.29 | 65.00 | 74.00 |
| Toxicological pathways | |||||
| nr-ahr | 91.00 | 0.90 | 0.75 | 74.00 | 97.00 |
| nr-ar | 86.00 | 0.73 | 0.73 | 81.00 | 91.00 |
| nr-ar-lbd | 83.00 | 0.75 | 0.72 | 76.00 | 90.00 |
| nr-aromatase | 89.00 | 0.75 | 0.79 | 79.00 | 97.00 |
| nr-er | 91.00 | 0.79 | 0.71 | 85.50 | 96.50 |
| nr-er-lbd | 89.00 | 0.80 | 0.75 | 79.50 | 97.50 |
| nr-ppar-gamma | 85.00 | 0.84 | 0.73 | 73.00 | 96.00 |
| sr-are | 87.00 | 0.79 | 0.72 | 73.00 | 97.00 |
| sr-hse | 86.00 | 0.87 | 0.75 | 80.00 | 92.00 |
| sr-mmp | 91.00 | 0.92 | 0.78 | 86.00 | 95.00 |
| sr-p53 | 89.00 | 0.87 | 0.79 | 80.50 | 96.00 |
| sr-atad5 | 84.00 | 0.80 | 0.78 | 73.00 | 95.00 |
Toxicological endpoints
Carcinogenicity
Chemicals that can induce tumors or increase the incidence of tumours are referred as carcinogens. (19). The data for the prediction of carcinogenicity are collected from the Carcinogenic Potency Database (CPDB) (20) and CEBS database (5). The ProTox-II carcinogenicity prediction model has a balanced accuracy of 81.24% on cross-validation and 83.30% on external validation. The AUC–ROC scores of cross-validation and external validation are 0.85 and 0.87 respectively. The kappa value is 0.69 for the model (Tables 1 and 2).
Mutagenicity
Chemicals that cause abnormal genetic mutations such as changes in the DNA of a cell are referred as mutagens (21). Such changes can cause harm to the cells and result in certain disease, e.g. cancer. ProTox-II, mutagenicity prediction is based on the benchmark data set from Ames test (22) as well as CEBS database (5). The ProTox-II mutagenicity prediction model has a balanced accuracy of 84.00% on cross-validation and 85.00% on external validation. The AUC–ROC scores of cross-validation and external validation are 0.90 and 0.91 respectively. The kappa value is 0.69 for the model (Tables 1 and 2).
Cytotoxicity
Prediction of cytotoxicity is important to screen compounds that can cause undesired and desired cell damage, the latter as in the case of the tumour cells (1). The ProTox-II cytotoxicity model is based on data extracted from the Chemical European Biology Laboratory (ChEMBL) database (23). All compounds with an IC50 value of less than or equal to 10 μM in the in vitro toxicity assay against HepG2 cells are considered as positively cytotoxic. The ProTox-II cytotoxicity prediction model has a balanced accuracy of 85.00% on cross-validation and 83.60% on external validation. The AUC–ROC scores of cross-validation and external validation are 0.89 and 0.90 respectively. The kappa value is 0.69 for the model (Tables 1 and 2).
Immunotoxicity
The adverse effect of xenobiotics on the immune system is called immunotoxicity (24). The immunotoxicity model is based on immune cell cytotoxicity data obtained from the U.S. National Cancer Institute's (NCI) public database. Growth inhibition (GI50) values from the B-cell line RPMI-8226 are used and compounds with GI50 values below 10 μM are defined as toxic (24). The ProTox-II immunotoxicity prediction model has a balanced accuracy of 74.00% on cross-validation and 70.00% on external validation. The AUC–ROC scores of cross-validation and external validation are 0.76 and 0.74 respectively. The kappa value is 0.35 for the model (Tables 1 and 2).
Toxicological pathways
Toxicology in the 21st Century (Tox21) platform, the US toxicology initiative which was started in 2008, provides a library of 10 000 chemical data, screened in high-throughput assays against a panel of 12 different biological target-based pathways, that involve two-major groups of adverse outcome pathways (AOPs): the nuclear receptor pathway and the stress response pathway. ProTox-II, prediction of chemical compounds active in toxicological pathways is based on the Tox21 dataset (11,25).
Nuclear receptor signaling pathways
There are seven target-pathway based models under nuclear receptor signaling pathways: aryl hydrogen receptor (AhR), androgen receptor (AR), androgen receptor ligand binding domain (AR-LBD), aromatase, estrogen receptor alpha (ER), estrogen receptor ligand binding domain (ER-LBD), and peroxisome proliferator activated receptor gamma (PPAR-Gamma). All the models have a balanced accuracy of >80% and AUC–ROC values within a range of 0.75–0.90 for both cross-validation and external validation. The kappa values are in a range of 0.60–0.80 (Tables 1 and 2).
Stress response pathways
There are five target-pathway based models under stress response pathways: Nuclear factor (erythroid-derived 2)-like 2/antioxidant responsive element (ARE), heat shock factor response element (HSE), mitochondrial membrane potential (MMP), phosphoprotein tumor suppressor (p53), and ATPase family AAA domain-containing protein 5 (ATAD5). All the models have a balanced accuracy of >80% and AUC–ROC values within the range of 0.80–0.90 for both cross-validation and external validation. Except for HSE the value for ROC–AUC is 0.79. The kappa values are in a range of 0.60–0.80 (Tables 1 and 2).
Prediction models
All the newly added prediction models on the ProTox-II platform are based on machine learning algorithms. A Random Forest (RF) algorithm (26) is used to construct the classification and prediction models for hepatotoxicity, cytotoxicity, mutagenicity, and carcinogenicity. The RF-based models are constructed using 500 decision trees and GINI index criterion. The advantage of using RF-based classifier is that it tends to avoid overfitting.
For the construction of theTox21 based toxicological pathway prediction, an ensemble approach is used including RF and Support Vector Machine (SVM) classifiers. The radial basis function (RBF) is used as kernel function for the SVM algorithm. Immunotoxicity prediction model is based on Bernoulli–Naive Bayes algorithm, as explained in the published work (24).
Here, two different fingerprints are used: MACCS molecular fingerprints-166 bits and Morgan circular fingerprints-2048 bits (http:/www.rdkit.org/). These two fingerprints have shown an optimal performance for prediction of chemical activity (11,24).
Additionally, a selective oversampling of minority class is introduced in the construction of the models. For each of the prediction end-points, the active (positive) and inactive (negative) data are fragmented using RECAP (27) and ROTBONDS (28) fragmentation methods. The propensity score (PS) (12) for each of the uniquely occurring fragments in both the sets is computed. Only those molecules having the highest propensity scores for fragments conserved for the active class are oversampled and added into model construction. The same ratio of active and inactive compounds was maintained for all the folds of cross-validation, using the fragment-based similarities between the compounds.
The prediction models are based on python programming language. Machine learning packages like scikit-learn (http:/scikit-learn.org) and cheminformatics package RDKit (http:/www.rdkit.org/) are used for the model implementation. All data are standardized using KNIME (29). A template script (Sample API script http://tox.charite.de/protox_II/simple_api.py) has been provided under the description ‘using the API’ on the FAQ section of the ProTox-II webserver.
METHODS VALIDATION
All the new models are validated using fragment-based CLUSTER 10-fold cross-validation. The data was divided using fragment similarity based sampling method and ensuring the active and inactive ratios are constant into 10 sets; 9 of which were used to train the model and 10th to validate the model. Additionally, external sets (which was previously not introduced in the training set) was used for external validation of the models. All the models are assessed on the following performance measures:
Balanced accuracy is defined as the (sensitivity + specificity)/2 which also equals to 1/2[true positive/(true positive + false negative) + true negative/(true negative + false positive)]
The area under the curve (AUC) of a receiver operating characteristic (ROC) curve plots sensitivity versus 1 – specificity at different thresholds. The AUC–ROC has been used as an effective measure for binary classifiers trained on imbalanced data set (having minority and majority class) (30).
The kappa index measures the quality of binary classification models. The kappa index ranges between 0 (less significant) to 1(perfect) (31).
Application case
To illustrate the functionalities as well as possible application of the ProTox-II webserver, we have selected two different examples: one is an approved drug (Etonogestrel) and another is a withdrawn drug (Tolcapone).
Etonogestrel is an approved drug and is used in hormonal contraceptives. Toxicology of progestogens has been reported in studies (32). Using our ProTox-II webserver, Etonogestrel has been predicted as Toxicity class 5 for acute oral toxicity with LD 50 value of 5000mg/kg, with an average similarity of 92.31% and prediction accuracy of 72.9%. The organ toxicity (hepatotoxicity) of etonogestrel is predicted with a confidence score of 0.68 and it is predicted active for five different toxicological pathways such AR-LBD, NR-AR, ER-LBD, ER and SR-ARE with high confidence scores of 0.94, 0.96, 0.73, 0.76 and 0.57 respectively. Additionally, three different toxicity targets (Androgen Receptor, Amine Oxidase A, Progesterone Receptor A) are predicted with probable binding. Etonogestrel has shown binding to Progesterone receptor and Androgen Receptor at 1uM. This is indeed interesting, as Etonogestrel is classified under relatively less harmful acute toxicity class; however, it has been predicted to be hepatotoxic and at the same time, active for five toxicological pathways and three toxicity targets (Supplementary material S3). Providing insight though etonogestrel is structurally safe compound, however, it's interaction with certain molecular targets might result in toxic response.
Tolcapone, a catechol o-methyltransferase inhibitor and a withdrawn drug serves as an interesting example of a chemical compound which tends to connect different prediction classes designed under ProTox-II method (16). Tolcapone is reported with a toxicity class 4 for acute oral toxicity with LD 50 value of 1600 mg/kg, with an average similarity of 100.00% and prediction accuracy of 100.00%. The organ toxicity (hepatotoxicity) of Tolcapone is predicted with a confidence score of 0.79 and toxicological endpoint (immunotoxicity) with a confidence score of 0.52 (33). Additionally, it is predicted to be active for MMP toxicological pathway with a confidence score of 0.99 (34). Amine Oxidase A and Prostaglandin G/H Synthase 1 are predicted as toxicity targets with low binding probability (Figure 1). This example indeed represents, that there is a possibility that a compound can be active for multiple toxicity endpoints and thereby resulting in severe toxic effects (Figure 1).
CONCLUSIONS AND FUTURE UPDATES
Here, we present ProTox-II which incorporates molecular similarity for acute toxicity prediction, pharmacophore-based models for 15 toxicity targets, fragment propensities and machine learning models for 17 different toxicity end points. To the best of our knowledge, ProTox-II is the freely available computational toxicity webserver enabling the prediction of the largest number of toxicity endpoints consisting of 33 models. A novelty of the updated ProTox-II webserver is that the prediction scheme is classified into different levels of toxicity such oral toxicity, organ toxicity (hepatotoxicity), toxicological endpoints (such as mutagenicity, carcinotoxicity, cytotoxicity and immunotoxicity), toxicological pathways (AOPs) and toxicity targets thereby providing insights into the possible molecular mechanism behind such toxic response. When compared with other standard published models, all the models of the ProTox-II platform performed from the range of comparatively good to better in some cases. Though, it is worth mentioning that due to differently processed training sets, and algorithm parameters as well as molecular descriptors selections and sampling methods considering different thresholds, a complete comparison of all the models are not feasible. However, a fair comparison using performance measures like accuracy and AUC–ROC has been provided as Supplementary Table S2.
We believe that in the process of toxicity analysis extending to drug discovery, ProTox-II in silico prediction platform will help to initiate focused experimental follow-up studies and to enhance hit selection and lead optimization process. Additionally, ProTox-II methods have the potential to support risk assessments for regulatory decisions such as to create novel hypotheses and get insights to the mechanisms of toxicity.
In future, ProTox-II will focus on methods development to foster better characterization of clinically relevant adverse effects based on published knowledge and relevant chemical-target-side effect and adverse outcome pathways networks. The next evolutionary step will be considering species and inter-individual genetic differences. Additionally, to maintain the high standard of the ProTox-II platform, regular updates are planned and will be executed on a quarterly basis. Furthermore, new data will be added to the existing models when available and new endpoints like genotoxicity, nephrotoxicity, neurotoxicity, and cardiotoxicity as well as food allergy prediction will be added.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the students of the Structural Bioinformatics Group at Charité for testing the ProTox-II webserver.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Berlin-Brandenburg research platform BB3R, Federal Ministry of Education and Research (BMBF), Germany [031A262C]; DKTK. Funding for open access charge: Charité – University Medicine Berlin.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zhang L., Mchale C.M., Greene N., Snyder R.D., Rich I.N., Aardema M.J., Roy S., Pfuhler S.. Commentary emerging approaches in predictive toxicology. 2014; 55:679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang Y., Xing J., Xu Y., Zhou N., Peng J., Xiong Z., Liu X., Luo X., Luo C., Chen K. et al. In silico ADME/T modelling for rational drug design. Q. Rev. Biophys. 2015; 48:488–515. [DOI] [PubMed] [Google Scholar]
- 3. Raies A.B., Bajic V.B.. In silico toxicology: computational methods for the prediction of chemical toxicity. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2016; 6:147–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Richard A.M., Williams C.R.. Distributed structure-searchable toxicity (DSSTox) public database network: a proposal. Mut. Res. 2002; 499:27–52. [DOI] [PubMed] [Google Scholar]
- 5. Lea I.A., Gong H., Paleja A., Rashid A., Fostel J.. CEBS: a comprehensive annotated database of toxicological data. Nucleic Acids Res. 2017; 45:D964–D971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thakkar S., Chen M., Fang H., Liu Z., Roberts R., Tong W.. The Liver Toxicity Knowledge Base (LKTB) and drug-induced liver injury (DILI) classification for assessment of human liver injury. Expert Rev. Gastroenterol. Hepatol. 2018; 12:31–38. [DOI] [PubMed] [Google Scholar]
- 7. Betts K.S. Tox21 to date: steps toward modernizing human hazard characterization. Environ. Health Perspect. 2013; 121:2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang R., Xia M., Nguyen D., Zhao T., Sakamuru S.. Tox21 challenge to build predictive models of nuclear receptor and stress response pathways as mediated by exposure to environmental chemicals and drugs. Front. Environ. Sci. 2016; 3:85. [Google Scholar]
- 9. Knudsen T.B., Keller D.A., Sander M., Carney E.W., Doerrer N.G., Eaton D.L., Fitzpatrick S.C., Hastings K.L., Mendrick D.L., Tice R.R. et al. FutureTox II: In vitro data and in silico models for predictive toxicology. Toxicol. Sci. 2015; 143:256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mayr A., Klambauer G., Unterthiner T., Hochreiter S.. DeepTox: toxicity prediction using deep learning. Front. Environ. Sci. 2016; 3:80. [Google Scholar]
- 11. Banerjee P., Siramshetty V.B., Drwal M.N., Preissner R.. Computational methods for prediction of in vitro effects of new chemical structures. J. Cheminformatics. 2016; 8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drwal M.N., Banerjee P., Dunkel M., Wettig M.R., Preissner R.. ProTox: a web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014; 42:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng F., Li W., Zhou Y., Shen J., Wu Z., Liu G., Lee P.W., Tang Y.. AdmetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inform. Model. 2012; 52:3099–3105. [DOI] [PubMed] [Google Scholar]
- 14. Schmidt U., Struck S., Gruening B., Hossbach J., Jaeger I.S., Parol R., Lindequist U., Teuscher E., Preissner R.. SuperToxic: a comprehensive database of toxic compounds. Nucleic Acids Res. 2009; 37:D295–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lounkine E., Keiser M.J., Whitebread S., Mikhailov D., Hamon J., Jenkins J.L., Lavan P., Weber E., Doak A.K., Côté S. et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012; 486:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siramshetty V.B., Nickel J., Omieczynski C., Gohlke B.-O., Drwal M.N., Preissner R.. WITHDRAWN—a resource for withdrawn and discontinued drugs. Nucleic Acids Res. 2016; 44:D1080–D1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J., Mansouri K., Judson R.S., Martin M.T., Hong H., Chen M., Xu X., Thomas R.S., Shah I.. Predicting hepatotoxicity using ToxCast in vitro bioactivity and chemical structure. Chem. Res. Toxicol. 2015; 28:738–751. [DOI] [PubMed] [Google Scholar]
- 18. Chen M., Suzuki A., Thakkar S., Yu K., Hu C., Tong W.. DILIrank: The largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov. Today. 2016; 21:648–653. [DOI] [PubMed] [Google Scholar]
- 19. Kroes R., Renwick A.G., Cheeseman M., Kleiner J., Mangelsdorf I., Piersma A., Schilter B., Schlatter J., van Schothorst F., Vos J.G. et al. Structure-based thresholds of toxicological concern (TTC): guidance for application to substances present at low levels in the diet. Food Chem. Toxicol. 2004; 42:65–83. [DOI] [PubMed] [Google Scholar]
- 20. Fitzpatrick R.B. CPDB: carcinogenic potency database. Med. Ref. Serv. Q. 2008; 27:303–311. [DOI] [PubMed] [Google Scholar]
- 21. Ames B.N., Durston W.E., Yamasaki E., Lee F.D.. Carcinogens are mutagens: a simple test system combining liver homogenates for activation and bacteria for detection. Proc. Natl. Acad. Sci. U.S.A. 1973; 70:2281–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hansen K., Mika S., Schroeter T., Sutter A., Laak A. Ter, Thomas S.H., Heinrich N., Müller K.R.. Benchmark data set for in silico prediction of Ames mutagenicity. J. Chem. Inform. Model. 2009; 49:2077–2081. [DOI] [PubMed] [Google Scholar]
- 23. Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M., Krüger F.A., Light Y., Mak L., McGlinchey S. et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014; 42:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schrey A.K., Nickel-Seeber J., Drwal M.N., Zwicker P., Schultze N., Haertel B., Preissner R.. Computational prediction of immune cell cytotoxicity. Food Chem. Toxicol. 2017; 107:150–166. [DOI] [PubMed] [Google Scholar]
- 25. Huang R., Xia M., Sakamuru S., Zhao J., Shahane S.A., Attene-Ramos M., Zhao T., Austin C.P., Simeonov A.. Modelling the Tox21 10 K chemical profiles for in vivo toxicity prediction and mechanism characterization. Nat. Commun. 2016; 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Breiman L. Random Forest. Mach. Learn. 2001; 45:5–32. [Google Scholar]
- 27. Lewell XQ, Judd DB, Watson SP, Hann M.M.. RECAP–retrosynthetic combinatorial analysis procedure: a powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J. Chem. Inf. Comput. Sci. 1998; 38:511–522. [DOI] [PubMed] [Google Scholar]
- 28. Ahmed J., Worth C.L., Thaben P., Matzig C., Blasse C., Dunkel M., Preissner R.. FragmentStore–a comprehensive database of fragments linking metabolites, toxic molecules and drugs. Nucleic Acids Res. 2011; 39:D1049–D1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berthold M.R., Cebron N., Dill F., Gabriel T.R., Kötter T., Meinl T., Ohl P., Sieb C., Thiel K., Wiswedel B.. KNIME: The Konstanz Information Miner. 2008; Berlin, Heidelberg: Springer; 319–326. [Google Scholar]
- 30. Bewick V., Cheek L., Ball J.. Statistics review 13: receiver operating characteristic curves. Crit. Care (London, England). 2004; 8:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gwet K.L. Variance estimation of Nominal-Scale Inter-Rater reliability with random selection of raters. Psychometrika. 2008; 73:407–430. [Google Scholar]
- 32. Jordan A. Toxicology of progestogens of implantable contraceptives for women. Contraception. 2002; 65:3–8. [DOI] [PubMed] [Google Scholar]
- 33. Desbonnet L., Tighe O., Karayiorgou M., Gogos J.A., Waddington J.L., O’Tuathaigh C.M.P.. Physiological and behavioural responsivity to stress and anxiogenic stimuli in COMT-deficient mice. Behav. Brain Res. 2012; 228:351–358. [DOI] [PubMed] [Google Scholar]
- 34. Haasio K., Koponen A., Penttilä K.E., Nissinen E.. Effects of entacapone and tolcapone on mitochondrial membrane potential. Eur. J. Pharmacol. 2002; 453:21–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.