Abstract
iPath3.0 (http://pathways.embl.de) is a web-application for the visualization and analysis of cellular pathways. It is freely available and open to everyone. Currently it is based on four KEGG global maps, which summarize up to 158 traditional KEGG pathway maps, 192 KEGG modules and other metabolic elements into one connected and manually curated metabolic network. Users can fully customize these networks and interactively explore them through its redesigned, fast and lightweight interface, which highlights general metabolic trends in multi-omics data. It also offers navigation at various levels of details to help users further investigate those trends and ultimately uncover novel biological insights. Support for multiple experimental conditions and time-series datasets, tools for generation of customization data, programmatic access, and a free user accounts system were introduced in this version to further streamline its workflow.
INTRODUCTION
Metabolic networks provide an intuitive and powerful framework for understanding cellular systems. With the availability of manually curated pathway diagrams of metabolic networks and the ever increasing volume of omics data to visualize, several tools (1–4) were developed to help scientists explore and visually integrate these data onto pathway diagrams. iPath (1,2), one of the first tools to popularize this concept online, proved useful in various contexts. For instance, it has been used to map differentially expressed functional transcripts between early and late plant development (5), and metabolic pathways that differentiate Crohn's disease form healthy phenotypes in human gut metaproteogenomic samples (6).
Here we present iPath3.0, the latest version of iPath, which greatly improves the user experience by speeding up the rendering of the maps and user interactions, by adding two new global maps and by enriching its biological knowledge base with 62 novel KEGG pathway modules (7), 1700 new reactions, 1500 new KEGG Orthology entries (KO) and close to 3000 new species.
USER INTERFACE AND PATHWAY CUSTOMIZATION
iPath3.0’s pathway explorer provides zooming and panning controls that allow the user to easily navigate the complex pathway maps. Clicking on nodes and edges in the map displays popup windows with detailed information about the associated data, such as enzymes, reactions, pathway maps and compounds involved, each hyper-linked to the original annotation source. With several built-in data mapping tools and extensive customization options, users can upload various types of data associated with enzymes or compounds to customize any overview or species-specific pathways map.
IMPROVEMENTS AND NEW FEATURES
A major advance in iPath3.0 is its new display engine. Implemented in Javascript and SVG, it greatly speeds up the rendering of the maps, brings faster and lighter user interactions, and therefore streamlines the whole user experience compared to the previous Flash-based engine. The performance of the search engine was greatly improved as well, and the results extended to display the number of map elements matched by each entry, and are emphasized upon selection.
Mapping conflicts, which occur when multiple IDs with different customization match the same element in the map, are now handled by a simple mechanism that allows the user to choose one of the available customizations, which not only helps in better exploring the maps but also reduces the amount of complex image post-processing that might be required for conveying a clear biological message.
In addition, support for time-series datasets was added using animated map transitions where users can toggle looping and control the transition speed and the color at each time-point. Interestingly, this feature could also be used for exploring differences in metabolic processes between two or more datasets, which is a common use case of iPath. To further simplify this task, we integrated a new tool to generate map customizations that reflect the intersection and the complements of two sets of identifiers. Similarly, a generator, based on numeric data, was also introduced to produce customizations based on the correlation coefficient or the average of two or more datasets.
Furthermore, a free user accounts system was implemented for saving customizations and organizing them in sharable workspaces and projects. This will help users keeping track and documentation of their customizations, and provide them instant restoration and switching between customizations as well as easy sharing with collaborators via shared links.
Finally, programmatic access to iPath3.0 is made available over HTTP to seamlessly integrate it into pipelines that rely on its pathway visualization (8) and also to visualize several customizations in one batch.
UNDERLYING DATA SET
iPath3.0 uses version 82.0 (15 May 2017) of KEGG, which in addition to the updated global maps for ‘Global metabolism’ and ‘Biosynthesis of secondary metabolites’, now includes two new global maps: ‘Microbial metabolism in diverse environments’ and ‘Biosynthesis of antibiotics’. The former should benefit microbiome research as it is focused on microbial metabolism, as opposed to the broad metabolic spectrum of the ‘Global metabolism’ map. Similarly, the latter could help exploring the link between the biosynthesis and the resistance to antibiotics. These global maps consist of 158, 48, 42, 47 summarized pathways, and 192, 38, 71, 55 modules, respectively.
INPUT AND OUTPUT DATA
Input data for map customization is space delimited, where the first element is a biological annotation followed by desired user customization. For example, ‘M00001 #FF0000 W20’ will highlight the glycolysis module in red and enlarge its edges to 20px. Supported KEGG annotations are pathways, modules, reactions, KOs, genes, compounds and EC numbers. Uniprot (9) and STRING (10) protein IDs, COGs (11), eggNOGs (12), NCBI (13) gene identifiers, ChEBI (14) and PubChem (13) identifiers and eight additional chemical compounds databases are also supported to appeal to a broader community of researchers. iPath also contains species information (NCBI taxonomy ID or KEGG organism code), that allows users to display only customized versions of species-specific pathways.
Customized maps can also be exported for further editing and publication. Available export formats are Scalable Vector Graphics (svg), Portable Network Graphics (png), Encapsulated Postscript (eps) and Portable Document Format (pdf).
ILLUSTRATIVE EXAMPLE
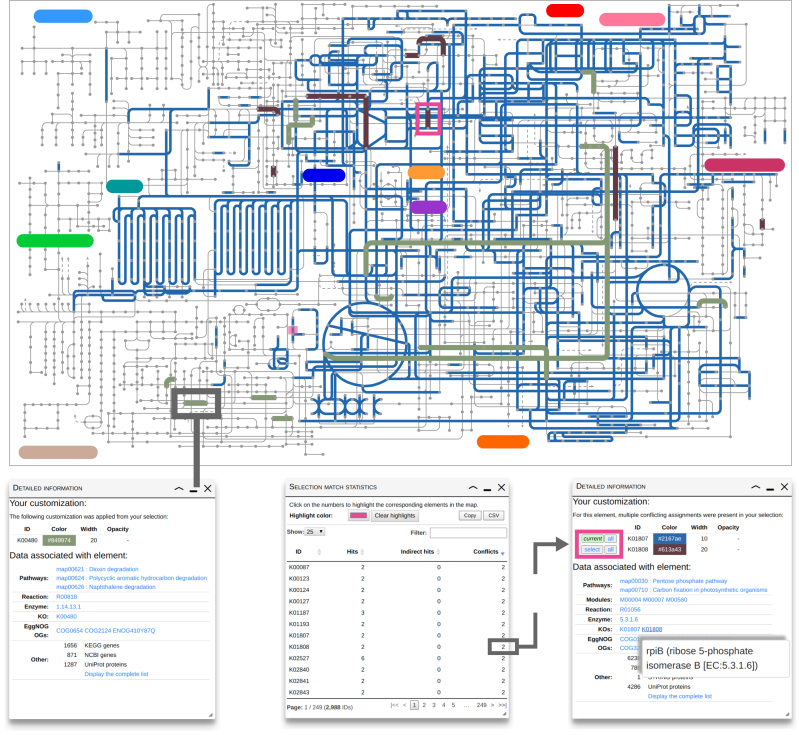
To illustrate new features of iPath3.0 we compared the metabolic repertoire of Escherichia coli O157:H7 Xuzhou21 (ELX) and E. coli K-12 MDS42 (ECOK). We used the new feature ‘Element selection based on ID overlap’ from the tools section to customize the ‘Global metabolism’ map based on their KOs. Reactions highlighted in blue (Figure 1) show that most enzymatic functions of these strains overlap, while those highlighted in brown are specific to ECOK and in green specific to ELX. By displaying detailed information about the ECOK-specific reaction highlighted in the Pentose phosphate pathway (Figure 1), we found that the both strains have the Ribose 5-phosphate isomerase (Rip) required for this reaction however ECOK has both RipA and RipB while ELX has only RipA. Using the conflict resolution tool (Figure 1) users could chose to color this match in blue if the finding is not of interest to their research question or keep it in brown to emphasis it. Resolving more conflicts is made easy by the ‘Selection match statistics’ panel which lists all conflicts and highlights them upon selection. Other hits were truly strain-specific as shown for K00480.
Figure 1.
Overlap and differences between the metabolic enzymes of Escherichia coli O157:H7 Xuzhou21 (green) and E. coli K-12 MDS42 (brown). The panel on the left shows detailed information about the element matched by K00480. The panel in the middle displays customization match statistics in a sortable and searchable table. Clicking the conflict number highlights (in pink) the elements associated with the conflict. The right panel shows details about the Ribose 5-phosphate isomerase and gives an example of conflict resolution, where clicking on the select button will change the color of the highlighted element to blue.
This example illustrates how iPath3.0 simplifies the navigation and the comparison of metabolic pathways profiles, but several other examples and video tutorials are also available in the online help section.
COMPARISON WITH OTHER TOOLS
KEGG Atlas (4) and Pathway projector (3) are web tools for pathway exploration. Both offer map navigation through zooming and panning, taxonomic filtering as well as limited map customizations. On top of these functionalities iPath3.0 offers a wide array of additional features covered in the previous section ‘Improvement and new features’, which sets it apart from these tools as not only a pathway explorer, but also an advanced visual analysis tool for metabolic data.
DISCUSSION
The new built-in color generation tools and the modernization of the web interface of iPath3.0 greatly improve and streamline the user experience. The addition of time-series visualization and programmatic access opens novel exploration perspectives. With all these improvements we hope that iPath3.0 will help scientists to comprehensively explore more cellular pathways with less effort and in a shorter time span. Meanwhile we will continue improving iPath by providing users with additional live editing features, an integrated viewer for traditional KEGG maps, and the possibility to upload user defined global maps.
FUNDING
European Molecular Biology Laboratory (EMBL); European Research Council Microbios [ERC-AdG-669830]; European Commission MetaCardis project [FP7-HEALTH-2012-INNOVATION-I-305312]; European Union Horizon 2020 research and innovation programme [686070]; Japan Science and Technology Agency PRESTO [JPMJPR1507]; Japan Agency for Medical Research and Development (AMED) [17ek0109187h0002]. Funding for open access charge: Tokyo Institute of Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Letunic I., Yamada T., Kanehisa M., Bork P.. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem. Sci. 2008; 33:101–103. [DOI] [PubMed] [Google Scholar]
- 2. Yamada T., Letunic I., Okuda S., Kanehisa M., Bork P.. iPath2.0: interactive pathway explorer. Nucleic Acids Res. 2011; 39:W412–W415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kono N., Arakawa K., Ogawa R., Kido N., Oshita K., Ikegami K., Tamaki S., Tomita M.. Pathway projector: web-based zoomable pathway browser using KEGG Atlas and Google Maps API. PLoS One. 2009; 4:e7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Okuda S., Yamada T., Hamajima M., Itoh M., Katayama T., Bork P., Goto S., Kanehisa M.. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008; 36:W423–W426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaparro J.M., Badri D.V., Vivanco J.M.. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014; 8:790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erickson A.R., Cantarel B.L., Lamendella R., Darzi Y., Mongodin E.F., Pan C., Shah M., Halfvarson J., Tysk C., Henrissat B. et al. Integrated metagenomics/metaproteomics reveals human Host-Microbiota signatures of Crohn's disease. PLoS One. 2012; 7:e49138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M.. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014; 42:D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leimena M.M., Ramiro-Garcia J., Davids M., van den Bogert B., Smidt H., Smid E.J., Boekhorst J., Zoetendal E.G., Schaap P.J., Kleerebezem M.. A comprehensive metatranscriptome analysis pipeline and its validation using human small intestine microbiota datasets. BMC Genomics. 2013; 14:530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bateman A., Martin M.J., O’Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R. et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Smirnov S., Nikolskaya A.N. et al. The COG database: an updated vesion includes eukaryotes. BMC Bioinformatics. 2003; 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M.C., Rattei T., Mende D.R., Sunagawa S., Kuhn M. et al. EGGNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016; 44:D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Resource Coordinators NCBI. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017; 45:D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C.. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016; 44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]