Abstract
Complex carbohydrates of plants are the main food sources of animals and microbes, and serve as promising renewable feedstock for biofuel and biomaterial production. Carbohydrate active enzymes (CAZymes) are the most important enzymes for complex carbohydrate metabolism. With an increasing number of plant and plant-associated microbial genomes and metagenomes being sequenced, there is an urgent need of automatic tools for genomic data mining of CAZymes. We developed the dbCAN web server in 2012 to provide a public service for automated CAZyme annotation for newly sequenced genomes. Here, dbCAN2 (http://cys.bios.niu.edu/dbCAN2) is presented as an updated meta server, which integrates three state-of-the-art tools for CAZome (all CAZymes of a genome) annotation: (i) HMMER search against the dbCAN HMM (hidden Markov model) database; (ii) DIAMOND search against the CAZy pre-annotated CAZyme sequence database and (iii) Hotpep search against the conserved CAZyme short peptide database. Combining the three outputs and removing CAZymes found by only one tool can significantly improve the CAZome annotation accuracy. In addition, dbCAN2 now also accepts nucleotide sequence submission, and offers the service to predict physically linked CAZyme gene clusters (CGCs), which will be a very useful online tool for identifying putative polysaccharide utilization loci (PULs) in microbial genomes or metagenomes.
INTRODUCTION
Importance of complex carbohydrates
Carbohydrates are one of the four major classes of large biopolymers found in all cells together with nucleic acids, proteins, and lipids. Carbohydrates include monosaccharides, oligosaccharides, and polysaccharides. Hybrid biopolymers with carbohydrates covalently linked to other biopolymers, such as glycoproteins and glycolipids, are called glycoconjugates. Complex carbohydrates and glycoconjugates are synthesized, degraded, and modified by carbohydrate active enzymes (CAZymes) in all organisms (1). Particularly, plants use photosynthesis to convert carbon dioxide and water into sugars, which are further turned into carbohydrates such as starches and celluloses with the help of CAZymes. Therefore, CAZymes are vitally important for plants and plant-associated animals and microbes, and not surprisingly CAZyme genes are particularly abundant in genomes of plants and plant-degrading microbes (2,3).
Importance of CAZymes
In addition to their significance in bioenergy and agricultural industries (4), CAZymes are also extremely important for human health (5). This is because humans and other animals depend on bacteria living in the digestive tracts to degrade various indigestible carbohydrates and salvage nutrients (6). It has been shown that the genomes of animal gut bacteria encode hundreds of carbohydrate-degrading GH (glycoside hydrolase) genes, in contrast to only 17 digestive GH genes encoded in the human genome (7). Recent research has suggested that altering the dietary carbohydrate composition has a profound impact on the gut microbiota structure, which further influence the human health (8,9).
CAZy database
Since 1990s over 360 CAZyme families have been defined and classified by the CAZy database (10), forming six major classes: glycosyltransferases [GTs], glycoside hydrolases [GHs], polysaccharide lyases [PLs], carbohydrate esterases [CEs], carbohydrate-binding module [CBM] and enzymes for the auxiliary activities [AAs]. CAZy also assigns GenBank proteins to CAZyme families and these CAZy pre-annotated proteins are the foundation for sequence similarity-based CAZyme annotation.
Methods for CAZyme annotation
Owing to the importance of CAZymes, newly sequenced genomes are often analyzed for putative CAZymes (collectively named CAZome). Two approaches of CAZome annotation exist in the literature:
Users contact the CAZy database for collaboration, who will perform semi-automatic CAZome annotation for the users (11); as expert manual curations are involved, CAZy annotation is regarded as the gold standard method.
Users run automatic tools such as HMMER (12) or BLAST (13) by themselves for CAZome annotation on their own computers or on the web (see below). Before 2012, BLAST was often used to search against CAZy pre-annotated proteins on users’ own computers.
In 2010, CAT (CAZyme Analysis Toolkit) was developed as a web server, which allows users to run both BLAST and HMMER searches remotely on the CAT web server (14). The HMMER search is run against Pfam HMMs (hidden Markov models) that are associated with CAZy pre-annotated CAZymes.
In 2012, we developed dbCAN, a database of HMMs for CAZyme family-specific signature domains (4). Different from CAT, for each CAZyme family we retrieved its signature domains from CAZy pre-annotated members, by searching against the CDD (conserved domain database of NCBI) database and manual literature curation; we then built our own HMMs for most CAZyme families instead of using Pfam HMMs.
We update dbCAN almost once a year, by creating HMMs for CAZyme families and subfamilies newly created in the CAZy database (Figure 1). Users can download our HMMs and run HMMER locally for automated CAZome annotation. We also provide a Perl script to help parse the HMMER output, which returns CAZyme signature domains, their boundaries, E-values, and HMM domain coverage. Such domain-based annotation is particularly useful for CAZymes, as they tend to be modular proteins with multiple CAZyme domains and sometime domain repeats (e.g. multiple CBMs of the same family).
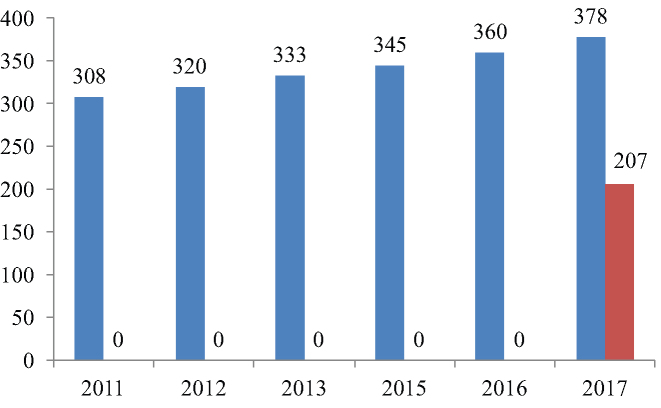
Figure 1.
dbCAN is updated every year and now has 575 HMMs. X-axis: year; Y-axis: number of HMMs of families (blue) and subfamilies (red).
To help users who do not have programming experience, we also developed a web server to allow users submit protein sequences and run HMMER on our server to identify CAZymes. With the CAT website no longer maintained since 2013 and eventually obsolete in 2017, dbCAN has become the only web server that is still actively updated and offering online CAZyme annotation service.
In 2017, a new tool named Hotpep (15) annotates CAZymes by searching against PPR (peptide pattern recognition) library for conserved short peptide motifs (16) present in different CAZyme families. In the PPR library, each CAZyme family has a set of 6-mer peptides that are conserved in that family, and Hotpep is used to scan new proteins for the presence of these peptides in order to assign the query proteins into existing CAZyme families.
Importance of automated CAZyme annotation
It should be mentioned that approach B is actually also included in approach A, but can be fully automated and carried out in the users’ own hands. Using CAZy already annotated CAZomes to benchmark the automated CAZyme annotation found >90% of accuracy typically for model bacterial genomes (3). Clearly, as more and more genomes and metagenomes becoming available, such automated CAZome annotation has a clear advantage over annotation by CAZy through collaboration, in that users can quickly obtain the candidate CAZyme gene list by themselves as part of their bioinformatics pipeline for genome annotation.
Indeed, the popularity of automated CAZome annotation can be manifested by citations of the two approaches. Specifically, ∼100 papers have been published since 2012 with CAZomes annotated by collaboration with CAZy (according to http://www.cazy.org/Genomes.html). As a comparison, more than 300 papers have been published since 2012 using dbCAN for CAZome annotation (according to Google Scholar: https://scholar.google.com/scholar?cites=5112424923296812233, only counted papers that used the tool for finding CAZymes), and more than 100 papers have been published since 2012 using CAT for CAZome annotation (according to Google Scholar: https://scholar.google.com/scholar?cites=12948408578800903520, also only counted papers that used the tool for finding CAZymes).
Lastly, the availability of dbCAN HMMs has also enabled other bioinformatics tools to incorporate CAZyme annotation step into their data analysis workflows, e.g., MOCAT2 (17), DemaDb (18), proGenomes (19) and SACCHARIS (20).
NEW FUNCTIONS AND UPDATES
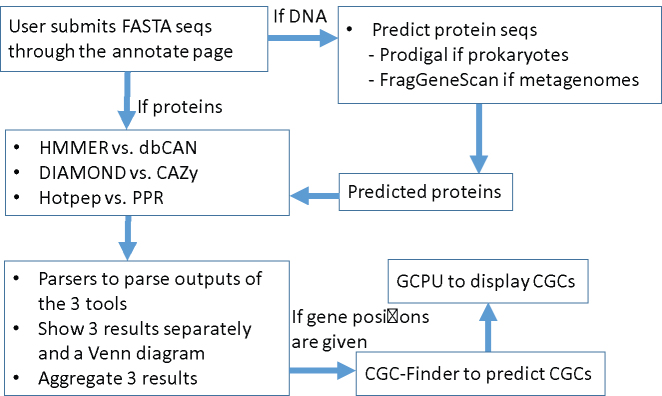
Figure 2 shows the overall design of dbCAN2, an updated meta server of dbCAN server, which has the following new functions: (i) allows submission of DNA sequences in addition to protein sequences; (ii) integrates three state-of-the-art tools/databases for automated CAZyme annotation; (iii) can identify transcription factors (TFs), transporters (TCs), and further CAZyme gene clusters (CGCs) using CGC-Finder (3); (iv) combines the results from the three tools, allows visualization as a Venn diagram and detailed results as graphs, and offers an easy solution to download results as text files.
Figure 2.
Overall design of dbCAN2 meta server. GCPU (gene cluster plot utility) and CGC-Finder (CAZyme gene cluster finder) are two tools developed for dbCAN2.
DNA sequence submission
In addition to protein submission, dbCAN2 now also accepts nucleotide sequences, e.g. the complete or draft genomes and metagenomes of prokaryotes. Protein sequences are predicted by calling Prodigal (21) if the query is genomes, or FragGeneScan (22) if the query is short DNAs from metagenomes or mRNAs or coding sequences of proteins. As eukaryotic gene prediction is more complex and often needs additional input data (e.g. transcriptome data), users should perform gene predictions for eukaryotic genomes elsewhere and only submit protein sequences to dbCAN2.
Meta server of three tools/databases
The dbCAN web server (http://csbl.bmb.uga.edu/dbCAN/) currently provides HMMER search against dbCAN HMM database, and also DIAMOND (23) search against CAZy pre-annotated CAZyme sequence database. However, the results from the two tools are presented on two separate pages and not integrated at any level. In dbCAN2, we have added the third tool: Hotpep search against the PPR short peptide library. We have also systematically compared the outputs of the three tools against the CAZy pre-annotated CAZomes (i.e. as the gold standard sets) of three bacterial genomes and three eukaryotic genomes (Supplementary Table S1), in order to: (i) find the best parsing thresholds (e.g. E-value) for each tool, (ii) evaluate the annotation performance of the three tools and (iii) find the best way to aggregate the three outputs to achieve the best annotation performance.
The accuracy is calculated as an F-score = 2 × (Recall × Precision)/(Recall + Precision) for the three tools on each examined genome, following the method presented in our previous papers (2,3). We removed unclassified CAZymes (e.g. GH0) and families not in the PPR library when calculating F-scores. Supplementary Table S1 presents the best parsing thresholds that we selected to use for the web server: (i) for HMMER+dbCAN, we use E-value <1e–15 and coverage >0.35; (ii) for DIAMOND+CAZy, we use E-value <1e–102 and (iii) for Hotpep+PPR, we use the number of conserved peptide hits >6 and the sum of conserved peptide frequencies >2.6. Table 1 shows that DIAMOND+CAZy has the highest F-score (0.89) for bacteria but the lowest F-score for eukaryotes (0.84); in contrast, Hotpep + PPR has the highest F-score (0.94) for eukaryotes but the lowest F-score for bacteria (0.80). HMMER + dbCAN performs very well for both eukaryotes (0.86) and bacteria (0.88) and a slightly higher overall F-score than the other two tools (Supplementary Table S1). In terms of running time, DIAMOND runs the fastest, followed by Hotpep and HMMER.
Table 1. Comparison of tools for automated CAZyme annotation.
| Accuracy (F-score) | |||||||
|---|---|---|---|---|---|---|---|
| Tools + databases | Bacteria | Eukaryotes | Subfamily | Multi-family proteins | Domain repeats | Domain positions | Speedc |
| HMMER+dbCAN | 0.88 | 0.86 | Yesa | Yes | Yes | Yes | 69 |
| DIAMOND+CAZy | 0.89 | 0.84 | Yesa | No | No | No | 4 |
| Hotpep+PPR | 0.80 | 0.94 | Yesb | Yes | No | No | 7 |
| Predicted by > = 2 tools | 0.93 | 0.92 | |||||
aTwenty four CAZyme families are classified into 207 subfamilies by phylogenetic clustering and CAZy expert curation (10).
cThe time is in seconds and calculated on Escherichia coli K-12 MG1655 proteome (4140 proteins). The detailed calculations on accuracy and speed are available in Supplementary Table S1. No correspondence has been established between PPR groups and CAZy subfamilies, and in dbCAN web server we only report CAZy subfamily annotation, whenever it is available.
More importantly, we found that the best performance of automated CAZyme annotation is to aggregate the outputs of the three tools and keep candidates found by at least two tools. Table 1 shows that the F-score can be increased to 0.93 when keeping proteins found by at least two tools.
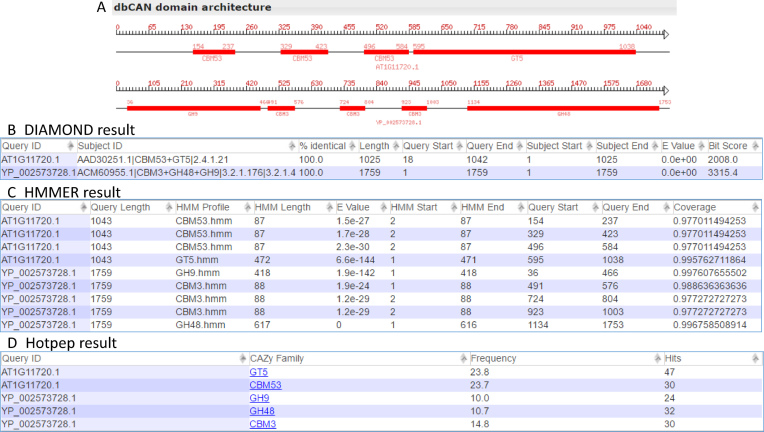
However, the above F-score calculation only considered whether a protein is found by any of the three tools. When considering if a protein is assigned to the correct family or families, we found that the F-scores for all the three tools had slightly dropped (Supplementary Table S2), with Hotpep + PPR dropped the most (dropped to 0.86 for eukaryotes and 0.70 for bacteria) and HMMER + dbCAN dropped the least (dropped to 0.85 for eukaryotes and 0.82 for bacteria). Additionally, proteins can have multiple CAZyme domains, and it is also interesting to know where the domain boundaries are. Figure 3 shows two example CAZyme proteins found by all the three tools. Both proteins have multiple CAZyme domains according to dbCAN annotation (Figure 3A). According to HMMER + dbCAN output (Figure 3C), AT1G11720.1 is annotated as CBM53(154–237) + CBM53(329–423) + CBM53(496–584) + GT5(595–1038) and YP_002573728.1 as GH9(36–466) +CBM3(491–576) + CBM3(724–804) + CBM3(923–1003) + GH48(1134–1753), i.e. all the CAZyme domains and domain repeats and their positions are reported (Table 1). However, according to both Hotpep + PPR and DIAMOND + CAZy, AT1G11720.1 is annotated as GT5 + CBM53 and YP_002573728.1 as GH9 + GH48 + CBM3, i.e. proteins are assigned to the multiple families correctly, though without reporting domain repeats and positions (Table 1).
Figure 3.
Comparison of annotation results for multi-domain CAZymes using three different tools. (A) Two example proteins (AT1G11720.1 and YP_002573728.1) are illustrated with their CAZyme domain architecture based on dbCAN search. (B) DIAMOND search result for the two proteins showing the best CAZy protein hit; (C) HMMER search result against dbCAN HMM database, from which (A) is derived; (D) Hotpep search result against PPR library; Frequency means the sum of conserved peptide frequencies and Hits means the number of conserved peptide hits (15).
It should be mentioned that DIAMOND + CAZy has a much higher risk than the other two tools to give wrong CAZyme family annotation. For example, if a query protein only has a GT5 domain and has AAD30251.1 as its best CAZy hit, transferring the family assignment of AAD30251.1 (GT5 + CBM53) to the query would be wrong (as no CBM53 in the query). However, such mistakes will not happen in HMMER and Hotpep searches, as they are conserved domain and motif-based methods.
CAZyme gene clusters (CGCs)
Another important new function of dbCAN2 is that it allows identification of CGCs, when the genomic locations of all genes of the query genome are given. In literature, CGCs are also known as polysaccharide utilization loci (PULs), which are defined as physically linked genes specializing in the degradation of various complex carbohydrates (24). Most experimentally characterized PULs are found in Bacteroidetes genomes (25), but have also been reported in Proteobacteria and Firmicutes of various carbohydrate-rich environments (26). The PULDB of CAZy initially focused on susCD (starch utilization system C and D transporters) associated PULs, and more recently expanded to present CAZyme clusters (3 and more CAZyme genes clustered in the genome) on its website (25). However, PULDB focuses on Bacteroidetes genomes and does not allow online genome submissions for PUL predictions. Recently, we defined CGCs as a more general term of PULs (3), which must contain three classes of signature genes: at least one CAZyme gene, one transporter (TC) gene, and one transcription factor (TF) gene. Between two adjacent signature genes, a certain number of non-signature genes can be inserted. We have developed a Python program (CGC-Finder) that can automatically identify CGCs (3).
In the dbCAN2 job submission page, we provide the ‘Find CAZyme gene clusters’ option. When users submit a protein query file, they must also provide a gene position file in order to predict CGCs. This gene position file is not required if users submit a nucleotide query file, because the gene prediction programs can generate the gene position file internally. With protein sequences, our server will predict TFs and TCs by DIAMOND search against TF and TC databases (explained in (3)), and then CGC-Finder will be called to locate genes of CAZymes, TFs, TCs in the genome, and identify CGCs.
Web design
For the job submission page, we have options to allow users to specify if they would: (i) use one of the three tools or all three tools for CAZyme annotation; (ii) use protein or nucleotide sequences as input; (iii) use CGC-Finder to predict CGCs. As shown in Figure 2, if nucleotide sequences are submitted, gene prediction programs will be first called to predict protein-coding genes and then protein sequences will be used for CAZyme annotation. If CGC-Finder option is selected, TFs and TCs will also be predicted and the gene location file will be used to predict CGCs.
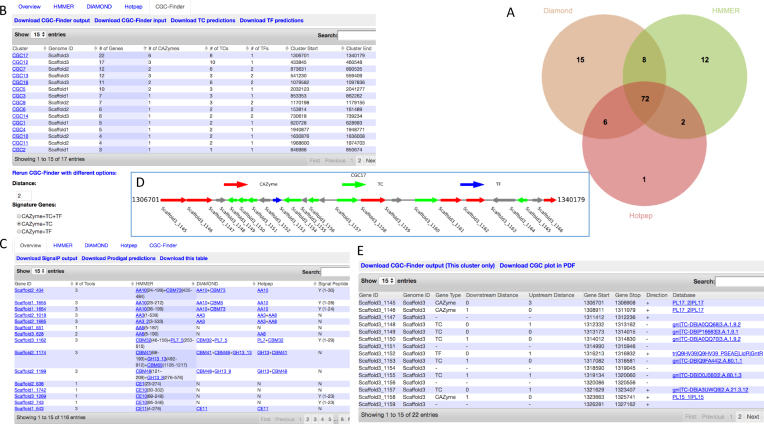
For the result page (Figure 4), five tabs are shown each with a data table: (i) HMMER result table; (ii) DIAMOND result table; (iii) Hotpep result table; (iv) Overview table; (v) CGC-Finder table. Above the tabs, a Venn diagram is shown to illustrate the overlaps among the outputs of the three tools (Figure 4A). Click on any numbers in the diagram will open a pop-out window displaying the protein IDs in that region.
Figure 4.
Screenshots of dbCAN2 result pages. (A) Venn diagram to show overlaps among the results of the three tools; (B) CGC-Finder result tab; (C) Overview tab combining results from the three tools and SignalP; (D) genomic location plot of an example CGC (signature genes are in red, green and blue colors, while non-signature genes are in gray); (E) detailed information of an example CGC.
The Overview tab combines the results of the three CAZyme annotation tools plus SignalP (27) prediction result (Figure 4C). The number of tools that find a CAZyme protein is also shown as a column, in addition to the CAZyme family assignment (for DIAMOND and Hotpep) and domain assignment (for HMMER). Users can sort the Table according to the number of tools column and easily filter out proteins found by only one tool to get the most accurate CAZyme list.
The CGC-Finder tab presents the CGCs identified in the query genome/proteome, with columns such as the genomic locations of the CGC and the three classes of signature genes in the CGCs (Figure 4B). The default parameters in running CGC-Finder include: (i) at least one CAZyme and one TC genes and (ii) the number of non-signature genes that are allowed to be inserted between two adjacent signature genes is ≤2. The two parameters can be changed underneath the CGC table to rerun CGC-Finder and then the CGC-Finder tab will be updated to display the new CGC list.
Clicking on each CGC opens a new page showing the CGC genomic context plot using GCPU (gene cluster plotting utility), a Python script we developed to plot the genes in the CGCs as arrows in different colors (Figure 4D). Below the plot is a Table (Figure 4E), which shows the detailed genomic location of each member gene in the CGC, including the distance of a signature gene from its upstream signature gene (Upstream distance) and the distance from its downstream signature gene (Downstream distance), as well as their best DIAMOND hits in the CAZy, TF and TC databases.
In all the five tabs and the individual CGC page, links to tab-delimited plain text files are provided for users to conveniently download and open in their local computers using Excel spreadsheet for further analysis. The Venn diagram and the CGC plot can also be downloadable as image files (e.g. SVG and PDF) and further edited by the users using Illustrator.
Lastly, we also provide a web page for each CAZyme protein to plot its dbCAN domains and PPR conserved peptides in the sequence. We also allow users to download a master script to run all tools as well as the CGC-Finder program on their local computers.
CONCLUSIONS
dbCAN2 is a web server for automated carbohydrate-active enzyme annotation. It is an updated version of the original dbCAN web server, and has the following new features:
dbCAN2 allows submission of nucleotide sequences: genomic sequences of prokaryotic draft genomes and metagenomes;
dbCAN2 integrates three state-of-the-art tools/databases for automated CAZyme annotation: (i) HMMER for annotated CAZyme domain boundaries determination according to the dbCAN CAZyme domain HMM database; (ii) DIAMOND for fast Blast hits in the CAZy database; (iii) Hotpep for short conserved motifs in the PPR library;
dbCAN2 can also identify transcription factors (TFs), transporters (TCs), and further CAZyme gene clusters (CGCs) using CGC-Finder if users submit protein sequences plus gene location files or genomic DNA sequence file;
dbCAN2 combines the results from the three tools and allows visualization of the overlaps as Venn diagram and the detailed results as graphs.
dbCAN2 meta server will be updated once a year to use the most updated CAZy database, dbCAN HMM database and Hotpep peptide database.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the Department of Computer Science of NIU for providing free access to the Linux computing cluster Gaea. We also thank our lab members for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (NSF) CAREER award [DBI-1652164]; National Institutes of Health (NIH) AREA award [1R15GM114706]; Research & Artistry Award of the NIU [2017-YIN to Y.Y.]; National Natural Science Foundation of China [31728013 to Y.Y. and H.Z.]. Funding for open access charge: NSF CAREER award [DBI-1652164].
Conflict of interest statement. None declared.
REFERENCES
- 1. Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B.. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009; 37:D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ekstrom A., Taujale R., McGinn N., Yin Y.. PlantCAZyme: a database for plant carbohydrate-active enzymes. Database. 2014; 2014:bau079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang L., Zhang H., Wu P., Entwistle E., Li X., Yohe T., Yi H., Yang Z., Yin Y.. dbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res. 2018; 46:D516–D521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yin Y., Mao X., Yang J., Chen X., Mao F., Xu Y.. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012; 40:W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cockburn D.W., Koropatkin N.M.. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J. Mol. Biol. 2016; 428:3230–3252. [DOI] [PubMed] [Google Scholar]
- 6. Flint H.J., Scott K.P., Duncan S.H., Louis P., Forano E.. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012; 3:289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. El Kaoutari A., Armougom F., Gordon J.I., Raoult D., Henrissat B.. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013; 11:497–504. [DOI] [PubMed] [Google Scholar]
- 8. Rogowski A., Briggs J.A., Mortimer J.C., Tryfona T., Terrapon N., Lowe E.C., Basle A., Morland C., Day A.M., Zheng H. et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat. Commun. 2015; 6:7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krumbeck J.A., Maldonado-Gomez M.X., Ramer-Tait A.E., Hutkins R.W.. Prebiotics and synbiotics: dietary strategies for improving gut health. Curr. Opin. Gastroenterol. 2016; 32:110–119. [DOI] [PubMed] [Google Scholar]
- 10. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B.. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014; 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Terrapon N., Lombard V., Drula E., Coutinho P.M., Henrissat B.. Aoki-Kinoshita KF. A Practical Guide to Using Glycomics Databases. 2017; Tokyo: Springer; 117–131. [Google Scholar]
- 12. Finn R.D., Clements J., Eddy S.R.. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011; 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park B.H., Karpinets T.V., Syed M.H., Leuze M.R., Uberbacher E.C.. CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology. 2010; 20:1574–1584. [DOI] [PubMed] [Google Scholar]
- 15. Busk P.K., Pilgaard B., Lezyk M.J., Meyer A.S., Lange L.. Homology to peptide pattern for annotation of carbohydrate-active enzymes and prediction of function. BMC Bioinformatics. 2017; 18:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Busk P.K., Lange L.. Function-based classification of carbohydrate-active enzymes by recognition of short, conserved peptide motifs. Appl. Environ. Microbiol. 2013; 79:3380–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kultima J.R., Coelho L.P., Forslund K., Huerta-Cepas J., Li S.S., Driessen M., Voigt A.Y., Zeller G., Sunagawa S., Bork P.. MOCAT2: a metagenomic assembly, annotation and profiling framework. Bioinformatics (Oxford, England). 2016; 32:2520–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuan C.S., Yew S.M., Chan C.L., Toh Y.F., Lee K.W., Cheong W.H., Yee W.Y., Hoh C.C., Yap S.J., Ng K.P.. DemaDb: an integrated dematiaceous fungal genomes database. Database. 2016; 2016:baw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mende D.R., Letunic I., Huerta-Cepas J., Li S.S., Forslund K., Sunagawa S., Bork P.. proGenomes: a resource for consistent functional and taxonomic annotations of prokaryotic genomes. Nucleic Acids Res. 2017; 45:D529–D534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones D.R., Thomas D., Alger N., Ghavidel A., Inglis G.D., Abbott D.W.. SACCHARIS: an automated pipeline to streamline discovery of carbohydrate active enzyme activities within polyspecific families and de novo sequence datasets. Biotechnol. Biofuels. 2018; 11:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J.. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010; 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rho M., Tang H., Ye Y.. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010; 38:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 24. Bjursell M.K., Martens E.C., Gordon J.I.. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J. Biol. Chem. 2006; 281:36269–36279. [DOI] [PubMed] [Google Scholar]
- 25. Terrapon N., Lombard V., Drula E., Lapebie P., Al-Masaudi S., Gilbert H.J., Henrissat B.. PULDB: the expanded database of Polysaccharide Utilization Loci. Nucleic Acids Res. 2018; 46:D677–D683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grondin J.M., Tamura K., Dejean G., Abbott D.W., Brumer H.. Polysaccharide utilization loci: fueling microbial communities. J.Bacteriol. 2017; 199:e00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petersen T.N., Brunak S., von Heijne G., Nielsen H.. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011; 8:785–786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.