Abstract
Epigenetic modifications, of which DNA methylation is the most stable, are a mechanism conveying environmental information to subsequent generations via parental germ lines. The paternal contribution to adaptive processes in the offspring might be crucial, but has been widely neglected in comparison to the maternal one. To address the paternal impact on the offspring’s adaptability to changes in diet composition, we investigated if low protein diet (LPD) in F0 males caused epigenetic alterations in their subsequently sired sons. We therefore fed F0 male Wild guinea pigs with a diet lowered in protein content (LPD) and investigated DNA methylation in sons sired before and after their father’s LPD treatment in both, liver and testis tissues. Our results point to a ‘heritable epigenetic response’ of the sons to the fathers’ dietary change. Because we detected methylation changes also in the testis tissue, they are likely to be transmitted to the F2 generation. Gene-network analyses of differentially methylated genes in liver identified main metabolic pathways indicating a metabolic reprogramming (‘metabolic shift’). Epigenetic mechanisms, allowing an immediate and inherited adaptation may thus be important for the survival of species in the context of a persistently changing environment, such as climate change.
Keywords: DNA methylation, exposure, wild mammal species, inheritance, plasticity, adaptation
Introduction
For (wild) species, environmental conditions are the pressures that define the ecological niche selecting for phenotypes with adaptive traits beneficial to survival and reproduction in that niche. Under different environmental conditions, a given genotype can give rise to different phenotypes [1, 2]. We still understand little of the underlying mechanisms providing the genetic plasticity for this phenotypic diversity. Insights into these mechanisms are crucial for understanding the diversity of species as well as their ability to adapt to external changes. Known mechanisms realizing genetic plasticity are epigenetic modifications [3], which alter in response to intrinsic (e.g. stress reaction, disease) and extrinsic (e.g. environmental factors, toxins, temperature) changes without altering the DNA sequence and regulate gene expression [4, 5]. Of the known epigenetic mechanisms such as histone modifications, non-coding RNAs and DNA methylation, the latter is the most stable one and can be transmitted to subsequent generations. Imprinted genes for example mediate paternal or maternal allelic transmission of specific DNA methylation patterns which is accompanied with a parent-of-origin gene expression [6].
In mammals, DNA methylation mostly occurs at cytosines in cytosine-phosphate-guanine dinucleotides context (CpG) [5]. CpG methylation at promoters [5] and enhancer regions [7] of genes silences gene transcription, whereas demethylation leads to transcriptional activation [5, 8]. Intragenic methylation can have both functions—acitivation and repression [9, 10]. Some DNA methylation patterns are set early in life and most of them remain stable throughout an individual’s lifespan and across generations (epigenetic stability), others are highly plastic and change in response to a changing environment (epigenetic plasticity) [11].
Epigenetic alterations in response to changing environmental conditions have mainly been studied in isogenic lab strains, having the advantage of a large number of individuals providing statistical robustness and an insight into the mechanism, but lacking the functional understanding of these responses to ecological cues (e.g. climate change) in naturally occurring, genetically heterogeneous species [12, 13]. Climate changes are changes in weather patterns such as temperature and rainfall, subsequently rearranging ecosystems by shifting the composition of floral communities, followed by changes in the composition of faunal communities. Surprisingly, epigenetic studies on wild species exposed to ecologically relevant impacts such as changes in ambient temperature and/or food quality are still rare (in plants [14], honey bees [15], fish [16], in baboons [17] and in guinea pig [18]), but mandatory to understand adaptation processes on a molecular level [12].
Medical reasons initiated studies on the epigenetic response to diet changes linking methylation changes to diabetes, obesity and other metabolic disorders [19–23]. However, such epigenetic modifications have mainly been studied with respect to maternal effects due to the close physical relationship of mother and offspring during and after pregnancy, and the reprogramming process in the blastocyst. In Agouti mice, e.g. fur colour and susceptibility to diabetes changed in offspring after feeding pregnant mice a diet rich in ascorbic and folic acid, which went along with a change in DNA methylation of the respective Agouti gene, and a change of its expression [20, 22]. At the end of World War II the Dutch suffered of a famine during winter (‘Dutch Hunger Winter’). Women exposed to the famine at an early stage of pregnancy gave birth to children with a stable, genome-wide change in methylation in genes important for growth and immunity (e.g. IGF2, INSIGF and IL10) [21]. Those children developed an increased rate of obesity altered lipid profiles, cardiovascular disease and schizophrenia [24, 25].
More recently the paternal impact on DNA methylation patterns in the offspring has gained more attention, in response to cocaine consumption by fathers [26], traumata experience by fathers [27], and in response to changes in the diet of fathers [23, 28, 29].
In wild mammal species, males predominantly disperse from their natal site, while females are often philopatric. Therefore males (the paternal line) are more frequently exposed to changing environments and need to react quicker to rapid changes than philopatric females.
This also hold true in Wild guinea pigs, Cavia aperea. They are living in harem structures with one dominant male, who mate-guards several females from other males [30]. The subordinate males disperse, and therefore need to adapt quite rapidly to novel habitats and available food sources in search of accepting female(s). Therefore, from an evolutionary point of view, we expect a paternal transmission of epigenetic responses to the offspring as it would contribute to a fitness increase of the offspring. Such fitness increase would be even more pronounced if the paternal response was transmitted over more than one generation.
As a small, mobile herbivore, the Wild guinea pig has high energy demands but only a small digestive tract. It compensates for its small gut capacity by (i) selection of high quality food, (ii) increased food intake, (iii) coprophagy and hindgut (caecum) fermentation [31–33] and (iv) an increased efficiency of digestion via symbiotic microorganisms fermenting even fibrous components [34–36].
Although dietary proteins are mandatory providers of nitrogen and essential amino acids for the animal’s protein synthesis, needed in many vital functions (e.g. cell membrane components, enzymes, hormones, carriers in active transport systems etc. [37]), it is a scarce component in their vegetarian food. Severe nutritive protein deficiency causes muscle and organ degradation associated with body mass reduction [38]. Moderate protein deficiencies will lead to compensatory food intake, lower growth rates [39, 40] and less reproductive success [41]. The compensatory increased food intake—known as ‘protein leverage effect’—is usually associated with an elevated metabolic rate—which attenuates ‘Luxus consumption’, meaning that the surplus of energy is released by heat production [42, 43].
Due to vegetation changes throughout seasons, herbivores regularly face varying and usually limited protein content in their food sources. Those effects may become more severe with global warming, as temperatures and reduced rainfall will cause long-term shifts in vegetation.
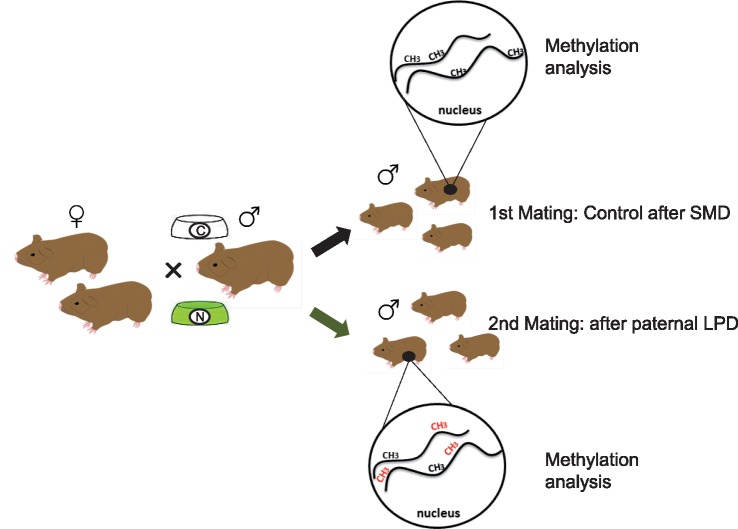
To test for a paternal contribution to an epigenetically adaptive response to diet changes, we temporally (2 months) exposed adult male Wild guinea pigs to a low protein diet (LPD) and allowed them to mate with the same females and sire offspring before and after LPD exposure. We then analysed and compared the two groups of 7-days old F1 sons—the ‘control group’ sired before (F1C, C = Control), and the ‘diet group’ sired after their fathers’ LPD treatment (F1D, D = diet)—for altered DNA methylation patterns in response to their father’s LPD (Fig. 1). We focussed on the liver (L), as the body’s main metabolic organ (F1LCvs F1LD), to assess the transmission of the father’s epigenetic responses to the F1 generation. To investigate a potential transmission to the F2 generation, we also studied the DNA methylation patterns of the testes of the two F1 son groups (F1TCvs F1TD).
Figure 1:
Experimental set-up. Male Wild guinea pigs (F0) were mated to the same two female Wild guinea pigs before and after a diet change to LPD (C, ‘control group’ fed SMD, and D, ‘diet group’, fed a LPD). Potential changes in methylation of nuclear DNA depict by red CH3-groups. Methylation patterns were analysed from DNA of whole livers (L) and testes (T) from sons sired in the first mating (‘control group’, F1LC, F1TC) and in the second mating (‘diet group’, F1LD, F1TD)
About 70% of mammalian promoter regions are associated with ‘CpG islands’ (CGIs), genomic regions rich in CG content and main regulatory sites for gene silencing [10]. Therefore, we focussed our search for methylation changes on CGIs and other elements with regulatory functions, such as coding regions, promoters.
Materials and Methods
Animal Housing and Treatment
All husbandry and experimental procedures were approved of by the German Committee of Animal Welfare in Research (permit no. V3-2347-35-2011). Wild guinea pigs (C. aperea) originating from Argentina and Uruguay [30] were obtained from F. Trillmich (University of Bielefeld) and housed at the IZW field station in Niederfinow, Germany. The animals were kept in combined indoor-outdoor enclosures with free access to both compartments at all times. Indoor enclosures measured 0.75 × 0.75 × 1 m and the outdoor enclosures 0.75 × 1 × 1 m (length × width × height). Animals were housed under natural photoperiod and temperature. To avoid male competition, males were held as singles and were always separated by females kept in separate cages between them in such a way that social interaction but no direct contact was possible between male and female cavies. Although kept solitary, the opportunity to freely move between short-tunnel-connected inside and outside enclosures optimized their welfare (Schumann et al. 2014). Both cages were filled with a bedding of wood shavings and large opaque tubes for cover.
Diets
To assess the paternal effect of a LPD on male offspring, five male Wild guinea pigs (F0 fathers, n = 5, labelled A–E) were fed a LPD with 42% less protein content (Supplementary Table S1), instead of standard maintenance diet (SMD). Pellets for both diets (Altromin Spezialfutter GmbH & Co. KG) were almost iso-energetic [dry matter (DM) energy], but differed in their protein to energy ratio and metabolizable energy, as protein content was partially made up with sucrose (Supplementary Table S1). Note that although the relevant dietary change in this experiment could be protein content, sucrose content, fat/protein ratio etc., for simplicity we refer to the diet as low protein henceforth.
LPD was fed for the duration of 62 days (April–June), which is the length of a full cycle of spermatogenesis in guinea pigs [44, 45]. The experimental set-up is illustrated in Fig. 1. During SMD, animals received SMD pellets and hay ad libitum and a supplement of 50 g (each) of minced apple, carrot and cucumber per individual (listed in Table 1). During the LPD period, animals received exactly 100 g of LPD pellets and exactly 100 g of fresh hay per day and individual. At each morning during both diets, the cages were cleaned to avoid consumption of left-over hay or faeces from the previous day.
Table 2:
Number of F1 sons per group, birth weights and times of birth
| Group of sons | n | Mean body mass [SD] | Born in |
|---|---|---|---|
| F1C | 15 | 65.44 ± 8.61 | February |
| F1D | 17 | 70.67 ± 12.13 | August |
Analysis of Faecal Samples and Food Components
During LPD, food intake and digestibility was measured every 9–11 days (8–11 a.m.). The animals were weighted and the daily food consumption was determined by collecting all faecal samples and leftovers 24 h (±30 min) after food supply. At measuring days, cages were cleaned and faeces and leftovers were collected. Leftovers were divided into the following components: dietary pellets and hay (in SMD and LPD), and apples, cucumbers and carrots (in SMD). Wet mass (±0.01g) was determined using a scale (Sartorius BP 1200). For the analysis of energy and macronutrients (protein, fiber), faeces and samples of all food components were freeze-dried (Lyovac GT2, GEA Lyophil GmbH, Hürth, Germany), weighted and ground (mill A11, IKA-Werke GmbH, Staufe, Germany). The dry mass of faeces was corrected for acid insoluble ash [46]. Ash content was determined by drying a subsample for 5 h at 525°C (Muffle kiln, Heraeus M110, Hanau, Germany). Energy content was determined using bomb calorimetry (IKA Kalorimetersystem C5000 control, IKA-Werke GmbH, Staufen, Germany), Protein content by using Dumas combustion (Elementar rapid N III; Elementar Analysesysteme GmbH, Hanau Germany). All analyses were performed in duplicates per food item and faecal sample.
Nutrition Data Calculations and Statistics
For nutrient content calculations, we used only DM corrected values. DM intake was calculated from the mass difference between dry mass of food offered and refused (leftovers). For net energy intake (NEI) and net protein intake (NPI), the energy and protein content of defaecated matter were subtracted from the respective content in ingested matter. Statistical analyses were performed within R (http://www.R-project.org). Consecutive values during treatment as well as values for maintenance diet were compared using a Wilcoxon rank sum test [47].
Parental Mating
Each F0 male (n = 5) was mated twice with the same two F0 females (n = 10), once before the LPD treatment (male ‘control group’; F0C), and once after (male ‘diet group’; F0D) (Fig. 2A). The five F0 males (A–E) were born in mid-November and the ten F0 females in April–May the year after. In order to achieve mating, males were introduced to the female’s cage, and after an observed mating, males were transferred back to their own cages. The ‘control’ mating took place from late December to early January and the mating after LPD treatment 6 months later in June. Because the males mated with the same two females before and after LPD exposure, the offspring produced (after gestation time of ∼62 days) per female were direct siblings and half-siblings.
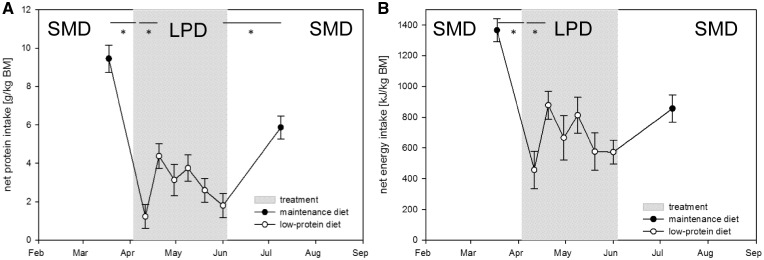
Figure 2:
Net intake before and after the LPD (white circles) are shown as mean values (with SEM) for five F0 male Wild guinea pigs at eight sampling time points, one before and after the LPD treatment (feeding SMD) and six during LPD from April to June (period shadowed in grey). Monitoring of (A) NPI in gram per kilogram of body mass, and (B) NEI in kilojoule per kilogram of body mass resulted in decreased protein and energy uptake with the start of LPD, which quickly re-adjusted with the reintroduction of SMD pellets. Significant differences among consecutive values are marked with stars (*P < 0.05, Wilcoxon sum rank test)
Tissue Sampling
F1C sons were born in February and F1D sons were born in August (Table 2). Seven days after their birth, we harvested whole livers (L) and testes (T) of all F1 sons from both groups (F1LC and F1TC D = 15; F1LD and F1TDn = 17). Livers and testes were homogenized, snapped frozen in liquid N2 and stored at −80°C until DNA isolation (see below).
Table 1:
Daily intake of Dry Matter, protein and ingested energy of F0 males
| Intake | Dry Matter [g/day/kg BW] | Crude protein [g/day/kg BW] | Energy [J/day/kg BW] | |||
|---|---|---|---|---|---|---|
| Diet | SMD | LPD | SMD | LPD | SMD | LPD |
| Pellets | 36.4 ± 15.8 | 31.5 ± 8.1 | 5.2 ± 2.9 | 2.1 ± 0.5 | 559.56 ± 313.7 | 553.1 ± 142.1 |
| Hay | 31.5 ± 15.6 | 30.5 ±13.2 | 3.7 ± 2.1 | 3.5 ± 1.6 | 525.7 ± 318.2 | 551.2 ± 242.6 |
| Carrot | 6.6 ± 2.9 | 0.4 ± 0.2 | 115.2 ± 52.7 | |||
| Apple | 8.1 ± 3.7 | 0.2 ± 0.1 | 146.2 ± 64.3 | |||
| Cucumber | 2.1 ± 1.2 | 0.5 ± 0.3 | 36.0 ± 21.1 | |||
| Sum | 79.2 ± 35.0 | 62.0 ±13.3* | 9.9 ± 4.3 | 5.6 ± 1.5*** | 1382.7 ± 606.5 | 1104.4 ± 242.9** |
| F0 body mass [g] | 659.9 ± 47.8 | 698.9 ± 66.3 | ||||
The table lists mean values of ingested food normalized per kg body mass during SMD and LPD period, and significant differences between both diets (*P < 0.05, **P < 0.001, ***P < 0.0001, unpaired T test).
Reduced Representation Bisulphite Sequencing
We performed reduced representation bisulphite sequencing (RRBS) [48] to profile DNA methylation changes among sons sired before and after the diet change of their fathers (Supplementary Table S2: F1LC [F1LC-A to F1LC-E, the last letter refers to the father’s ID] and F1LD [F1LD-A to F1LD-E] and Supplementary Table S3: F1TC [F1TC-A to F1TC-E] and F1TD [F1TD-A to F1TD-E]. The F1 liver and testis samples, respectively, were pooled by father (‘son groups’) before being sequenced [F1LC (pool of n = 5 sample pools) vs F1LD (n = 5) and F1TC (n = 5) vs F1TD (n = 5)]. RRBS was performed and data was analysed as previously described in [49]. Reads obtained from a HiSeq2000 (Illumina) were mapped against an in-house-generated C. aperea reference sequence (http://www.ncbi.nlm.nih.gov/biosample/2252454; Acc. No. AVPZ00000001-AVPZ00003131) using BismarkMapper [50, 51].
Differentially Methylated Regions
Differentially methylated regions (DMRs) were calculated as described previously [49] using the software MethPipe [52]. DMRs were assessed in pairwise comparisons of ‘control’ and ‘diet’ samples for F1 sons grouped according to their father (A–E). We selected for DMRs located in CGIs, promoter regions and CDS, which occurred in ‘son groups’ pooled by father: F1LCvs F1LD and F1TCvs F1TD (henceforth called ‘annotated DMRs’). Results were visualized using R (with functions customized by T. Girke; http://faculty.ucr.edu/∼tgirke/Documents/R_BioCond/My_R_Scripts/overLapper.R). The gene-network analysis of hypomethylated and hypermethylated genes was performed using the web-based String database, (https://string-db.org/) [53, 54].
qPCR and Relative Quantification
Gene expression of two genes with key function in physiological pathways identified by the String network (Adcy9, Stat3) was measured by qPCR. Expression values were normalized by relative quantification of expression values using the three most stable expressed reference genes (Hmbs, Gapdh and B2m) as described previously (Weyrich et al. 2016b). RIN values were between 6.0 and 9.2. The run efficiency for Adcy9 was 2.03 and for Stat3 1.84. RNA of one sample of the control group was of low quality and thus excluded from statistical analysis. Amplicons were Sanger sequenced and verified by similarity control using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Statistical analyses were performed on F1LC= 17 and F1LD= 16 with R (http://www.R-project.org) using a Wilcoxon rank sum test and an unpaired Ttest.
Results
Physiological Effects of the LPD on Fathers
Surprisingly, with the start of the LPD, the male guinea pigs changed their feeding behaviour. They consumed 13.5% less pellets and 59.6% less protein (Table 1). This was also illustrated by their net protein (Fig. 2A) and NEI (Fig. 2B), which dropped significantly with the start of LPD. However, male Wild guinea pigs seemed to have compensated their energy needs, because male F0 body mass did not change significantly (Table 1).
Total DNA Methylation Changes in Sons
The overall single cytosine methylation level did not significantly vary between the compared control and treatment groups, neither for F1LCvs F1LD, nor for F1TCvs F1TD (schemata of experimental set-up, Fig. 1). On average, ∼10% of all cytosines were methylated in liver, out of which ∼70% were located in CpGs. In F1 (immature) testis of both son groups ∼9% of all cytosines occurred in a methylated stage, out of which ∼66% were in a CpG context. Only 1% was in CHGs (H can be C, A or T) and 1% a CHHs context, as expected from our former studies [18, 50].
Epigenetic Inheritance
By comparing liver samples of sons sired before (F1LC) and after their fathers’ LPD (F1LD), we identified 2796 DMRs of which 946 were located in annotated regions [promoters, coding sequences (CDSs), CGIs], hereafter referred to as ‘annotated DMRs’ (Fig. 3). These results indicated a ‘heritable epigenetic response’ (or ‘epigenetic inheritance’) to the LPD of the sires. With 4572 DMRs identified in the comparison of methylation patterns in testis (1420 of which were in annotated regions; Fig. 3), the F1TCvs F1TD comparison yielded a much larger number of DMRs than detected in the liver (F1LCvs F1LD), which reveals from a greater coverage of CpG positions of testis data in comparison to liver.
Figure 3:
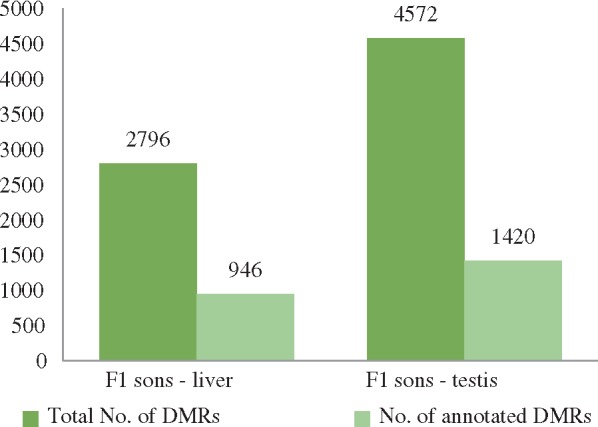
Tissue-specific differential methylated regions (DMRs) after paternal LPD. Bars depict the number of ‘total’ (black) and ‘annotated DMRs’ (grey) in F1Cvs F1D sons grouped by organ
We verified the robustness of our results by performing a randomization approach, shuffling the methylation ratios of F1LCvs F1LD and F1TCvs F1TD, respectively, 100 times per father (total 500 times). For F1LCvs F1LD the approach resulted in only one DMR in 24 calculations and zero in the remaining 476 random calculations. No DMR was detected when shuffling F1TCvs F1TD. The observed DNA methylation changes in F1 sons clearly indicated an effect on the male offspring by the paternal epigenetic response to the diet change.
General Response
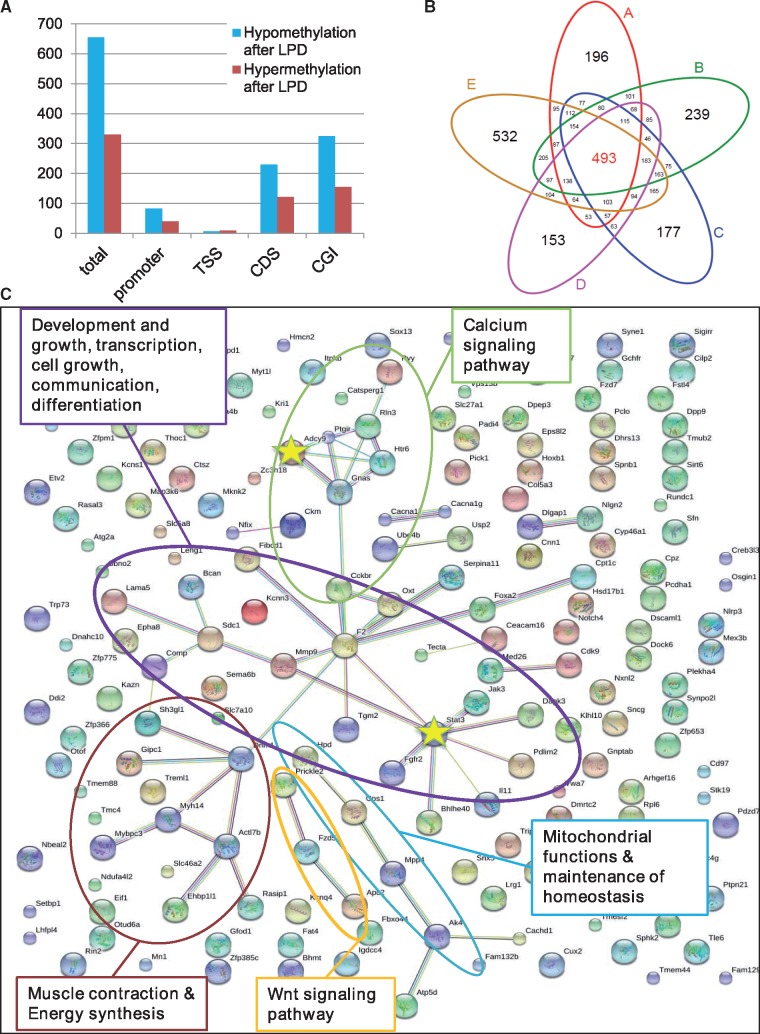
The LPD significantly affected the feeding behaviour of the adult male Wild guinea pig and thus their physiology (Table 1, Fig. 2). To evaluate if these physiological changes were reflected on the molecular level as a shift in the regulation of metabolic gene pathways, we further focussed on the liver as the main metabolic organ maintaining and regaining the body’s homeostasis. In annotated regions in liver cells, we detected twice as many hypomethylated (n = 655) than hypermethylated DMRs (n = 330). Those annotated DMRs were overlapping with one or several annotated genomic regions, incl. promoters, transcription start sites (TSSs), CDSs and CGI (Fig. 4A). Of those hyper- and hypomethylated annotated DMRs, 493 were shared among the five son groups pooled by father (Fig. 4B). As these genomic regions are modified and thus react among the five son groups we interpret them as ‘general response’ to the environmental alteration (diet). Of the 493 annotated DMRs, 152 were overlapping with CDSs, 56 with promoter regions, 2 with TSSs and 283 with CGIs.
Figure 4:
Metabolic shift after LPD. (A) Number of hypomethylated (blue) and hypermethylated (red) annotated DMRs identified in the comparison of F1LCvs F1LD both in total and overlapping with specific annotated regions: promoter, TSS, transcription start side; CDS, coding sequence; CGI, CpG Islands. (Note that some DMRs overlapped with more than one annotated region leading to more than one annotation.) (B) Venn diagram showing the number of shared and explicitly annotated DMRs per pairwise comparison of F1 groups of sons (F1LCvs F1LD) of the same father (A–E) in liver after paternal LPD. Son groups (grouped by father) are colour-coded, overlapping areas indicate numbers of shared annotated DMRs among the respective groups in liver. (C)String gene network of genes from DMRs detected in livers of all five son groups (reference: M. musculus). Gene-network analysis identified genes (dots) important in main metabolic pathways (colours of dots are chosen by the string database and are of no account for certain gene function). Genes marked with stars have been investigated for expression changes (see Fig. 5)
To evaluate which genes were affected by a change in composition of diet and to elucidate the physiological gene-pathways they were part of, we performed a gene-network analysis of the genes detected to have been epigenetically influenced in all F1C/F1N pairs (Fig. 4C). We therefore combined hypomethylated and hypermethylated DMRs with at least one annotated gene (total 210) and submitted them to the String database for analysis. Out of the 210 genes, 185 were recognized by the String database using gene function data of Mus musculus as a reference. The network analysis resulted in significantly more connections than expected by chance [nobs = 106, nexp = 68, P = 0.000367, increasing the stringency of the interaction score to 0.5 (vs the default setting of 0.4)]. The network identified genes in pathways with main metabolic functions such as development and growth, transcription, cell growth, communication, differentiation, muscle contraction and energy synthesis, Wnt-signalling pathway, Calcium signalling pathway, mitochondrial functions and maintenance of homeostasis.
Gene Expression
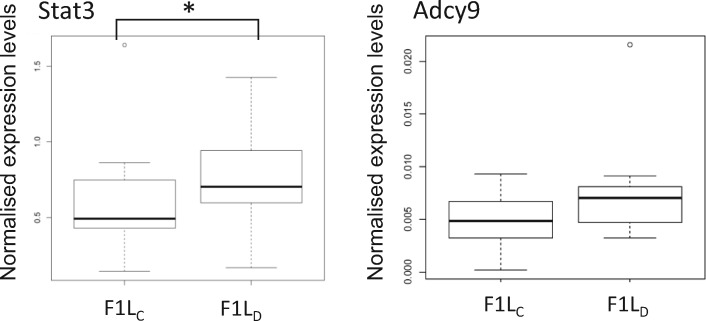
We tested two key genes (Stat3 and Adcy9, marked with stars in Fig. 4C and Supplementary Fig. S1) of the two major physiological pathways (development and growth, transcription, cell growth, communication, differentiation (highlighted in violet) and Calcium signalling pathway (highlighted in green)) for potential changes in gene expression by qPCR (Fig. 5). The Signal transducer and activator of transcription 3 (Stat3) expression was changed according to its methylation changes; Stat3 was hypermethylated in its CDS (in F1N sons) and accordingly significantly changed its expression in liver samples of 15 F1Cvs 17 F1N (fold-change = 1.2, P = 0.05). Adenylate cyclase (Adcy9) was hypomethylated in its CDS in sons. With a P = 0.058 expression change of Adcy9 was slightly above the significance threshold of 5%.
Figure 5:
Box plot of total gene expression changes in Stat3 (left) and Adcy9 (right). Expression of Stat3 but not of Adcy9 was significantly changed in F1 sons after LPD of their fathers [P = 0.058, signed Wilcoxon rank test (unpaired)]. Stat3 was 1.2-fold up-regulated after the diet. C, control [F1LC]; N, sons after LPD of fathers [F1LD]. Vertical lines in boxes depict median. Dots, outliers
Treatment-Dependent Response
Stat3 showed a response dependent to specific environmental change, because its expression decreased after a change in diet (Fig. 5, left), but increased after a change in temperature [55] (Fig. 4).
Due to a change in season between the first and the second mating, ambient temperature change may have acted as a cofounding environmental effect (Table 1 and Supplementary Table S4). To further distinguish nutritional from temperature effects, we compared the ‘annotated DMRs’ obtained here to previously published results having a very similar experimental set-up. During the experiments we exposed five fathers of the same age to an increased temperature of 30°C (10°C more than ambient temperature) for also 2 months [generating F1 sons prior (control: F1LC) and after heat (H) exposure of fathers (F1LH); Supplementary Table S4] [18].
We identified overlapping annotated DMRs in livers of F1LD sons (this experiment) and F1LH sons (previous experiment; Weyrich et al. 2016b). Annotated DMRs were only partially overlapping in response to the two environmental effects (Table 3). Gene-network analysis of genes with methylation changes occurring only after paternal LPD resulted in the identification of same metabolic pathways (compare Supplementary Fig. S1 to Fig. 4), despite the removal of genes that overlapped in F1LD (this study) and F1LH (the ‘heat experiment’).
Table 3:
Annotated DMRs from all five son groups (grouped by father), sorted according to genomic region and experiment
| DMRs from ‘Diet’ experiment | DMRs overlapping between ‘Diet’ and ‘Heat’ experiments | |
|---|---|---|
| CGI | 202 | 39 |
| CDS | 106 | 18 |
| Promoter region | 45 | 3 |
| TSS | 2 | 0 |
Discussion
Paternal nutrition alterations lead to treatment-specific changes in DNA methylation patterns, which were partially translated into changes in gene expression in the Wild guinea pig male offspring. This is the first study detecting a ‘heritable epigenetic response’ (or ‘epigenetic inheritance’) to diet changes in a wild, genetically heterogeneous mammal species. Epigenetic mechanisms, such as DNA methylation might therefore enable genetic plasticity in response to intrinsic and extrinsic factors, to better prepare the offspring for potential environmental changes and as such closes the gap between quick (and short-lasting) physiological responses and reaction shifts via very long-lasting mutational changes.
Diet-Related Paternal Epigenetic Effects
So far only a few studies investigated paternal epigenetic effects of DNA methylation changes studying highly inbred lab animals [23, 26, 28, 29, 56, 57]. For example, a high fat diet of male rats leads to changes in expression in the pancreatic ß-cells in their female offspring and hypomethylation of the Il13ra2 genes [28]. Although we did not detect hypomethylation of Il13ra2, similar pathways to ours were changed in their gene expression incl. Calcium-, Wnt-signalling pathways, apoptosis and the cell cycle. Pathway similarity between both studies point to a conserved evolutionary response to nutrition change.
In another study, male mice were fed a LPD from weaning up to 12 week of age. Their offspring had numerous DNA methylation changes in their livers. Among those loci, an enhancer of the lipid regulatory protein PPARa was increased in expression [23]. As this study’s experimental set-up is similar to ours, we aimed to compare those results to ours, by investigating potential alterations of PPARa expression in F1 male Wild guinea pigs, but neither detected significant expression changes (data not shown) nor alterations in methylation.
In wild mammals, so far only one study investigated ‘resource base’ DNA methylation patterns in baboons (Lea et al. 2015) [58]. The authors compared the genome-wide methylation in both lodge and wild-feeding baboons, and also recorded physiological parameters (serum insulin, cholestrol and body fat). They detected feeding resource dependent genome-wide differential methylation patterns mainly in promoter and enhancer regions close to metabolism-related genes [58]. Our study is the first investigating inherited epigenetic effects of nutrition changes and the second study investigating DNA methylation changes to diet changes in a wild mammal.
Paternal Behavioural Change Due to Low Protein
We expected male Wild guinea pigs to ingest more pellets during the LPD than during the SMD to satisfy their daily supply of protein. This, however, was not the case. Instead they consumed less pellets (Table 1). With the ingestion of caecal faeces, caecotrophy guinea pigs can increase their protein uptake, due to ingestion of bacterial proteins [59, 60]. By this they may have partially compensated for the lower protein levels in the LPD to perform amino acid synthesis. In addition, an alteration in regulation (type and amount) of protein-transporting channels of the gastro intestinal tract is essential for homeostasis [61] and might have enhanced protein uptake, allowing the male Wild guinea pigs to supplement their total protein intake. The latter is supported by the detected epigenetic change of the Calcium signalling and maintenance of homeostasis pathways in their male offspring (Fig. 4C and Supplementary Fig. S1).
Effects of Paternal Diet on DNA Methylation in Sons
Mounting evidence supports the hypothesis that diet influences DNA methylation patterns via three mechanisms: (i) the provision of substrates necessary for DNA methylation, and/or (ii) the provision of cofactors modulating the enzymatic activity of DNA methyltransferases, and/or (iii) a changing activity of the enzymes regulating the one-carbon cycle [62]. The universal methyl-donor for DNA and protein methyltransferases [63], S-adenosylmethionine (SAM) is synthesized in the methionine cycle from precursors present in the diet [64, 65]. All these precursors, including methionine, folate, choline, betaine and vitamins B2, B6 and B12, enter at different sites in the methionine pathway and contribute to the net synthesis of SAM.
We are uncertain whether the supply of methyl-donors is reduced in our studies, but as it is commonly accepted that undernutrition correlates with reduced methyl-donor availability (Zhang 2015), we assume that the protein reduction may have reduced the supply of methyl-donors such as Vitamin B and SAM, leading to an over-all hypomethylation (as shown in Fig. 4A) indicating a genome-wide activation.
Among all five son groups we detected changes in functional regions of the same genes, indicating a similar functional response to the paternal diet change despite the animals’ genetic heterogeneity. We call this a ‘general response’.
Metabolic Shift in Livers of Sons after LPD of Fathers
The gene-network analysis of ‘general response’ genes identified several main metabolic pathways in F1LCvs F1LD sons whose genes had been impacted in their methylation patterns by the diet change of their fathers. These genes were relevant in development and growth, transcription, cell growth, communication, differentiation, muscle contraction and energy synthesis, the canonical Wnt-signalling pathway, the Calcium signalling pathway, mitochondrial functions and the maintenance of homeostasis. The involvement of these pathways implies a general metabolic shift, which may change in the organisms’ physiology in response to a longer-lasting environmental change. An environmental stimulus is sensed and initially processed by specific brain areal, releasing neurotransmitters and hormones. For example the Wnt-signalling pathway is important for reacting to outer signals by its Wnt (Wingless) and Integrator Complex Subunit 2 (Int-2) signal proteins [66]. These signals convey the information to the organs triggering metabolic processes, which stabilize or regain homeostasis [67]. The liver is the main metabolic organ, which also stores energy in form of glycogen, whose hydrolysis to glucose subsequently generates the energy required for a systemic response. The metabolic pathways detected here to be involved in the epigenetic response are part of this hepatic response. As this response was detected in F1LCvs F1LD sons, we hypothesize that the DNA-methylation patterns transmitted by the father to the sons pre-set a regulatory programme for the offspring to increase fitness by enabling to cope more rapidly with an environmental condition similar to the one the fathers had experienced (here LPD). If this holds true, the epigenetic response may enable the metabolic shift on the genetic level and thus a long-term memory set within a life-span and transmittable to the next generation(s). Further studies will be needed addressing this in the Wild guinea pig.
Epigenetic Response Depends on the Type of Environmental Change
Although Wild guinea pigs breed throughout the year, their body growth rate is somewhat greater in spring than in autumn, indicating seasonal dependency [68]. In our study control sons (F1LC) were born in spring and sons sired after paternal diet change (F1LD, Table 2) and temperature change (F1LH, Supplementary Table S4) were born in autumn. To identify potential seasonal changes in body weight we compared birth weights of sons born in February to that of sons born in August (F1Cvs F1D/H; Table 3 and Supplementary Table S4), but did not find significant weight differences at that early age (7 days). In addition, as both experiments (‘diet change of fathers’ and ‘heat exposure of fathers’) were run in the same season, ‘season-based DMRs’ would be identical in both experiments, resulting in overlapping DMRs (in the respective sons) between the two experiments. Assuming that ‘season’ had a great impact on the experiments, we would expect a large proportion of DMRs to overlap between the two experiments due to ‘different season’. Even though the total number of overlapping DMRs may consist of DMRs both from (i) the different ambient temperatures the fathers were exposed to before and after their treatment (either LPD or heat) and (ii) from a general response to the changed season, the total number of overlapping DMRs between the two experiments was low (Table 1). In combination with the finding that the birth weights of sons in the two seasons did not significantly differ, we reject the hypothesis that the different seasons the sons had been born in had a significant impact on the epigenetic response measured.
Gene expression analysis of Stat3 supported the treatment-dependent response, because we detected a reverse pattern of response for diet and heat. Although in sons sired after F0 heat exposure Stat3 was hypermethylated and expression repressed compared with control sons (F1LCvs F1LH) [55], in sons sired after LPD of fathers, Stat3 was hypermethylated and enhanced in expression compared with control sons (F1LCvs F1LD).
DNA Methylation Regulation of Gene Expression
As visualized in the STRING network (Fig. 4C) Stat3 and Adcy9 are interacting with several other proteins, indicating their key function in those metabolic pathways. The Stat3 is a transcription factor which is activated by interleukins and growth factors. STAT3 mediates the expression of a variety of genes in response to cell stimuli and is essential for embryogenesis, cell survival and proliferation [69], the regulation of body weight [70] and many cellular processes [71] (GO terms listed in Supplementary Table S5).
Adenylyl cyclase 9 (Adcy9) is required for cellular signal transduction and has been associated with psychological diseases incl. bipolar disorder and depression [72]. Adcy9 gene encodes for an enzyme that catalyses Cyclic adenosine monophosphate (cAMP) to Adenosine triphosphate (ATP) and as such is crucial for cellular energy production, and as such necessary to cope with changing outer stressors (Supplementary Table S5) [73].
Even though Stat3 was changed in methylation in its CDS and not in its promoter regions, gene expression was detected to have changed. Although promoter methylation is strongly associated with gene repression and silencing [5, 8] intragenic methylation can have both activating and repressive effects [74]. The methylation changes in the Adcy9 did not (yet) lead to changes in expression. Several hypotheses can be postulated to explain these findings: (i) the degree of methylation depends on the length of the period of environmental change and has (time-dependently) to reach a certain gene-specific level to become relevant for gene-expression, but here that level has not yet been reached because the length of the period of environmental change was too short (there were still non-methylated Cs), (ii) the genes are pleiotropically regulated and thus their regulation needs to be balanced among the different functions and pathways, (iii) [in conjunction with hypothesis i] methylation only serves a ‘priming purpose’ for a faster response should the sons experience a similar environment as their fathers.
To our knowledge, there is no prediction tool available at the moment to translate ‘degree of methylation changes’ into ‘degree of gene expression changes’.
Tissues-Specific Methylation Effects
We measured the change in DNA methylation patterns in two organs of sons, the liver (F1LCvs F1LD) and the testis (F1TCvs F1TD). The presence of DMRs in both organs indicated a systemic, yet organ specific transmission of the epigenetic response to the environmental change the fathers had experienced. Such a response has a strong ecological relevance. The changes we expected in testes of sons (F1TCvs F1TD; Fig. 3) have the potential to be transmitted to the F2 generation if germ cells are affected. However, because we had to homogenize the testis tissue we could not distinguish between somatic cells and germ cells. In addition, DNA methylation is a regulator of cell differentiation [75] and at an age of 7-day post-partum testis cells were still differentiating [76]. Thus, what proportion of DMRs detected in testes was due to ongoing cellular differentiation and what proportion was due to paternal transmission of the epigenetic response could not be disentangled here. There is a growing evidence for tissue-specific epigenetic regulation, such in case of a-Actin (Acta1) increased in methylation within pancreas but in liver in mice fed a diet containing both daidzein and genistein [77]. Also the gene encoding the oestrogen receptor-α (Erα), which show a defined methylation profile in liver, but is not methylated in pancreas [77].
Advantage and Limitation of Using Whole Liver as Sample
As for our research focus we were not interested in determining liver cell-type specific methylomes, we homogenised the whole liver of each son to avoid the results to be biased by unequal distribution of tissue cell type composition. This way we obtained liver-wide methylation patterns, which we then analysed, assuming that the cellular composition of the livers was very similar across individuals.
The differentiation of cells into functionally different cell types is regulated by methylation, wherefore different cell types have different methylomes [75, 78]. Thus, a cell-type specific analysis would provide much more detailed results. However, with whole liver samples such detailed analysis is not possible. But because our aim was to investigate a liver-wide response (the liver being the main metabolic and thermo-regulating organ), and because cell-type specific analyses would have increased the costs drastically (without results adding equally to our research focus), we decided to use homogenized whole liver samples.
Epigenetic Plasticity during Puberty
Because in wild mammal species, males predominantly disperse from their natal site, they have to respond faster to outer changes. We hypothesise that the generally observed roaming behaviour of mammal males has led to a greater reactivity of epigenetic mechanisms in males during puberty, realized by a dynamic epigenetic-endocrine interplay. This is supported by the fact that the onset of puberty (and fertility) is accompanied by a major shift in endocrine levels as well as an overall genomic re-methylation [79]. Furthermore, because epigenetic factors are changed with altering environments roaming animals are exposed to, the mechanistic reactivity might be increased in comparison to females of the same age. To test this hypothesis, experiments in which both females and males are exposed to identical environmental factors would have to be set-up in parallel and samples would need to be collected along a time span before and after puberty to assess changes in hormone levels and epigenetic patterns.
Ecological Relevance
The experimentally applied reduction of 42% in protein content (LPD vs SMD) is a realistic scenario, caused by temporal resource limitations, for instance due to seasonal effects on the vegetation or dispersal to new habitats. Nevertheless, these changes are gradual. In our experiment, however, adult males were exposed not only to a sudden environmental change but also for a prolonged time span, mimicking global changes. Wild guinea pigs are generalists—they live in a wide range of habitats and across great heights and depths of South-America—and as such they are well equipped with an epigenetic ‘tool box’ to respond adequately to nutritional challenges [30, 80]. Future research needs to focus on habitat specialists and their responses to environmental (e.g. dietary) changes.
Conclusion
Our results show that a changing environment procures a diet change-specific epigenetic response, which is transmitted to the next generation (and maybe beyond). The epigenetic regulation may therefore increase the genotypically encoded adaptive plasticity to a widened epigenetically modified phenotypic plasticity.
Dispersing male may not only increase genetic, but also epigenetic diversity. Those epigenetic processes in response to diet changes might be crucial for species health and survival. We conclude that DNA methylation is an essential epigenetic mechanism that provides (i) non-random and (ii) heritable genetic plasticity for wild species to quickly respond to environmental conditions (adaptability) which are likely to be beneficial for species survival and reproduction in a long term (adaptation).
Data Accessibility
Next generation-sequencing data were uploaded to the National Centre for Biotechnology Information Short Reads Archive (http://www.ncbi.nim.nih.gov/sra) and are publicly accessible under the SRA study accession number SRP048942 in .fastq file format. Raw data, coverage, methylation ratios (file names: F1L_and F1T_coverage_MethLevels) are accessible on Dryad doi:10.5061/dryad.7bg3t6s.
Supplementary data
Supplementary data are available at EnvEpig online.
Supplementary Material
Acknowledgements
We thank Prof Trillmich and Dr Anja Guenther from the University of Bielefeld for the provision of C. aperea and the staff of the IZW—field research station for their support. This project was funded by the Leibniz Competition Fund (SAW-2011-IZW-2) and further supported by Leibniz Competition Fund (SAW-2018-IZW-3-EpiRank).
Conflict of interest statement. None declared.
References
- 1. West-Eberhard MJ. Phenotypic plasticity and the origins of diversity. Annu Rev Ecol Syst 1989;20:249–78. [Google Scholar]
- 2. Stearns SC. . The Evolution of Life Histories. Oxford: Oxford University Press; 1992, 249. [Google Scholar]
- 3. Jablonka E, Raz G.. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol 2009;84:131–76. [DOI] [PubMed] [Google Scholar]
- 4. Szyf M. Epigenetics, DNA methylation and chromatin modifying drugs. Annu Rev Pharmacol Toxicol 2009;49:243–63. [DOI] [PubMed] [Google Scholar]
- 5. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002;16:6–21. [DOI] [PubMed] [Google Scholar]
- 6. Kaneda M. Genomic imprinting in mammals-Epigenetic parental memories. Differentiation 2011;82:51–6. [DOI] [PubMed] [Google Scholar]
- 7. Aran D, Sabato S, Hellman A.. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol 2013;14:R21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics 2011;6:838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He HY, Chen Cm, Xie Y, Asea A, Calderwood SK.. HSP70 and heat shock factor 1 cooperate to repress Ras-induced transcriptional activation of the c-fos gene. Cell Stress Chaperones 2000;5:406–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deaton AM, Bird A.. CpG islands and the regulation of transcription. Genes Dev 2011;25:1010–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szyf M. The early life social environment and DNA methylation DNA methylation mediating the long-term impact of social environments early in life. Epigenetics 2011;6:971–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kilvitis HJ, Alvarez M, Foust CM, Schrey AW, Robertson M, Richards CL.. Ecological epigenetics. Ecol Genomics 2014;781:191–210. [DOI] [PubMed] [Google Scholar]
- 13. Penuelas J, Sardans J, Estiarte M, Ogaya R, Carnicer J, Coll M, Barbeta A, Rivas-Ubach A, Llusia J, Garbulsky M.. Evidence of current impact of climate change on life: a walk from genes to the biosphere. Glob Change Biol 2013;19:2303–38. [DOI] [PubMed] [Google Scholar]
- 14. Herrera Cm, Bazaga P.. Untangling individual variation in natural populations: ecological, genetic and epigenetic correlates of long-term inequality in herbivory. Mol Ecol 2011;20:1675–88. [DOI] [PubMed] [Google Scholar]
- 15. Kucharski R, Maleszka J, Foret S, Maleszka R.. Nutritional control of reproductive status in honeybees via DNA methylation. Science 2008;319:1827–30. [DOI] [PubMed] [Google Scholar]
- 16. Varriale A, Bernardi G.. DNA methylation and body temperature in fishes. Gene 2006;385:111–21. [DOI] [PubMed] [Google Scholar]
- 17. Lea AJ, Altmann J, Alberts SC, Tung J.. Resource base influences genome-wide DNA methylation levels in wild baboons (Papio cynocephalus). Mol Ecol 2016;25:1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weyrich A, Lenz D, Jeschek M, Chung Th, Rubensam K, Goritz F, Jewgenow K, Fickel J.. Paternal intergenerational epigenetic response to heat exposure in male Wild guinea pigs. Mol Ecol 2016;25:1729–40. [DOI] [PubMed] [Google Scholar]
- 19. Godfrey KM, Sheppard A, Gluckman PD, Lillycrop KA, Burdge GC, McLean C, Rodford J, Slater-Jefferies JL, Garratt E, Crozier SR, et al. Epigenetic gene promoter methylation at birth is associated with child’s later adiposity. Diabetes 2011;60:1528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolff GL, Kodell RL, Moore SR, Cooney Ca.. Maternal epigenetics and methyl supplements affect agouti gene expression in A(vy)/a mice. Faseb J 1998;12:949–57. [PubMed] [Google Scholar]
- 21. Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH.. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA 2008;105:17046–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klebig ML, Wilkinson JE, Geisler JG, Woychik RP.. Ectopic expression of the agouti gene in transgenic mice causes obesity, features of type-II diabetes, and yellow fur. Proc Natl Acad Sci USA 1995;92:4728–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 2010;143:1084–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Susser E, Lin SP.. Schizophrenia after prenatal exposure to the dutch hunger winter of 1944-1945 - reply. Arch Gen Psychiatry 1994;51:333–4. [DOI] [PubMed] [Google Scholar]
- 25. Kyle UG, Pichard C.. The Dutch Famine of 1944-1945: a pathophysiological model of long-term consequences of wasting disease. Curr Opin Clin Nutr 2006;9:388–94. [DOI] [PubMed] [Google Scholar]
- 26. Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC.. Epigenetic inheritance of a cocaine-resistance phenotype. Nat Neurosci 2013;16:42–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gapp K, Soldado-Magraner S, Alvarez-Sanchez M, Bohacek J, Vernaz G, Shu H, Franklin TB, Wolfer D, Mansuy IM.. Early life stress in fathers improves behavioural flexibility in their offspring. Nat Commun 2014;5:5466.. [DOI] [PubMed] [Google Scholar]
- 28. Ng SF, Lin RCY, Laybutt DR, Barres R, Owens JA, Morris MJ.. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature 2010;467:963–U103. [DOI] [PubMed] [Google Scholar]
- 29. Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY.. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A 2014;111:1873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Asher M, Lippmann T, Epplen JT, Kraus C, Trillmich F, Sachser N.. Large males dominate: ecology, social organization, and mating system of wild cavies, the ancestors of the guinea pig. Behav Ecol Sociobiol 2008;62:1509–21. [Google Scholar]
- 31. Holtenius K, Bjornhag G.. The colonic separation mechanism in the guinea-pig (Cavia-porcellus) and the chinchilla (Chinchilla-laniger). Comp Biochem Physiol A Comp Physiol 1985;82:537–42. [DOI] [PubMed] [Google Scholar]
- 32. Sakaguchi E. Digestive strategies of small hindgut fermenters. Anim Sci J 2003;(74:): 327–37. [Google Scholar]
- 33. Sakaguchi E, Itoh H, Uchida S, Horigome T.. Comparison of fiber digestion and digesta retention time between rabbits, guinea-pigs, rats and hamsters. Br J Nutr 1987;58:149–58. [DOI] [PubMed] [Google Scholar]
- 34. Naya DE. Gut size flexibility in rodents: what we know, and don’t know, after a century of research. Rev Child Hist Nat 2008;81:599–612. [Google Scholar]
- 35. Rechkemmer G, Ronnau K, von Engelhardt W.. Fermentation of polysaccharides and absorption of short chain fatty acids in the mammalian hindgut. Comp Biochem Physiol A Comp Physiol 1988;90:563–8. [DOI] [PubMed] [Google Scholar]
- 36. Hagen P, Robinson KW.. The production and absorption of volatile fatty acids in the intestine of the guinea-pig. Aust J Exp Biol Med Sci 1953;31:99–103. [DOI] [PubMed] [Google Scholar]
- 37. Havera SP, Robbins CT.. Wildlife feeding and nutrition - robbins, Ct. J Wildlife Manage 1984;48:664. [Google Scholar]
- 38. Deitch EA, Winterton J, Li M, Berg R.. The gut as a portal of entry for bacteremia - role of protein-malnutrition. Ann Surg 1987;205:681–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Claeyssens S, Lavoinne A, Fresel-Ragot M, Bois-Joyeux B, Chanez M, Peret J.. Metabolic changes in rats fed a low protein-diet during post-weaning growth. Metab Clin Exp 1990;39:676–81. [DOI] [PubMed] [Google Scholar]
- 40. Darmon N, Pelissier MA, Heyman M, Albrecht R, Desjeux JF.. Oxidative stress may contribute to the intestinal dysfunction of weanling rats fed a low-protein diet. J Nutr 1993;123:1068–75. [DOI] [PubMed] [Google Scholar]
- 41. Lister D, McCance RA.. The effect of two diets on the growth, reproduction and ultimate size of guinea-pigs. Br J Nutr 1965;19:311–9. [DOI] [PubMed] [Google Scholar]
- 42. Raubenheimer D, Simpson SJ.. Nutrient transfer functions: the site of integration between feeding behaviour and nutritional physiology. Chemoecology 1998;8:61–8. [Google Scholar]
- 43. Rothwell NJ, Stock MJ.. Diet-Induced Thermogenesis. Adv Nutr Res 1983;5:201–20. [DOI] [PubMed] [Google Scholar]
- 44. Hingst O, Blottner S.. Quantification of apoptosis (programmed cell-death) in mammalian testis by DNA-fragmentation elisa. Theriogenology 1995;44:313–9. [DOI] [PubMed] [Google Scholar]
- 45. Holt WV. Postnatal development of the testes in the cuis, Galea musteloides. Lab Anim 1977;11:87–91. [DOI] [PubMed] [Google Scholar]
- 46. Van Keulen J, Young BA.. Evaluation of acid-insoluble ash as a natural marker in ruminant digestibility studies. J Anim Sci 1977;44:282–7. [Google Scholar]
- 47. Mann H, Whitney D.. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Statist 1947;18:50–60. [Google Scholar]
- 48. Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R.. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 2005;33:5868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weyrich A, Lenz D, Jeschek M, Chung TH, Rübensam K, Göritz K, Jewgenow K, Fickel J.. Paternal intergenerational epigenetic response to heat exposure in male Wild guinea. Mol Ecol 2016;25:1729–40. [DOI] [PubMed] [Google Scholar]
- 50. Weyrich A, Schullermann T, Heeger F, Jeschek M, Mazzoni CJ, Chen W, Schumann K, Fickel J.. Whole genome sequencing and methylome analysis of the wild guinea pig. Bmc Genomics 2014;15:1036–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krueger F, Andrews SR.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Song Q, Decato B, Hong EE, Zhou M, Fang F, Qu JH, Garvin T, Kessler M, Zhou J, Smith AD.. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. Plos One 2013;8:e81148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Snel B, Lehmann G, Bork P, Huynen MA.. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res 2000;28:3442–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weyrich A, Benz S, Karl S, Jeschek M, Jewgenow K, Fickel J.. Paternal heat exposure causes DNA methylation and gene expression changes of Stat3 in Wild guinea pig sons. Ecol Evol 2016;6:2657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dias BG, Ressler KJ.. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci 2014;17:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vyssotski DL. Transgenerational epigenetic compensation. Evolocus 2011;6:838–6. [Google Scholar]
- 58. Lea AJ, Altmann J, Alberts SC, Tung J.. Resource base influences genome-wide DNA methylation levels in wild baboons (Papio cynocephalus). Mol Ecol 2016;25:1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hintz HF. Effect of coprophagy on digestion and mineral excretion in the guinea pig. J Nutr 1969;99:375–8. [DOI] [PubMed] [Google Scholar]
- 60. Franz R, Kreuzer M, Hummel J, Hatt JM, Clauss M.. Intake, selection, digesta retention, digestion and gut fill of two coprophageous species, rabbits (Oryctolagus cuniculus) and guinea pigs (Cavia porcellus), on a hay-only diet. J Anim Physiol Anim Nutr 2011;95:564–70. [DOI] [PubMed] [Google Scholar]
- 61. Brini M, Cali T, Ottolini D, Carafoli E.. The plasma membrane calcium pump in health and disease. Febs J 2013;280:5385–97. [DOI] [PubMed] [Google Scholar]
- 62. Zhang N. Epigenetic modulation of DNA methylation by nutrition and its mechanisms in animals. Anim Nutr 2015;1:144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Loenen WAM. S-adenosylmethionine: jack of all trades and master of everything? Biochm Soc Trans 2006;34:330–3. [DOI] [PubMed] [Google Scholar]
- 64. Mckay JA, Mathers JC.. Diet induced epigenetic changes and their implications for health. Acta Physiol (Oxf) 2011;202:103–18. [DOI] [PubMed] [Google Scholar]
- 65. Feil R, Fraga MF.. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 2012;13:97–109. [DOI] [PubMed] [Google Scholar]
- 66. Logan CY, Nusse R.. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 2004;20:781–810. [DOI] [PubMed] [Google Scholar]
- 67. Tortora G, Derrickson B.. Principles of Anatomy and Physiology. New Jersey, USA: Wiley, 2012. [Database]. [Google Scholar]
- 68. Guenther A, Palme R, Dersen M, Kaiser S, Trillmich F.. Photoperiodic effects on reproductive development in male cavies (Cavia aperea). Physiol Behav 2014;123:142–7. [PubMed] [Google Scholar]
- 69. Geissler EN, Liao M, Brook JD, Martin FH, Zsebo KM, Housman DE, Galli SJ.. Stem-cell factor (Scf), a novel hematopoietic growth-factor and ligand for C-kit tyrosine kinase receptor, maps on human-chromosome 12 between 12q14.3 and 12qter. Somat Cell Mol Genet 1991;17:207–14. [DOI] [PubMed] [Google Scholar]
- 70. Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014;13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yuan ZL, Guan YJ, Wang LJ, Wei WY, Kane AB, Chin YE.. Central role of the threonine residue within the p+1 loop of receptor tyrosine kinase in STAT3 constitutive phosphorylation in metastatic cancer cells. Mol Cell Biol 2004;24:9390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Toyota T, Yamada K, Saito K, Detera-Wadleigh SD, Yoshikawa T.. Association analysis of adenylate cyclase type 9 gene using pedigree disequilibrium test in bipolar disorder. Mol Psychiatry 2002;7:450–2. [DOI] [PubMed] [Google Scholar]
- 73. Taussig R, Gilman AG.. Mammalian membrane-bound adenylyl cyclases. J Biol Chem 1995;270:1–4. [DOI] [PubMed] [Google Scholar]
- 74. Hahn MA, Wu X, Li AX, Hahn T, Pfeifer GP.. Relationship between gene body DNA methylation and intragenic H3K9me3 and H3K36me3 chromatin marks. Plos One 2011;6:e18844.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Khavari DA, Sen GL, Rinn JL.. DNA methylation and epigenetic control of cellular differentiation. Cell Cycle 2010;9:3880–3. [DOI] [PubMed] [Google Scholar]
- 76. Kubo N, Toh H, Shirane K, Shirakawa T, Kobayashi H, Sato T, Sone H, Sato Y, Tomizawa S-I, Tsurusaki Y, et al. DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. Bmc Genomics 2015;16:624.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Guerrero-Bosagna CM, Sabat P, Valdovinos FS, Valladares LE, Clark SJ.. Epigenetic and phenotypic changes result from a continuous pre and post natal dietary exposure to phytoestrogens in an experimental population of mice. BMC Physiol 2008;8:17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Michalowsky LA, Jones PA.. DNA methylation and differentiation. Environ Health Perspect 1989;80:189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Almstrup K, Johansen ML, Busch AS, Hagen CP, Nielsen JE, Petersen JH, Juul A.. Pubertal development in healthy children is mirrored by DNA methylation patterns in peripheral blood. Sci Rep 2016;6. doi:10.1038/srep28657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dunnum J. Family Caviidae G. Fischer, 1817. In: Patton JL, Pardiñas UFJ, D'Elía G (eds), Mammals of South America, The Chicago Press, 2015; 690–716. http://dx.doi.org/10.2305/IUCN.UK.2016-2.RLTS.T86257782A22189256.en. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next generation-sequencing data were uploaded to the National Centre for Biotechnology Information Short Reads Archive (http://www.ncbi.nim.nih.gov/sra) and are publicly accessible under the SRA study accession number SRP048942 in .fastq file format. Raw data, coverage, methylation ratios (file names: F1L_and F1T_coverage_MethLevels) are accessible on Dryad doi:10.5061/dryad.7bg3t6s.