Abstract
CavityPlus is a web server that offers protein cavity detection and various functional analyses. Using protein three-dimensional structural information as the input, CavityPlus applies CAVITY to detect potential binding sites on the surface of a given protein structure and rank them based on ligandability and druggability scores. These potential binding sites can be further analysed using three submodules, CavPharmer, CorrSite, and CovCys. CavPharmer uses a receptor-based pharmacophore modelling program, Pocket, to automatically extract pharmacophore features within cavities. CorrSite identifies potential allosteric ligand-binding sites based on motion correlation analyses between cavities. CovCys automatically detects druggable cysteine residues, which is especially useful to identify novel binding sites for designing covalent allosteric ligands. Overall, CavityPlus provides an integrated platform for analysing comprehensive properties of protein binding cavities. Such analyses are useful for many aspects of drug design and discovery, including target selection and identification, virtual screening, de novo drug design, and allosteric and covalent-binding drug design. The CavityPlus web server is freely available at http://repharma.pku.edu.cn/cavityplus or http://www.pkumdl.cn/cavityplus.
INTRODUCTION
Binding cavities on protein surfaces are important for protein function because they are usually the sites at which a protein binds to other biological macromolecules such as nucleic acids and proteins, or small molecules such as metabolites and drugs. Computational detection of protein cavities has been considered a vital step for functional annotation of proteins and structure-based drug design. Various geometry- and energy-based methods have been developed to address this problem with high detection accuracies (1,2). It is also crucial to characterize the detected binding cavities to understand the principles of molecular binding (2). A comprehensive analysis of these cavities is very important.
According to the official definition by the International Union of Pure and Applied Chemistry (IUPAC), a pharmacophore model is “An ensemble of steric and electronic features that is necessary to ensure the optimal supramolecular interactions with a specific biological target and to trigger (or block) its biological response.” (3). A pharmacophore model within a protein cavity can be used for virtual screening of bioactive molecule databases, de novo design of novel candidate molecules, and potential drug target identification (4). Allostery is the regulation of a macromolecule by binding a ligand at one site then affecting the function of other sites. Allostery regulation has been proved to participate in various biological processes (5). Designing allosteric drugs is a future direction of drug discovery because allosteric drugs have many advantages compared with traditional orthosteric drugs (those that affect the site at which they bind), including fewer side effects, higher specificity and easier regulation (6). It is a common phenomenon for covalent binding of ligands in complex biological processes. Cysteine usually has the highest nucleophilic reactivity among the natural amino acid residues (7). Recently, covalent ligands with high potency and selectivity have attracted a resurgence of interest. About one third of the FDA approved drugs have been reported to act through covalent mechanisms (7,8). In summary, pharmacophore modelling, allosteric site identification, and covalent ligand-binding prediction are important issues for drug design related to protein cavities.
We collected previously developed web services for cavity or pocket detection, pharmacophore modelling, allosteric site prediction and guiding covalent ligand design in Supplementary Table S1. However, there is no integrated web server reported to allow for systematic analyses of protein cavities and functions. Here, we developed an interactive and easy-to-use web server, CavityPlus (the computing page of CavityPlus is shown in Supplementary Figure S1), for precise and robust protein cavity detection and further functional analyses. Four original toolkits—CAVITY (9), CavPharmer (10,11), CorrSite (12) and CovCys (13)—are integrated in CavityPlus to provide online identification of cavities on protein surfaces and, for a given cavity, pharmacophore modelling, allosteric site identification, and covalent ligand-binding ability prediction. Using this integrated platform, comprehensive knowledge of a protein cavity can be mined, e.g. CavityPlus can identify novel binding sites for designing covalent allosteric ligands. Overall, CavityPlus aims to offer a comprehensive platform for use in many aspects of drug design and discovery, such as target selection and identification, virtual screening, de novo drug design, design of allosteric and covalent-binding drugs.
MATERIALS AND METHODS
CAVITY
CAVITY adopts a structural geometry-based method for ligand-binding site detection and analysis. Compared to other binding site detection methods, CAVITY yields high prediction accuracies both in the bound and unbound protein test sets (9). A unique feature of CAVITY is that it uses CavityScore and CavityDrugScore to quantitatively calculate the ligandability and druggability, respectively, of a detected binding cavity. The ligandability value represents the possibility of designing small ligands with high binding affinities to a certain cavity, and the Druggability value reflects the possibility of a cavity being a good target for binding drug-like molecules. The CavityScore is influenced by cavity volume, pocket lip size, hydrophobic volume, cavity surface area, and hydrogen-bond-forming surface area. Using the data from the refined set of PDBBind (14,15), linear relationships between the maximal experimental binding affinity and CavityScore were found. Druggability is a more complicated metric that is related to higher-level properties of ligands (ADME/T) as well as to the role of the macromolecules acting in cellular pathways. CAVITY used the NRDLD dataset (16) to train and validate the CavityDrugScore, and this score was able to successfully separate druggable and undruggable proteins.
CavPharmer
We developed a new update of the Pocket method (10,11) for CavityPlus, CavPharmer (Pocket version 4.0), which can rapidly derive receptor-based pharmacophores from the structures of specific protein binding cavities. CavPharmer outputs six types of pharmacophore features, including hydrophobic center, hydrogen bond donor and acceptor, positive- and negative-charge center and exclude volume. The method automatically analyses the input cavity by scored grids and reduces the crucial features in the pharmacophore model to a reasonable number. A set of excluded volumes are defined to characterize the shape and boundary of the cavity.
We tested CavPharmer using a refined DUD database (17), in which all 40 protein targets were analysed by CAVITY with default parameters to detect potential ligand-binding sites. CAVITY output more than one cavities for each protein, but only the cavities bound to real ligands were used for pharmacophore modelling. Thirty nine real binding sites were successfully detected by CAVITY and were used to generate pharmacophore models with CavPharmer, which were then taken by a pharmacophore-match program PharmFit (18) for virtual screening. Supplementary Figure S2 shows that the average area under the curve (AUC) for virtual screening with the CavPharmer pharmacophore models is 0.69, which outperforms the average AUC of 0.63 with the pharmacophore models yielded by a pharmacophore modelling software LigandScout V2.02 (19).
CorrSite
Given a non-fibrous protein structure, it is important to identify the location of allosteric sites before beginning structure-based allosteric drug design. Based on the hypothesis that the motions of orthosteric and allosteric sites are highly correlated, the CorrSite method (12) was developed to rapidly predict allosteric sites with the presumed correlation. The Gaussian network model, which is a minimalist Normal-Mode Analysis model, was used to calculate the correlations, and then the correlations were normalized using the Z-score (12,20). Allosteric proteins from the Core-Diversity set of ASBench (21) are collected to build a testing dataset with 23 known monomeric allosteric proteins, which included 24 known allosteric sites. CorrSite identifies 23 out of the 24 known allosteric sites (12), which outperforms other established allosteric site prediction servers using the same dataset (Shown in Supplementary Table S2).
CovCys
CovCys was developed based on a comprehensive statistical analysis of covalently modified cysteine residues in protein structures. Compared to unmodified cysteine residues in the same protein structure, the covalently modified cysteine residues showed lower pKa values and higher solvent exposure. Such cysteine residues are usually located within or near a pocket detected by a cavity-detection program (13).
CovCys methodology was trained on the data set ‘Covalent’ (Cov) by exploring the Protein Data Bank (PDB) for complexes with covalent ligands binding to cysteine residues. Structures and corresponding literature reports were checked to ensure the accuracy of the data set. Another data set, ‘Non-covalent’ (Non-Cov), was composed of all the other cysteine residues from the same structure that did not form covalent bonds with the ligand in that crystal structure. The details about data set preparation were referred to the reference (13). An external test set from the Cysteinome database (1377 Cov and 5185 Non-Cov) was used to evaluate CovCys. Support vector machine was used to construct the predictive model to evaluate the potential that a cysteine residue could be covalently bonded, with a predicting accuracy of 0.85 for internal cross-validation and 0.73 for external validation.
CavityPlus web server
Input
CAVITY is always the first step to run when using the CavityPlus web server, and it uses protein three-dimensional (3D) structural information as the input. Users can use CavityPlus by providing valid PDB IDs and our web server will automatically load the protein structures from RCSB (http://www.rcsb.org/), or they can upload their own protein structure files in the PDB format. All three modules, CavPharmer, CorrSite and CovCys, use CAVITY outputs as their input. For the CavPharmer module, at least one cavity should be selected as input; the CorrSite module needs an orthosteric site as input and users can provide the orthosteric site by selecting a detected cavity or by uploading a custom orthosteric site with two specific formats: a) PDB format; b) text format as 46:A;47:A;49:A. The CovCys module uses all of the cavity results as input, and no extra information is required.
Output
The CAVITY module outputs all potential ligand-binding sites on the protein surfaces. The predicted maximum pKd, DrugScore and Druggability of all detected cavities are given in table format. The CavPharmer module outputs pharmacophore features within given cavities. We assigned different colors to different feature types to facilitate the graphical view. The CorrSite module outputs the correlation between the orthosteric site and other detected cavities, and the cavities are sorted by Z-score, which represents the possibility of a cavity being an allosteric binding site. Cavities with a Z-score larger than 0.5 are regarded as potential allosteric sites. The CovCys module provides a list of potentially covalently druggable cysteine residues in protein cavities, with important supplementary features of these cysteine residues also listed. All graphical viewing is implemented by using the JSmol applet (22) on the website. Users can also download the result files and view them via third-party software such as Pymol (23) or LigandScout (19).
TEST CASE
Case 1: Cysteine protease domain
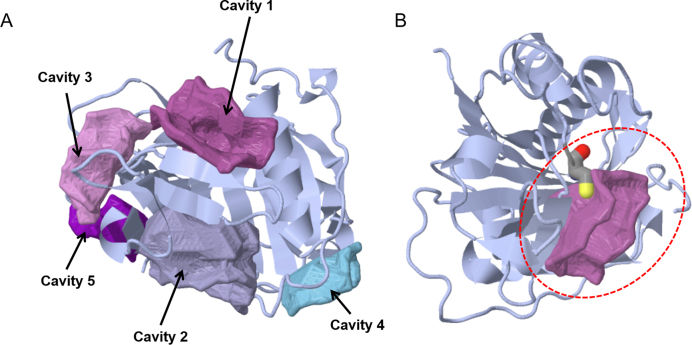
Cysteine protease domain (CPD) was demonstrated to proteolytically activate the Vibrio cholerae MARTX toxin (24). We used CavityPlus to evaluate the CPD structure (PDB ID: 3GCD). Note that though CPD is monomeric, in this crystal structure, four chains are packed in one asymmetric unit. For CAVITY analysis, only one chain should be used. We selected chain A in 3GCD as input. The CAVITY module was performed with default parameters and “No Ligand” mode. The five output cavities are shown in Table 1 and Figure 1A, in which Cavity_1 is actually the active site of CPD. With Cavity_1 as the orthosteric site, Cavity_2, with a Z-score of 1.64, was predicted as a potential allosteric binding pocket by the CorrSite module. Cavity_2 was previously proved to be an allosteric switch that efficiently activate CPD (25). We then used the CovCys module to analyse these cavities. Figure 1B shows that Cys140 of CPD in Cavity_1 is predicted as a covalent binding site in this protein, which agrees well with previous discovery (26).
Table 1. Output of the CAVITY module with PDB ID of 3GCD and 2OU7 as input.
| PDB ID | Cavity No. | Predicted Maximal pKd | Predicted Average pKd | DrugScore | Druggability |
|---|---|---|---|---|---|
| 3GCD | 1 | 11.77 | 6.81 | 8 | Less druggable |
| 2 | 10.87 | 6.34 | −477 | Undruggable | |
| 3 | 8.03 | 5.37 | −1142 | Undruggable | |
| 4 | 5.66 | 4.56 | −1306 | Undruggable | |
| 5 | 5.61 | 4.54 | −1223 | Undruggable | |
| 2OU7 | 1 | 11.55 | 6.92 | 989 | Druggable |
| 2 | 11.54 | 6.57 | −105 | Less druggable | |
| 3 | 8.87 | 5.66 | −606 | Undruggable | |
| 4 | 8.54 | 5.55 | −973 | Undruggable | |
| 5 | 8.37 | 5.49 | −299 | Undruggable | |
| 6 | 7.57 | 5.22 | −967 | Undruggable | |
| 7 | 7.23 | 5.10 | −456 | Undruggable | |
| 8 | 6.63 | 4.89 | −868 | Undruggable | |
| 9 | 6.49 | 4.84 | −1065 | Undruggable |
Figure 1.
The detected cavities and a covalent-binding site of the cysteine protease domain (PDB ID: 3GCD) shown in JSmol. (A) Cavities detected by the CAVITY Module; (B) a cysteine covalent-binding site within Cavity_3.
Case 2: Polo-like kinase 1
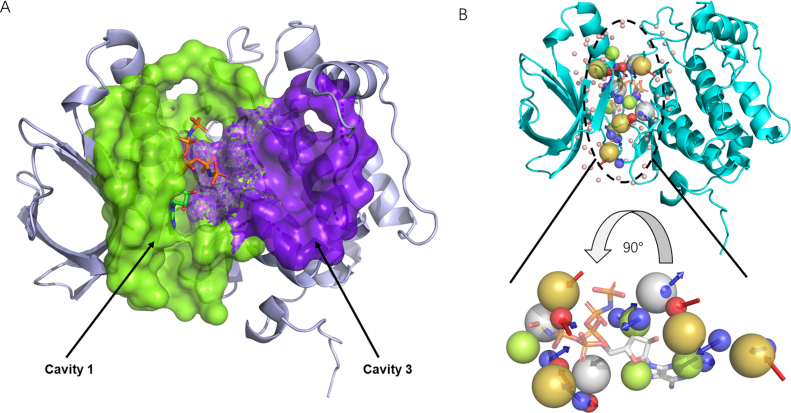
Polo-like kinase 1 (PLK1) is a serine/threonine kinase in mammalian cells that participates in multiple steps in mitosis (27). The crystal structure of the T210V mutant PLK1 kinase domain (KD) in complex with phosphoaminophosphonic acid-adenylate ester (ANP) (PDB ID: 2OU7) was used as input. The CAVITY module was run with the default parameters. The “With Ligand” mode of CAVITY was used and ANP was provided as the ligand. The program output nine cavities and only one cavity named Cavity_1 was predicted to be druggable, which was the ATP-binding site, shown in Table 1. Next, we ran CorrSite by choosing Cavity_1 as the orthosteric site. Four potential allosteric sites were predicted and Cavity_3 ranked first with a Z-score of 0.96. Cavity_3 (shown in Figure 2A in purple) has been shown to be a binding site in PLK1 and had been successfully used to discover non-ATP-competitive PLK1 inhibitors (27). We also used the CovCys module and identified two potential covalent binding-sites in this protein, Cys67 and Cys133 (Table 2), which are both inside the ATP-binding site of Cavity_1. Cys67 in PLK1 is a conserved Cys among various human kinases and covalent ligand has been reported for this site (28). These two Cys sites may attract a renewed interest in developing cysteine-targeted irreversible protein kinase inhibitors (29).
Figure 2.
Detected cavities and a pharmacophore model for Polo-like kinase 1 (PDB ID: 2OU7) shown in Pymol. (A) Two cavities detected by CAVITY. Green: active site; purple: potential allosteric site. (B) The pharmacophore model of the active ATP binding site. Blue and red arrows represents the H-bond donor center and h-bond acceptor, respectively; green spheres: hydrophobic center; olive spheres: positive electrostatic center; grey spheres: negative electrostatic center.
Table 2. Output of the CovCys module for test case 1(PDB ID: 3GCD) and test case 2 (PDB ID: 2OU7).
| PDB ID | ResID | CavID | Cov | Probability | pKa | QSASA | pKdAve |
|---|---|---|---|---|---|---|---|
| 3GCD | CYS_A_140 | cavity_1 | Yes | 0.90 | 7.70 | 0.23 | 6.81 |
| CYS_A_140 | cavity_3 | No | 0.17 | 7.70 | 0.23 | 5.37 | |
| 2OU7 | CYS_A_67 | cavity_1 | Yes | 0.91 | 9.24 | 0.16 | 6.92 |
| CYS_A_67 | cavity_8 | No | 0.19 | 9.24 | 0.16 | 4.89 | |
| CYS_A_133 | cavity_1 | Yes | 0.92 | 11.33 | 0.16 | 6.92 | |
| CYS_A_133 | cavity_9 | No | 0.09 | 11.33 | 0.16 | 4.84 | |
| CYS_A_212 | cavity_1 | No | 0.28 | 7.34 | 0.59 | 6.92 | |
| CYS_A_212 | cavity_2 | No | 0.49 | 7.34 | 0.59 | 6.57 | |
| CYS_A_212 | cavity_4 | No | 0.29 | 7.34 | 0.59 | 5.55 | |
| CYS_A_239 | cavity_3 | No | 0.21 | 11.15 | 0.06 | 5.66 | |
| CYS_A_239 | cavity_5 | No | 0.38 | 11.15 | 0.06 | 5.49 | |
| CYS_A_164 | None | undetermined | 0.00 | 7.84 | 0.09 | None | |
| CYS_A_255 | None | undetermined | 0.00 | 5.15 | 0.30 | None | |
| CYS_A_318 | None | undetermined | 0.00 | 10.68 | 0.09 | None |
Note. ResID: cysteine residues ID of the protein; CavID: the cavity ID detected by the CAVITY; Cov: the predicted annotation of being a covalent targetable cysteine; Probability: the probability of being a covalent targetable cysteine. pKa: the pKa value predicted by the PROPKA program (30); QSASA: the exposure degree of the cysteine, which is calculated by dividing the solvent accessible surface area (SASA) of this cysteine with the theoretical SASA, which is calculated by the Pops program (31). pKdAve: the average binding affinity of the associated pocket, which is calculated by CAVITY.
DISCUSSION
Identifying binding cavities suitable for drug design is the key step for successful drug discovery efforts. CavityPlus is a freely accessible, interactive and easy-to-use web server that offers protein cavity detection and evaluation. In addition to druggable cavity identification, pharmacophore features can be automatically derived even without known binding ligand, which can be used in pharmacophore based virtual screening, ligand selection from docking results, or scaffold-hopping for known active compounds. Potential allosteric ligand-binding sites can be predicted based on the dynamic correlations between an indicated orthosteric site and other potential binding sites predicted from the cavity detection step. In addition, druggable covalent ligand-binding cysteine residues can be predicted for covalent drug discovery. Other residues suitable for covalent drug design will also be included in the future. Users only need to provide the 3D structure data for the protein of interest or even its PDB code, plus the location of the orthosteric site in the case of allosteric site prediction, CavityPlus will do the rest. To our knowledge, CavityPlus is the first web server for protein binding site detection with comprehensive function analyses of detected cavities. Such analyses are useful for many aspects of drug design and discovery, such as target selection and identification, virtual screening, de novo drug design, and allosteric and covalent-binding drug design. All the methods used in CavityPlus were originally developed in the authors’ group, and we expect to develop and integrate more functions for analysing protein cavities in the future, such as cavity or pharmacophore similarity analyses for protein function annotation and target identification for endogenous ligands.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members in Professor Luhua Lai's group, and thank Professor Honglin Li at East China University of Science and Technology and Professor Huanhuan Liang at Sun Yat-sen University for their helpful discussion and test on the CavityPlus web server.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China (in part) [2016YFA05023032, 2015CB910302]; National Natural Science Foundation of China [21673010, 21633001]. Funding for open access charge: Ministry of Science and Technology of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zheng X., Gan L., Wang E., Wang J.. Pocket-based drug design: exploring pocket space. AAPS J. 2013; 15:228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nisius B., Sha F., Gohlke H.. Structure-based computational analysis of protein binding sites for function and druggability prediction. J. Biotechnol. 2012; 159:123–134. [DOI] [PubMed] [Google Scholar]
- 3. Wermuth C.G. Pharmacophores: historical perspective and viewpoint from a medicinal chemist. Methods Principles Med. Chem. 2006; 32:3. [Google Scholar]
- 4. Yang S.-Y. Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov. Today. 2010; 15:444–450. [DOI] [PubMed] [Google Scholar]
- 5. Nussinov R., Tsai C.-J.. Allostery in disease and in drug discovery. Cell. 2013; 153:293–305. [DOI] [PubMed] [Google Scholar]
- 6. Peracchi A., Mozzarelli A.. Exploring and exploiting allostery: models, evolution, and drug targeting. Biochim. Biophys. Acta (BBA)-Proteins Proteomics. 2011; 1814:922–933. [DOI] [PubMed] [Google Scholar]
- 7. Weerapana E., Wang C., Simon G.M., Richter F., Khare S., Dillon M.B., Bachovchin D.A., Mowen K., Baker D., Cravatt B.F.. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010; 468:790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singh J., Petter R.C., Baillie T.A., Whitty A.. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 2011; 10:307. [DOI] [PubMed] [Google Scholar]
- 9. Yuan Y., Pei J., Lai L.. Binding site detection and druggability prediction of protein targets for structure-based drug design. Curr. Pharmaceut. Des. 2013; 19:2326–2333. [DOI] [PubMed] [Google Scholar]
- 10. Chen J., Lai L.. Pocket v. 2: further developments on receptor-based pharmacophore modeling. J. Chem. Inform. Model. 2006; 46:2684–2691. [DOI] [PubMed] [Google Scholar]
- 11. Chen J., Ma X., Yuan Y., Pei J., Lai L.. Protein-protein interface analysis and hot spots identification for chemical ligand design. Curr. Pharmaceut. Des. 2014; 20:1192–1200. [DOI] [PubMed] [Google Scholar]
- 12. Ma X., Meng H., Lai L.. Motions of allosteric and orthosteric ligand-binding sites in proteins are highly correlated. J. Chem. Inform. Model. 2016; 56:1725–1733. [DOI] [PubMed] [Google Scholar]
- 13. Zhang W., Pei J., Lai L.. Statistical analysis and prediction of covalent ligand targeted cysteine residues. J. Chem. Inform. Model. 2017; 57:1453–1460. [DOI] [PubMed] [Google Scholar]
- 14. Wang R., Fang X., Lu Y., Wang S.. The PDBbind database: collection of binding affinities for protein−ligand complexes with known three-dimensional structures. J. Med. Chem. 2004; 47:2977–2980. [DOI] [PubMed] [Google Scholar]
- 15. Wang R., Fang X., Lu Y., Yang C.-Y., Wang S.. The PDBbind database: methodologies and updates. J. Med. Chem. 2005; 48:4111–4119. [DOI] [PubMed] [Google Scholar]
- 16. Coleman R.G., Salzberg A.C., Cheng A.C.. Structure-based identification of small molecule binding sites using a free energy model. J. Chem. Inform. Model. 2006; 46:2631–2637. [DOI] [PubMed] [Google Scholar]
- 17. Jahn A., Hinselmann G., Fechner N., Zell A.. Optimal assignment methods for ligand-based virtual screening. J. Cheminform. 2009; 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mattes A., Witte K., Hohmann W., Lemmer B.. PHARMFIT—a nonlinear fitting program for pharmacology. Chronobiol. Int. 1991; 8:460–476. [DOI] [PubMed] [Google Scholar]
- 19. Wolber G., Langer T.. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inform. Model. 2005; 45:160–169. [DOI] [PubMed] [Google Scholar]
- 20. Haliloglu T., Bahar I., Erman B.. Gaussian dynamics of folded proteins. Phys. Rev. Lett. 1997; 79:3090. [Google Scholar]
- 21. Shen Q., Wang G., Li S., Liu X., Lu S., Chen Z., Song K., Yan J., Geng L., Huang Z.. ASD v3. 0: unraveling allosteric regulation with structural mechanisms and biological networks. Nucleic Acids Res. 2015; 44:D527–D535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L.. JSmol and the next‐generation web‐based representation of 3D molecular structure as applied to proteopedia. Isr. J. Chem. 2013; 53:207–216. [Google Scholar]
- 23. DeLano W.L. Pymol: an open-source molecular graphics tool. CCP4 Newslett. Protein Crystallogr. 2002; 40:82–92. [Google Scholar]
- 24. Shen A., Lupardus P.J., Albrow V.E., Guzzetta A., Powers J.C., Garcia K.C., Bogyo M.. Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nat. Chem. Biol. 2009; 5:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lupardus P.J., Shen A., Bogyo M., Garcia K.C.. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science. 2008; 322:265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scheer J.M., Romanowski M.J., Wells J.A.. A common allosteric site and mechanism in caspases. Proceedings of the National Academy of Sciences. 2006; 103:7595–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yun T., Qin T., Liu Y., Lai L.. Discovery of Non‐ATP‐Competitive Inhibitors of Polo‐like Kinase 1. ChemMedChem. 2016; 11:713–717. [DOI] [PubMed] [Google Scholar]
- 28. Miller R.M., Paavilainen V.O., Krishnan S., Serafimova I.M., Taunton J.. Electrophilic fragment-based design of reversible covalent kinase inhibitors. Journal of the American Chemical Society. 2013; 135:5298–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Q., Sabnis Y., Zhao Z., Zhang T., Buhrlage S.J., Jones L.H., Gray N.S.. Developing irreversible inhibitors of the protein kinase cysteinome. Chemistry & Biology. 2013; 20:146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bas D.C., Rogers D.M., Jensen J.H.. Very fast prediction and rationalization of pKa values for protein–ligand complexes. Proteins: Structure, Function, and Bioinformatics. 2008; 73:765–783. [DOI] [PubMed] [Google Scholar]
- 31. Cavallo L., Kleinjung J., Fraternali F.. POPS: a fast algorithm for solvent accessible surface areas at atomic and residue level. Nucleic Acids Research. 2003; 31:3364–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.