Abstract
The unprecedented growth of high-throughput sequencing has led to an ever-widening annotation gap in protein databases. While computational prediction methods are available to make up the shortfall, a majority of public web servers are hindered by practical limitations and poor performance. Here, we introduce PANNZER2 (Protein ANNotation with Z-scoRE), a fast functional annotation web server that provides both Gene Ontology (GO) annotations and free text description predictions. PANNZER2 uses SANSparallel to perform high-performance homology searches, making bulk annotation based on sequence similarity practical. PANNZER2 can output GO annotations from multiple scoring functions, enabling users to see which predictions are robust across predictors. Finally, PANNZER2 predictions scored within the top 10 methods for molecular function and biological process in the CAFA2 NK-full benchmark. The PANNZER2 web server is updated on a monthly schedule and is accessible at http://ekhidna2.biocenter.helsinki.fi/sanspanz/. The source code is available under the GNU Public Licence v3.
INTRODUCTION
Proteins with accurate functional annotations are vital to biological research. Unfortunately, the vast majority of protein sequences are functionally uncharacterized, i.e. they have no experimentally-verified annotations. While advances in high-throughput sequencing ensures the continued growth of sequence data, there is no scalable means to annotate these sequences experimentally. Computational annotation has, therefore, emerged as a necessary alternative; substituting computational inference for experimental evidence. Protein function prediction uses data-intensive computational methods to assign Gene Ontology (GO) terms to proteins, specifying molecular functions (MF), involvement in biological processes (BP) and subcellular localizations (CC) (1). In addition to GO annotations, some methods predict free text descriptions (DE) that are required for the submission of new sequences to databases.
Functional annotation involves integrating many data sources, correlating GO annotations with, for example, sequence similarity, gene expression or biomedical literature, to make predictions. Managing a comprehensive annotation pipeline involves keeping databases up-to-date and ensuring that growing disk space and memory requirements are met. This suggests that a majority of users will use public web servers for annotation. Unfortunately, public annotation servers tend to be slow, infrequently updated and overly restrictive in the number of queries that can be submitted at once. When this time-consuming process is complete, the results may lack predictions for a substantial proportion of queries, not provide uncertainty estimates and very few annotation servers output DE predictions (see Table 1 for examples).
Table 1. Feature comparison between selected annotation servers. DE prediction stands for free text protein descriptions. Last database update is taken from explicit statements on annotation servers (at time of writing 22/03/18).
| Server | GO prediction | DE prediction | >1000 query sequences | Prob. estimate | Open source | Last database update/update schedule |
|---|---|---|---|---|---|---|
| PANNZER2 | Yes | Yes | Yes | Yes | Yes | Monthly (synchronised with UniProt) |
| ARGOT | Yes | No | Yes | No | No | 11/2016 |
| PFP | Yes | No | No | yes | No | Unknown |
| FunFam | Yes | No | No | No | Data can be downloaded | Daily |
| INGA | Yes | No | No | Yes | No | 04/2015 |
| eggNOG | Yes | Keyword | Yes | No | Yes | 11/2017 |
| dcGO | Yes | No | Error | Yes | No | 06/2016* |
An asterisk (*) following the last database update indicates that timestamps for database files were used instead. Timestamps are conservative because the data might be older than the timestamp suggests.
PANNZER2 remedies these issues by providing a fast, publically accessible web server for functional annotation. PANNZER was not previously available as a web server and was slow due to its use of BLAST. PANNZER2, however, is built using SANSparallel (2), a protein homology search tool thousands of times faster than BLAST. This allows PANNZER2 to analyze tens of thousands of queries in batch mode. Like PANNZER, PANNZER2 outputs both GO and DE predictions that can either be downloaded or explored via a web application. The web application displays predictions together with color-coded probabilities. We provide links to homology search results for each query sequence, enabling users to see how predictions were derived. The databases used by PANNZER2 are updated on a monthly schedule, ensuring that predictions benefit from new data. Finally, users can select from multiple alternative scoring functions in order to see which predictions are robust across different predictors.
MATERIALS AND METHODS
PANNZER2 overview
PANNZER2 is a weighted k-nearest neighbour classifier based on sequence similarity and enrichment statistics. PANNZER2 is implemented using three separate servers: the frontend web server–containing the interface, the SANSparallel server—for fast homology search and the DictServer—for managing associated metadata, for example, the GO structure, GO annotations and background frequencies of annotations. PANNZER2 implements the following annotation pipeline.
Homology search
For each query sequence, we use SANSparallel to find homologous sequences in the UniProt database (3). We refer to homology search results as the sequence neighbourhood. By default, PANNZER2 uses a maximum of 100 database hits. As we are transferring annotations based on sequence similarity, it is necessary for sequence matches to meet several criteria for inclusion in the sequence neighbourhood. Search results must have at least 40% sequence identity and 60% alignment coverage of both the query and target sequences. We refer to this step as sequence filtering.
The sequence neighbourhood will contain a subset of sequences associated with GO annotations and all results will have a free text description of variable quality. Both annotations and descriptions are gathered for each search result by calling the DictServer.
Gene ontology annotation
All GO predictors implemented by PANNZER2 are based on enrichment statistics from the sequence neighborhood of the query sequence, we refer to these as scoring functions. All scoring functions use the same filtered sequence neighborhood as input, but differ in how the score is calculated. PANNZER2 includes implementations of the scoring functions from ARGOT (4), BLAST2GO (5) and PANNZER (6), as well as hypergeometric enrichment and best informative hit. By default, PANNZER2 uses the ARGOT scoring function which was found to work best overall. We performed experiments to validate both the selection of the ARGOT scoring function and sequence filtering parameters in supplementary methods, where all scoring functions available in PANNZER2 are described in detail.
DE prediction
PANNZER2 reimplements the DE prediction method from PANNZER. In brief, descriptions from the sequence neighbourhood are clustered and a weighted average of several statistics used to identify overrepresented words occurring in those descriptions (6).
Test set preparation
Evaluating annotation predictions requires a test set of manually annotated target sequences. It is important to remove poor quality and overrepresented sequences, while retaining as many sequences as possible. Starting with all proteins from SwissProt (3), (downloaded 6 February 2017), we excluded sequences whose descriptions included the following words: putative, uncharacterized, probable, fragment or potential. Next we removed GO annotations (downloaded 16 January 2017) with certain non-experimental evidence codes (IEA, ISS and ND), known uninformative annotations and highly prevalent GO classes (those found in >5% of proteins annotated with the same ontology, see supplemental data for a complete list of all GO terms excluded). The remaining set of annotations form a set of small informative GO classes. We created separate protein sets for each GO ontology containing only those proteins with one or more annotations from that ontology.
Finally, we clustered each test set at 70% similarity using CD-hit (7) to remove highly similar sequences. We selected the protein with the most GO annotations from each CD-hit cluster for inclusion in the final test sets. After all filtering steps, the size of the final test sets used to assess prediction accuracy were 50994 proteins in BP, 44 405 in MF and 33739 in CC. Speed measurements used smaller test sets containing 500 and 2000 proteins from the BP set. All test sets are available on our supplementary web page (http://ekhidna2.biocenter.helsinki.fi/sanspanz/NAR_supplementary_data/).
In all experiments, we removed the query sequence from SANSparallel search results (i.e. the self-hit), prior to running any scoring functions in PANNZER2. This simulates the situation where the user is annotating a sequence with unknown function.
RESULTS
Feature comparison
As table 1 shows PANNZER2 has the best feature coverage of all surveyed annotation web servers. We compared web servers that were shown to have good performance in the recent CAFA2 competition (8). We note that only 5 out of the top 10 performing methods from CAFA2 are currently available as public web servers (ARGOT (4), PFP (9), INGA (10), dcGO (11) and FunFam (12)), though these may differ to some extent from what was used in CAFA. Despite not being entered into CAFA, we included eggNOG-mapper due to its reported high-performance and scalability (13).
A majority of public annotation web servers severely restrict the number of query sequences. Indeed, only ARGOT2, eggNOG-mapper and PANNZER2, are suitable for high-throughput data analysis (a server error prevented us from testing batch submission for the dcGO server). Of these methods, only PANNZER2 and eggNOG-mapper accept raw sequences at this scale; ARGOT2 allows up to 5000 queries to be submitted, but requires that BLAST results are precomputed by the user. In addition to GO annotations, PANNZER2 and eggNOG-mapper are the only methods to output DE predictions. Surprisingly, only PANNZER2, dcGO and INGA attempt to quantify the uncertainty in their predictions with per-annotation probabilities. PFP does not output probabilities, but reports score ranges where its annotation confidence is very strong, strong, moderate, etc.
Speed comparison
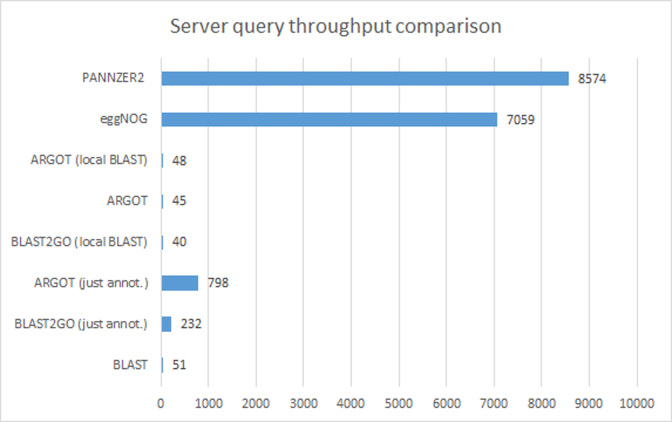
We benchmarked the performance of all high-throughput methods identified in Table 1, in addition to BLAST2GO, which was included due its popularity (Figure 1). Using each method, we annotated two subsets of 100 and 2000 sequences extracted from the BP test set and averaged the results. We made two exceptions to this procedure: eggNOG-mapper was not run with the 100 sequence benchmark due to its use of DIAMOND which has poor performance for lower numbers of queries (14) and ARGOT2 was not run with the 2000 sequence benchmark to avoid congesting a slower public server. For ARGOT2 and BLAST2GO we additionally included performance measurements that excluded BLAST (15) runtime, i.e. using precomputed BLAST results.
Figure 1.
Comparison of query throughput. We first show query throughput for combined sequence search and annotation steps for PANNZER2, eggNOG-mapper, ARGOT2 and BLAST2GO. We also show separate speeds for the annotation and BLAST steps for ARGOT2 and BLAST2GO. Notice that PANNZER2 and eggNOG-mapper outperform even the annotation step in ARGOT2 and BLAST2GO.
In our benchmark, PANNZER2 was the fastest annotation method, processing 21% more queries per hour than eggNOG-mapper, which was the second fastest method. However, with larger data sets (∼50K sequences used in the prediction accuracy experiment), eggNOG-mapper was faster than PANNZER2. The two annotation methods that use BLAST: ARGOT2 and BLAST2GO, were 178 and 214x slower than PANNZER2, respectively. BLAST, however, was not the sole reason for poor performance. Even if we exclude the BLAST runtime, ARGOT2 is still an order of magnitude slower than PANNZER2. BLAST2GO was the slowest method tested, but it is not advertised as a high-throughput method.
Prediction accuracy
We compared the GO prediction accuracy for the high-throughput annotation methods identified previously. Due to the scale of our test data, only eggNOG-mapper and PANNZER2 could make predictions for all queries. To include ARGOT2 in the evaluation, we extracted subsets of 5000 proteins from each test set and analysed these separately with each method. Prediction accuracy was evaluated with Fmax and Smin (see supplemental methods for details) both with and without annotations with non-experimental evidence codes (i.e. IEA, ISS and ND). EggNOG-mapper and ARGOT2 were run with default parameters. As eggNOG-mapper predicts annotations, but does not provide probabilities, we used −log10(max(E value, 10−200)) as a proxy with which to base the calculation of Fmax and Smin. Fmax and Smin were calculated using evaluation functions available in PANNZER2 (equivalent to full mode in CAFA). We want to stress that while we can exclude self-hits from ARGOT2 and PANNZER2, we cannot do so with the public eggNOG-mapper web server. Therefore, this experiment might give eggNOG-mapper an advantage. All results are shown in Table 2.
Table 2. Comparison of PANNZER2, using the ARGOT scoring function, ARGOT2 and eggNOG-mapper. Tests were repeated by a) omitting annotations with IEA, ISS and ND evidence codes and b) using all GO annotations. Evaluation was repeated using 5000 query subsets of the test data to allow for comparison with ARGOT2. We show results with Fmax and with Smin. Note that higher values of Fmax and lower values of Smin show better performance. PANNZER2 outperforms both eggNOG-mapper and ARGOT2 methods consistently.
| Comparisons with the whole dataset | Comparisons with subsets of the data | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Truth Set = > | No IEA, ISS, ND | All evidence codes | No IEA, ISS, ND | All evidence codes | |||||||
| Ontology | Metric | PANZ2 | eggNOG | PANZ2 | eggNOG | PANZ2 | eggNOG | ARGOT2 | PANZ2 | eggNOG | ARGOT2 |
| BP | Fmax | 0.699 | 0.615 | 0.786 | 0.640 | 0.700 | 0.613 | 0.608 | 0.784 | 0.629 | 0.682 |
| MF | Fmax | 0.708 | 0.640 | 0.867 | 0.591 | 0.708 | 0.641 | 0.649 | 0.858 | 0.591 | 0.777 |
| CC | Fmax | 0.823 | 0.752 | 0.863 | 0.774 | 0.820 | 0.749 | 0.757 | 0.853 | 0.773 | 0.776 |
| BP | Smin | 31.401 | 45.918 | 27.643 | 45.376 | 30.264 | 45.920 | 38.375 | 27.474 | 46.408 | 42.483 |
| MF | Smin | 9.597 | 12.942 | 6.701 | 15.995 | 9.682 | 12.890 | 11.609 | 7.196 | 16.06 | 11.946 |
| CC | Smin | 9.415 | 14.053 | 7.917 | 14.114 | 9.645 | 14.184 | 13.418 | 8.692 | 14.401 | 15.587 |
With the complete test sets, PANNZER2 outperforms eggNOG-mapper in terms of both Fmax and Smin in all three ontologies. The differences in performance can be extreme, for example, PANNZER2’s Fmax score in MF (all evidence codes) is almost 46.7% higher than eggNOG-mapper (27.6 percentage points). Furthermore, the difference is consistent across all ontologies, both with and without non-experimental annotations. This poor performance is partially due to coverage: eggNOG-mapper outputs empty GO predictions for 8–10% more query sequences than PANNZER2 (data not shown). Furthermore, for each query eggNOG-mapper outputs a set of annotations with only a single E-value (between query sequence and orthology group). The results from eggNOG-mapper might, therefore, be improved if separate probabilities or scores could be assigned to each annotation.
With the smaller test sets, both PANNZER2 and eggNOG-mapper exhibited similar Fmax and Smin scores compared to their respective performances on the complete test sets. This suggests that the smaller test sets are representative of the complete test sets. Interestingly, there is a difference in performance between PANNZER2 (which uses the ARGOT scoring function) and ARGOT2. This is likely due to our sequence filtering (see supplementary text) and the older databases used by the ARGOT2 web server. ARGOT2 outperforms eggNOG-mapper in all cases with the exception of Fmax score for BP (no non-experimental evidence codes) and Smin for CC (all evidence codes). Our supplementary web page includes test sequence sets and prediction tables generated by PANNZER2, ARGOT2 and eggNOG-mapper.
CAFA2 benchmark
We evaluated PANNZER2 using the CAFA2 no knowledge (NK) benchmark containing 3,681 proteins (8). In terms of Smin score, PANNZER2 ranked in the top 10 methods for MF and BP. We repeated the CAFA2 experiment using archived versions of the UniProt database (dated December 2013) and GOA database (dated 11 December 2013) to annotate the set of NK genes. We scored predictions using the CAFA2 Matlab scripts in full mode. Results are shown in Supplementary Tables S4 and S5.
PANNZER2 performed well in MF and BP evaluations, ranking in third and seventh place, respectively, for Smin. For Fmax, PANNZER2 ranked in 20th place for MF and 22nd place for BP. Smin emphasizes predictions of smaller, more informative GO classes, whereas Fmax treats broadly-defined classes as equally meaningful as more specialized ones. The difference in rankings highlights the tradeoff made by PANNZER2 to predict the most informative annotations possible as these are more relevant to biologists.
Our ranking for CC was 35th for Fmax and 31st for Smin. It was shown, however, that the CAFA2 CC evaluation set was biased to larger GO classes (8), penalizing methods like PANNZER2 that predict smaller, more informative GO classes.
PANNZER2 web server
Input
The PANNZER2 web server is available at http://ekhidna2.biocenter.helsinki.fi/sanspanz/. Protein sequences can be annotated by either copying protein sequences in FASTA format into a text box or by uploading a FASTA file. Users then enter the scientific name of the organism being analysed or, if novel or not found in the list, a closely-related species (this is a necessary parameter for the original PANNZER scoring function). Users initiate annotation by selecting either interactive or batch mode. A number of advanced parameters are available to set the values of sequence filtering, DE prediction and select between GO scoring functions.
Annotation time varies with the number of sequences uploaded, but in our experience a bacterial proteome will take several minutes to process, whereas a eukaryotic proteome will take about an hour. A comprehensive user manual is available from the PANNZER2 website and for developers we have included details on the SANSPANZ framework that was used to implement PANNZER2.
Output
If users selected batch mode when uploading their sequences, once annotation is complete, PANNZER2 gives links to download the complete results table and to view the prediction summary pages. If interactive mode was selected, users are taken directly to the summary page for the individual scoring function selected.
Each output summary is a table containing the sequence identifier, description predictions and GO predictions for biological process, molecular function and cellular component. All predictions, whether for descriptions or GO annotations, include colour-coded probabilities from green (high confidence predictions) to red (low confidence). Predictions are ranked in descending order of probability. Underneath each sequence identifier, we provide a ‘search’ link to access the output of SANSparallel on which predictions were based. Example outputs of reannotating 77 reference proteomes from Ensembl are available from the website.
New features in PANNZER2
PANNZER2 is the first PANNZER web server, reimplementing PANNZER1 with a modular architecture. PANNZER relied on BLAST and only included a single scoring function for GO annotation. In contrast, PANNZER2 is an interactive web server that uses SANSparallel (2) instead of BLAST, making it orders of magnitude faster. PANNZER2 contains implementations of several scoring functions and provides a probability estimate for each prediction along with associated KEGG reaction identifiers and EC numbers (downloaded from http://www.geneontology.org/external2go/). These features were unavailable in PANNZER. PANNZER2 additionally contains evaluation metrics (Fmax, Smin, weighted Fmax) that can be accessed programmatically to ease the development of novel scoring functions.
DISCUSSION
PANNZER2 provides a fast, interactive web server for functional annotation of protein sequences based on sequence homology and multiple annotation predictors. PANNZER2 provides an interactive interface for users to browse and interpret predictions. Here, we introduced the PANNZER2 web server, demonstrating its speed, scalability and features versus other high-throughput annotation methods. In terms of prediction accuracy PANNZER2 outperforms eggNOG-mapper, the only other method capable of operating at the same scale, in Fmax and Smin, in all three ontologies. It also outperforms ARGOT2, a similar annotation server that has lower throughput. Using the data from the CAFA2-NK-full benchmark we showed that PANNZER2 would have ranked in third place in the Smin evaluation of molecular function and seventh for biological process. In terms of practicalities, PANNZER2 is not only faster than other annotation servers, but allows users to submit up to 100000 query sequences at once. PANNZER2 provides predictions for free text descriptions as well as GO predictions, providing a comprehensive pipeline for proteome annotation projects.
DATA AVAILABILITY
Our supplementary web page (http://ekhidna2.biocenter.helsinki.fi/sanspanz/NAR_supplementary_data/) includes links to used test sequence sets and predictions from PANNZER2, ARGOT and eggNOG-mapper. These were not made available as supplementary data due to large file sizes.
Supplementary Material
ACKNOWLEDGEMENTS
Authors would like to thank Yuxiang Jiang (Indiana University) for help with CAFA2 evaluation pipe and Stefano Toppo (University of Padova) for ARGOT server support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academy of Finland [292589]; University of Helsinki Institute of Life Sciences (HiLife). Funding for open access charge: HiLife.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. Gene Ontology: tool for the unification of biology. Nature Genet. 2000; 25:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Somervuo P., Holm L.. SANSparallel: interactive homology search against Uniprot. Nucleic Acids Res. 2015; 43:W24–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2014; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Falda M., Toppo S., Pescarolo A., Lavezzo E., Di Camillo B., Facchinetti A., Cilia E., Velasco R., Fontana P.. Argot2: a large scale function prediction tool relying on semantic similarity of weighted Gene Ontology terms. BMC Bioinformatics. 2012; 13:S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M.. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005; 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- 6. Koskinen P., Törönen P., Nokso-Koivisto J., Holm L.. PANNZER: high-throughput functional annotation of uncharacterized proteins in an error-prone environment. Bioinformatics. 2015; 31:1544–1552. [DOI] [PubMed] [Google Scholar]
- 7. Li W., Godzik A.. CD-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006; 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- 8. Jiang Y., Oron T.R., Clark W.T., Bankapur A.R., D’Andrea D., Lepore R., Funk C.S., Kahanda I., Verspoor K.M., Ben-Hur A. et al. An expanded evaluation of protein function prediction methods shows an improvement in accuracy. Genome Biol. 2016; 17:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khan I.K., Wei Q., Chitale M., Kihara D.. PFP/ESG: automated protein function prediction servers enhanced with Gene Ontology visualization tool. Bioinformatics. 2014; 31:271–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piovesan D., Giollo M., Leonardi E., Ferrari C., Tosatto S.C.. INGA: protein function prediction combining interaction networks, domain assignments and sequence similarity. Nucleic Acids Res. 2015; 43:W134–W140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang H., Gough J.. DcGO: database of domain-centric ontologies on functions, phenotypes, diseases and more. Nucleic Acids Res. 2012; 41:D536–D544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Das S., Sillitoe I., Lee D., Lees J.G., Dawson N.L., Ward J., Orengo C.A.. CATH FunFHMMer web server: protein functional annotations using functional family assignments. Nucleic Acids Res. 2015; 43:W148–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huerta-Cepas J., Forslund K., Coelho L.P., Szklarczyk D., Jensen L.J., von Mering C., Bork P.. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017; 34:2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59. [DOI] [PubMed] [Google Scholar]
- 15. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our supplementary web page (http://ekhidna2.biocenter.helsinki.fi/sanspanz/NAR_supplementary_data/) includes links to used test sequence sets and predictions from PANNZER2, ARGOT and eggNOG-mapper. These were not made available as supplementary data due to large file sizes.