Abstract
Salivary gland tumor (SGT) is a rare tumor type, which exhibits broad-spectrum phenotypic, biological, and clinical heterogeneity. Currently, the molecular mechanisms that cause SGT pathogenesis remain poorly understood. A lack of animal models that faithfully recapitulate the naturally occurring process of human SGTs has hampered research progress on this field. In this report, we developed an inducible keratin 5-driven conditional knockout mouse model to delete gene(s) of interest in murine salivary gland upon local RU486 delivery. We have deleted two major tumor suppressors, Pten, a negative regulator of the PI3K pathway, and Smad4, the central signaling mediator of TGFβ pathway, in the murine salivary gland. Our results have shown that deletion of either Pten or Smad4 in murine salivary gland resulted in pleomorphic adenomas, the most common tumor in human SGT patients. Deletion of both Pten and Smad4 in murine salivary gland developed several malignancies, with salivary adenoid cystic carcinoma (SACC) being the most frequently seen. Molecular characterization showed that SACC exhibited mTOR activation and TGFβ1 overexpression. Examination of human SGT clinical samples revealed that loss of Pten and Smad4 is common in human SACC samples, particularly in the most aggressive solid form, and is correlated with survival of SACC patients, highlighting the human relevance of the murine models. In summary, our results offer significant insight into synergistic role of Pten and Smad4 in SGT, providing a rationale for targeting mTOR and/or TGFβ signaling to control SGT formation and progression.
Abbreviations: CxPA, carcinoma ex PA; GEMM, genetically engineered mouse model; HNSCC, head and neck squamous cell carcinomas; IF, immunofluorescence; K5, keratin 5; PI3K, phosphatidylinositol 3-kinase; SACC, salivary adenoid cystic carcinoma; SDC, salivary ductal carcinoma; SG, salivary gland; SGT, salivary gland tumor; SPA, salivary pleomorphic adenoma; SMA, smooth muscle actin; TGFβ, transforming growth factor β; YFP, yellow fluorescence protein
Introduction
Salivary gland tumors (SGTs), although uncommon, account for approximately 5% of all head and neck tumors and are composed of more than 20 histopathological subtypes with widely varied clinical outcomes [1], [2]. Both rarity of incidence and heterogeneity of pathology pose challenges for SGT studies, resulting in SGT being one of the least studied tumor types [3]. The most common subtype of SGT is salivary pleomorphic adenoma (SPA). Although benign, about 20% of SPAs will eventually progress to malignancy such as carcinoma ex PA (CxPA) or salivary ductal carcinoma (SDC) [4]. Salivary adenoid cystic carcinoma (SACC) is among the most common malignant SGTs; it exhibits slow but persistent tumor progression and frequent recurrence and metastasis [5]. Current therapeutic options for human SGT are limited. Depending on their location, some SGTs are difficult to remove completely. Radiation therapy is less effective for clinical treatment. The effects of current chemotherapies have been disappointing [6]. Thus, there is a need to acquire understanding of the molecular mechanisms of SGT pathogenesis that can be used toward the development of novel therapeutic approaches and that will allow clinicians to improve survival and quality of life for patients with SGT. Roadblocks impeding progress toward the realization of clinical diagnostics and therapeutics in this field are a lack of characterization of SGTs at the molecular level and a lack of research tools such as mouse models that mimic human SGTs.
Molecular characterization of human SGTs has been made recently. Of note, the application of exome sequencing on samples from the SGT biorepository has shown that, in addition to commonly seen chromosome translocations, molecular alterations of Pten and/or Smad4 occur in human SGTs [7], [8], [9], [10]. Pten, best known as a lipid phosphatase, negatively regulates the phosphatidylinositol 3-kinase (PI3K) pathway, which is activated in multiple human cancers [11]. Alteration of Pten has been reported in several subtypes of human SGTs. For example, we and others have recently shown that loss of Pten expression is common in human SACC patients, with the solid form most frequent [12], [13]. Loss of heterozygosity and a germline mutation of Pten have been reported in human epithelial myoepithelial carcinomas of salivary gland [14] and a case of acinic cell carcinoma patients [15], respectively. Moreover, recent studies showed that loss of Pten is fairly common in multiple human SGTs and is associated with a worse prognosis, relapse, and metastasis [10], [16], [17], [18], [19]. Smad4, the central signaling mediator for the transforming growth factor β (TGFβ) pathway, is a tumor suppressor for multiple human cancers [20], including its mutations in human SGTs [8]. In addition, TGFβ signaling has been known to regulate development and differentiation of salivary gland [21], [22]. And overexpression of TGFβ1 ligand has been reported in human CxPAs [23] and promotes migration and invasion of SACCs [24].
We have seen murine SGTs development in our Pten and/or Smad4 conditional deletion mouse models for head and neck squamous cell carcinoma driven by the Keratin 5 (K5) promoter [25]. We further modified RU486 delivery specifically into murine salivary glands and noticed that deletion of either Pten or Smad4 in murine salivary gland leads to the development of SPAs and that combined deletion of both Pten and Smad4 results in malignant progression of murine SGT, with SACC the most common. Examination of human SGT samples showed that reduced expression of Pten and Smad4 is common in human SACC clinical samples, particularly in the most aggressive solid form, and are correlated with survival of SACC patients, validating the clinical relevance of the murine SGT models. Furthermore, the murine SGTs exhibit activation of mTOR and elevated TGFβ1 ligand. Thus, our study will not only reveal molecular mechanisms of Pten and Smad4 in tumor formation and malignant progression of SGT but also identify novel targets for SGT therapy.
Material and Methods
Generation and Characterization of the Inducible Head and Neck–Specific Pten and/or Smad4 Genetically Engineered Mouse Model
All animal experiments were performed in accordance with protocols approved by the Institutional Animal Cancer and Use Committees of the University of Colorado Anschutz Medical Campus. All mouse experiments were performed in a C57BL/6 background. The inducible K5CrePR1 mouse was reported previously [26]. The Pten-floxed (Ptenf/f) mouse has been originally generated by Dr. Hong Wu, from University of California at Los Angeles [27], and was purchased from Jackson Laboratory (Bar Harbor, ME, USA). The Smad4-floxed mice (Smad4f/f) have been provided by Dr. Chuxia Deng at the National Institutes of Health [28]. The K5CrePR1 mice were mated with homozygous Ptenf/f mice or Smad4f/f mice. The offspring, K5CrePR1.Ptenf/wt or K5CrePR1.Smad4f/wt hemizygous mice, were cross-bred to Ptenf/f or Smad4f/f mice, respectively, to generate K5CrePR1.Ptenf/f, K5CrePR1.Smad4f/f mice or K5CrePR1.Ptenf/f.Smad4f/f mice. PCR genotyping was performed at 3 weeks of age using genomic DNA isolated from tail biopsies. The primers used to detect the K5CrePR1 transgene were forward CGGTCGATGCAACGAGTGAT and reverse CCACCGTCAGTACGTGAGAT [26], primers to detect Pten-floxed gene were forward ACTCAAGGCAGGGATGAGC and reverse AATCTAGGGCCTCTTGTGCC [27], and primers to detect Smad4-floxed gene were forward GGGCAGCGTAGCATATAAGA and reverse GACCCAAACGTCACCTTCAC [25].
To delete Pten and/or Smad4 in murine head and neck epithelia, 100 μl of RU486, dissolved in sesame oil (0.2 μg/μl), was applied in the oral cavity of mouse once a day for 5 days as described previously [29]. The resulting mouse lines K5CrePR1.Ptenf/f, K5CrePR1.Smad4f/f, and K5CrePR1.Ptenf/f.Smad4f/f were selected by using genotyping primers specific for K5CrePR1, Pten-floxed, and Smad4-floxed mice as described above.
Generation and Characterization of the Inducible Salivary Gland–Specific Knockout Mouse Model
Mice was anesthetized and placed onto a plate for fixation. We then shaved the neck skin and injected either 0.01% Evans blue or RU486 (100 μg in 50 μl sesame oil, twice a week for consecutive 2 weeks) to the anesthetized mice. Salivary gland–specific delivery was confirmed by visualization of Evans blue dyes. To determine the extent of Cre recombinase activity in murine salivary gland after local delivery of RU486, we crossed the K5CrePR1 mice with the Rosa26tdTomato reporter mouse (Jackson Lab, Bar Harbor, ME), which harbors a lox-stop-lox sequence upstream of a constitutively expressed yellow fluorescence protein (YFP) reporter. Bigenic mice harboring both K5CrePR1 and the Rosa26 reporter were treated with salivary gland delivery of RU486 (100 μg in 50 μl sesame oil) or vehicle twice a week for consecutive 2 weeks and sacrificed 1 month later to examine the YFP activity using a fluorescence scope (Leica, Wetzlar, Germany). To delete Pten and/or Smad4 in murine salivary glands, 100 μg RU486 dissolved in 50 μl sesame oil (2 μg/μl) was applied in the oral cavity of mouse once a day for 5 days as described previously [29]. The resulting mouse lines K5CrePR1.Ptenf/f, K5CrePR1.Smad4f/f, and K5CrePR1.Ptenf/f.Smad4f/f were selected by using genotyping primers specific for K5CrePR1, Pten-floxed, and Smad4 floxed mice as described above. Detection of recombinant PCR products upon Pten or Smad4 deletion was performed using primers for Pten: forward ACTCAAGGCAGGGATGAGC and reverse GCTTGATATCGAATTCCTGCAGC, [27] and primers for Smad4: forward AAGAGCCACAGGTCAAGCAG and reverse GACCCAAACGTCACCTTCAC [25].
Mouse MRI and Harvest
Mice underwent gadolinium-enhanced MRI using the Bruker 4.7-T Pharmacan scanner. Mice were injected with 0.2 mmol/kg gadolinium contrast (Gadobenate Dimeglumine, MultiHance) in volumes not to exceed 0.2 ml. Animals was anesthetized and placed onto a Bruker animal bed and inserted into a Bruker 4.7-T Pharmascan/38-mm whole-body coil [30]. All images/spectra were obtained and processed using Bruker ParaVision 3.2 software.
The general health condition of mice was monitored twice weekly, and their body weight was measured once weekly. Mice were given softened food when there were signs of food intake difficulty or decreased body weight. At the end of animal experiment, mice were euthanized. Murine salivary glands, buccal mucosa, tongue, lymph node, and lung tissues were harvested for histological analysis and immunostaining.
Histology and Immunostaining
Tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned to 5-μm thickness. Sections were stained with H&E and examined for the pathological diagnosis by the head and neck cancer pathologist (S.S.). Immunohistochemistry was performed as described previously [29]. Incubation with primary antibodies was as follows: Keratin 5 (PRB-160P, Convance), smooth muscle actin (M0851, DAKO), Ki67 (ab16667, Abcam, Cambridge, UK), p63 (ab124762, Abcam, Cambridge, UK), and p-mTOR (#2976, Cell Signaling, Danvers, MA). Slides were examined with a Leica microscope, and images were taken using the Q Capture Pro software (Q Imaging).
Double immunofluorescence (IF) was performed as previously described [31] using primary antibodies Pten (#18-0652, Zymed Laboratories, San Francisco, CA) and Smad4 (#9515, Santa Cruz Biotechnology, Santa Cruz, CA) and secondary antibodies Alexa Fluor 488–conjugated goat anti-rabbit IgG (H + L) and Alexa Fluor 594–conjugated goat anti-mouse IgG (H + L) (Molecular Probes, Invitrogen, Carlsbad, CA). Nuclei were labeled with 4,6′-diamidino-2-phenylindole (DAPI, Beyotime Biotech, Jiangsu, China).
RNA Isolation and Quantitative Reverse-Transcription PCR (qRT-PCR)
RNA isolation and cDNA synthesis were performed as described previously [29]. cDNAs were subjected to qRT-PCR using either the SYBR mix (Biorad) or the TaqMan Assays-on-Demand probes (Applied Biosystems, Foster City, CA). qRT-PCR was run on the CFX connect qPCR machine (Biorad, Hercules, CA). β-Actin was used as an internal control. Primer sequences for murine Pten qRT-PCR are forward AATTCCCAGTCAGAGGCGCTATGT and reverse GATTGCAAGTTCCGCCACTGAACA, and primers for murine Smad4 qRT-PCR are forward ACTGCTCAGCCAGCTACTTAC and reverse GGCTGGAATGCAAGCTCATTGTGA. Each sample was examined in triplicate. The relative RNA expression levels were determined by normalizing to the internal control, the values of which were calculated using the comparative Ct method.
Protein Analysis
Whole salivary gland tissues and SGT tissues were lysed using a homogenizer (Pro Scientific, Oxford, CT) in lysis buffer (Roche, Basel, Switzerland) containing a pellet of protease inhibitor cocktail. Protein concentration was measured using Pierce 660-nm protein assay reagent (Thermo, Waltham, MA), and 40 μg of protein lysates was analyzed using the standard Western protocol we have described previously. [32] Blots were incubated with primary antibodies at 4°C overnight and secondary antibodies for 1 hour at room temperature. Western blots were imaged by ECL (Biorad, Hercules, CA) and normalized with respect to GAPDH (#5174, Cell Signaling, Danvers, MA) expression. The primary antibodies were Pten (#9559, Cell Signaling, Danvers, MA) and Smad4 (#9515, Cell Signaling, Danvers, MA). TGFβ1 ELISA was performed as described previously using the R&D systems kit [31].
Patients
All patient samples were collected from the Dalian Medical University and the 9th People's Hospital of Shanghai Jiao Tong University during 2005 to 2013 under the approvals of the protocol by both Institutional Review Boards. Diagnosis of the SGTs in this study was evaluated by head and neck cancer pathologists (J. Li and J. Xiao), and informed consent was obtained from each patient. The samples comprised of a total of 55 SACCs and 24 SPAs. Among the 55 SACC cases, there were 17 tubular, 16 cribriform, and 22 solid forms of SACC cases. Ten normal salivary gland tissues were also included in this study.
Statistical Analysis
Statistical analyses were performed by IBM SPSS Statistics 19.0 package program. For analysis of follow-up data, overall survival were evaluated using the Kaplan-Meier curve, and the differences among the levels of possible prognostic factors were compared by the log-rank test in the univariate analyses. A two-tailed P < .05 was considered statistically significant.
Results
Spontaneous Development of Pleomorphic Adenomas in Salivary Glands of the PtenHN-KO or the Smad4HN-KO Mice
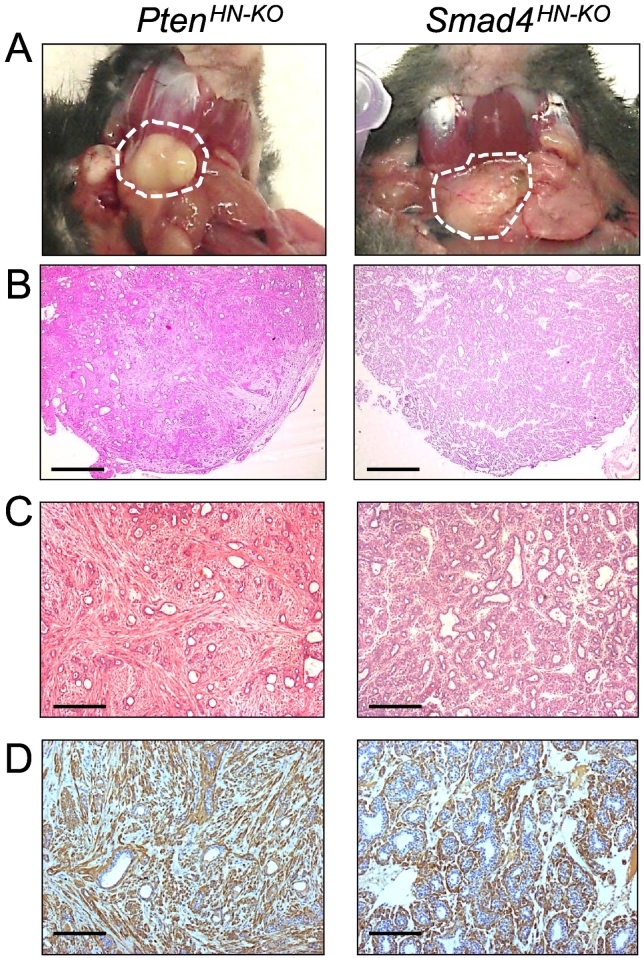
Our initial insight into SGT came from a spontaneous occurrence of SGT in an inducible head and neck–specific knockout mouse model which we used to delete target genes in head and neck epithelia [25], [29], [33]. This model uses a basal epithelial marker, K5 promoter, to drive expression of CrePR1, a fusion protein comprised of Cre recombinase fused to a truncated progesterone receptor ligand binding domain (ΔPR-LBD). Upon RU486 application, the CrePR1 fusion protein translocates into the nucleus where it excises DNA sequences that have been flanked by loxP sites (“floxed”). Thus, this system will delete target gene in the K5-positive basal epithelial cells in the region exposed to RU486 application (Supplementary Figure S1). We have used this system to develop a number of murine models for head and neck squamous cell carcinomas (HNSCCs), including the TGFβ type II receptor [29], its downstream signal mediator Smad4 [25], and a negative regulator of the PI3K pathway, Pten. In addition to HNSCC formation upon oral application of RU486, we observed that some neck nodules developed from the K5CrePR1.Ptenf/f (hereafter referred to as PtenHN-KO) or the K5CrePR1.Smad4f/f (hereafter referred to as Smad4HN-KO) mice. These were originally suspected to be lymph node metastases, but pathological examination showed that they were SGTs. As shown Figure 1A, the neck masses as discrete nodules were observed in the murine salivary glands from either the PtenHN-KO or the Smad4HN-KO mice. Histological examination showed that these tumors are salivary pleomorphic adenomas (SPAs) (Figure 1, B and C). Both SPAs are characterized by their morphological diversities composed of multiple cell types. The SPAs from the PtenHN-KO mice contain more myoepithelial, spindle cells, and mesenchymal components, as exemplified by more smooth muscle actin (SMA) staining, while the SPAs from the Smad4HN-KO mice contain more epithelial components, (Figure 1, C and D).
Figure 1.
Development of SGTs in the PtenHN-KO or the Smad4HN-KO mice.
(A) Representative gross pictures of salivary gland tumors (dotted circles) developed in head and neck–specific Pten knockout mice (left, PtenHN-KO, n = 5) or Smad4 knockout mice (right, Smad4HN-KO, n = 3).
(B) Representative H&E staining at lower magnification of salivary pleomorphic adenomas developed in salivary glands of PtenHN-KO mice (left, n = 5) or Smad4HN-KO mice (right, n = 3). Scale bar: 200 μm.
(C) Representative HE staining at higher magnification of salivary pleomorphic adenomas developed in salivary glands of PtenHN-KO mice (left, n = 5) or Smad4HN-KO mice (right, n = 3). Scale bar: 100 μm.
(D) Representative immunohistochemistry staining of smooth muscle actin (brown) in murine salivary pleomorphic adenomas of PtenHN-KO mice (left, n = 5) or Smad4HN-KO mice (right, n = 3). Scale bar: 50 μm.
Establishment of an Inducible Salivary Gland–Specific Knockout Mouse Model
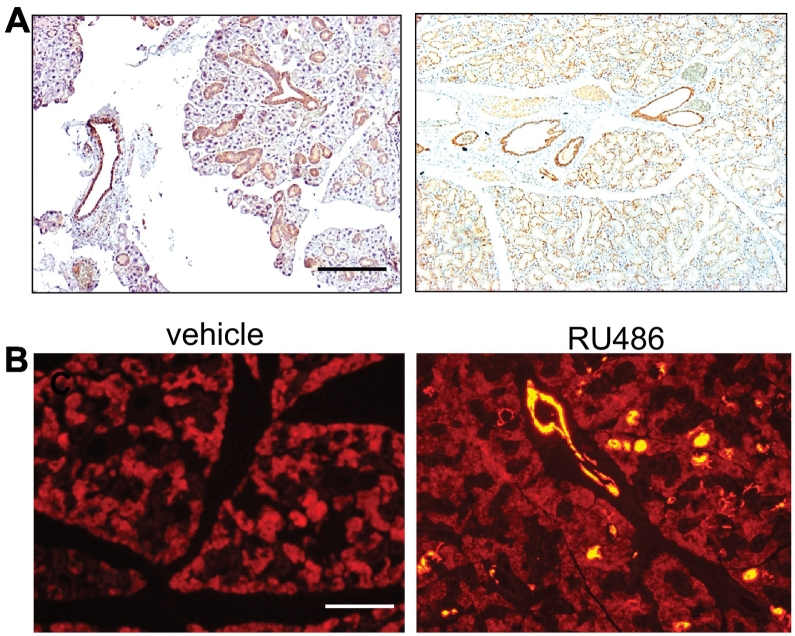
It has been shown that salivary glands, particularly their ductal cells, are lined with K5-positive cells [34]. We suspect that the spontaneous SGTs developed from the PtenHN-KO or Smad4HN-KO mice are due to a leak of RU486 into murine salivary glands through their openings in the oral cavity. This leaked RU486 in murine salivary glands activates the Cre recombinase activity and deletes target gene expression in the K5-positive cells of salivary glands. Our suspicion was further supported by a report that SGTs developed from a conditional expression of Kras driven by K5 promoter [34]. We then examined K5 expression in normal murine salivary glands. As shown in Figure 2A, K5 expression is strongest in the ductal cells and the epithelial cells of mucous salivary gland and is weakest in the epithelial cells from serous salivary gland and acinar cells of salivary gland. We thus postulate that local delivery of RU486 into salivary gland will induce Cre recombinase activity specifically in the K5-positive cells in salivary gland. In mouse, the largest salivary gland is the submandibular gland, which is located just below the skin of murine neck region. We autopsied a few mice to determine the location of submandibular gland. We then shaved the neck skin and injected the anesthetized mice with 0.01% Evans blue to confirm the injection into the salivary gland (Supplementary Figure S2). To determine the extent of Cre recombinase activity in murine salivary gland after local delivery of RU486, we crossed the K5CrePR1 mice with the Rosa26tdTomato reporter mouse, which harbors a lox-stop-lox sequence upstream of a constitutively expressed yellow florescence protein (YFP) reporter. Bigenic mice harboring both K5CrePR1 and the Rosa26 reporter were treated with salivary gland delivery of RU486 or vehicle twice a week for 2 consecutive weeks and sacrificed 1 month later to evaluate the YFP activity. As shown in Figure 2B, mice after salivary gland delivery of RU486 exhibit significantly higher Cre recombinase activity than the vehicle controls.
Figure 2.
Establishment of an inducible salivary gland–specific knockout mouse model.
(A) Immunohistochemistry staining of keratin 5 (brown) in mucous (left) and serous (right) salivary glands of control mice. n = 3, scale bar: 100 μm.
(B) Fluorescence images showing Cre recombinase activity (yellow for YFP fluorescence) in the salivary glands of K5CrePR1.Rosa26.tdTomato reporter mice after salivary gland–specific RU486 delivery (right) compared to vehicle control (left). n = 3 of each, scale bar: 50 μm.
Development of Salivary Pleomorphic Adenoma upon Deletion of Pten or Smad4 in Murine Salivary Gland
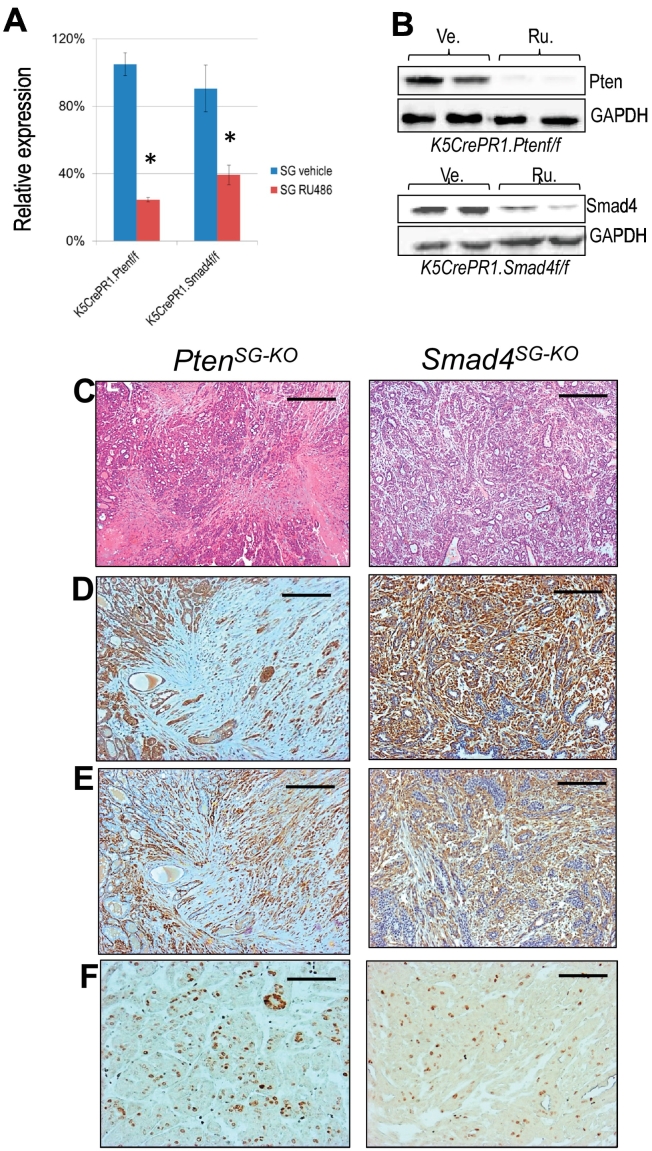
The spontaneous SGT formation from the PtenHN-KO or the Smad4HN-KO mice suggested that Pten or Smad4 may suppress tumor formation in murine salivary gland. However, the fact that ~90% of head and neck tumors are squamous cell carcinomas upon oral application of RU486 forced us to explore the feasibility of salivary gland delivery of RU486 to generate high frequency of SGTs. After establishing the technique and validating the induced Cre recombinase activity upon salivary gland delivery of RU486 as shown in Figure 2, we injected RU486 into the salivary glands of 1-month-old K5CrePR1.Ptenf/f (hereafter referred to as PtenSG-KO) and K5CrePR1.Smad4f/f mice (hereafter referred to as Smad4SG-KO). Mice were euthanized 1 month later, and the salivary glands were collected to evaluate Pten or Smad4 deletion. Upon salivary gland delivery of RU486, we detected the recombinant Pten or Smad4 gene product in the salivary glands from the PtenSG-KO or the Smad4SG-KO mice (Supplementary Figure S3). The mRNA expression level of Pten or Smad4 was significantly reduced in the salivary glands from the PtenSG-KO or the Smad4SG-KO mice upon salivary gland delivery of RU486, respectively (Figure 3A). Western blotting further validated the reduced protein expression of these samples (Figure 3B). These results demonstrate the feasibility of using K5-driven CrePR1 construct and salivary gland delivery of RU486 to delete gene(s) of interest in murine salivary gland. Around 8 to 10 months after salivary gland delivery of RU486, the PtenSG-KO or the Smad4SG-KO mice started to develop palpable neck masses. We performed MRI scan to evaluate its value for SGT diagnosis. As shown in the Supplementary Figure S4, enlarged and hyperintense SGTs were clearly detectable by the axial proton density MRI. We euthanized these tumor-bearing mice and found similar pathological findings as those SGTs developed from the PtenHN-KO or the Smad4HN-KO mice (Figure 3C). Deletion of Pten or Smad4 in murine SG resulted in SPA formation as revealed by both epithelial maker K5 (Figure 3D) and myoepithelial marker SMA (Figure 3E). Although histologically benign, the SPAs from the PtenSG-KO mice tend to contain more stromal and myoepithelial components, with more aggressive histological parameters, such as mitoses and necrosis (not shown), and higher Ki67 index compared to the SPAs from the Smad4SG-KO mice (Figure 3F).
Figure 3.
Deletion of Pten or Smad4 alone in murine salivary gland developed salivary pleomorphic adenoma.
(A) qRT-PCR measurement of Pten or Smad4 mRNA levels in salivary glands of either the K5CrePR1.Ptenf/f mice or the K5CrePR1.Smad4f/f mice treated by vehicle or RU486, n = 3 of each. *P < .01.
(B) Western blotting of Pten or Smad4 in salivary glands of either the K5CrePR1.Ptenf/f mice or the K5CrePR1.Smad4f/f mice treated by vehicle or RU486. n = 2 of each. Ve: vehicle control, Ru: RU486.
(C) Representative H&E staining of salivary pleomorphic adenomas developed in salivary glands of PtenSG-KO mice (left, n = 7) or Smad4SG-KO mice (right, n = 5). Scale bar: 100 μm.
(D) Representative immunohistochemistry of keratin 5 in salivary pleomorphic adenomas developed in salivary glands of PtenSG-KO mice (left, n = 7) or Smad4SG-KO mice (right, n = 5). Scale bar: 50 μm.
(E) Representative immunohistochemistry of smooth muscle actin in salivary pleomorphic adenomas developed in salivary glands of PtenSG-KO mice (left, n = 7) or Smad4SG-KO mice (right, n = 5). Scale bar: 50 μm.
(F) Representative immunohistochemistry of Ki67 in salivary pleomorphic adenomas developed in salivary glands of PtenSG-KO mice (left, n = 7) or Smad4SG-KO mice (right, n = 5). Scale bar: 25 μm.
Development of Malignant SGTs upon Combined Deletion of Pten and Smad4 in Murine Salivary Gland
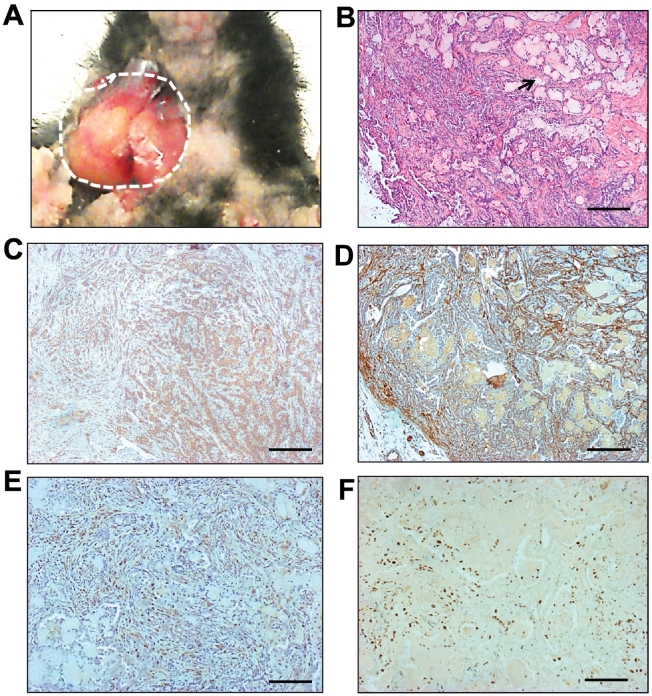
Although the SPAs developed from the PtenSG-KO or the Smad4SG-KO mice represent the most common SGT, it is a benign tumor with favorable prognosis. The malignancies of salivary gland are the main cause for the poor prognosis and survival of SGT patients [1], [35]. Smad4 has been shown to suppress Pten loss–induced prostate cancer progression [36]. This synergistic suppression effect of Pten and Smad4 on tumor progression has also been observed in other cancer types, such as pancreas [37], forestomach [38], and liver [39]. Thus, we predict that combined deletion of Pten and Smad4 may promote malignant progression of murine SGT. Mice harboring K5CrePR1.Ptenf/f.Smad4f/f alleles received salivary gland delivery of RU486 (hereafter referred to as PtenSmad4SG-KO). About 8 to 10 months after the salivary gland delivery of RU486, the PtenSmad4SG-KO mice started to develop palpable neck masses. Dissection of these mice showed tumors occurring from murine salivary gland (Figure 4A). Tumor histology revealed several subtypes of malignant SGTs, with SACCs being the most frequently seen (Table 1). As shown in Figure 4B, the SACCs composed of both epithelial (revealed as K5+ staining in Figure 4C) and myoepithelial cells (revealed as SMA+ staining in Figure 4D) were arranged in a cribriform pattern, with nests of cells containing small, circular cyst-like space, or so-called “Swiss cheese” (arrow in Figure 4B), which is a special pathological characteristic found in human SACCs. The SACC was also positive for the basal cell marker p63 staining (Figure 4E) and exhibited massive Ki67 staining (Figure 4F). In addition to SACC, we also observed salivary ductal carcinoma (Supplementary Figure S5A) and salivary adenosquamous carcinoma (Supplementary Figure S5B) in salivary glands from the PtenSmad4SG-KO mice. The tumor types developed by far from the PtenSG-KO, Smad4SG-KO, and PtenSmad4SG-KO mice as well as the control mice are summarized in Table 1.
Figure 4.
Combined deletion of Pten and Smad4 in murine salivary glands caused malignant SGTs.
(A) Gross picture of a salivary gland tumor (dotted circle) developed in a Pten.Smad4SG-KO mouse.
(B) H&E staining of a murine SACC developed in a Pten.Smad4SG-KO mouse. Arrow showed the “Swiss cheese” change, which is a special pathological characteristic found in human SACCs. Scale bar: 100 μm.
(C) Representative immunohistochemistry staining of keratin 5 (brown) in the murine SACCs (n = 6). Scale bar: 50 μm.
(D) Representative immunohistochemistry staining of smooth muscle actin (brown) in the murine SACCs (n = 6). Scale bar: 50 μm.
(E) Representative immunohistochemistry staining of p63 (brown) in the murine SACCs (n = 6). Scale bar: 50 μm.
(F) Representative immunohistochemistry staining of Ki67 (brown) in the murine SACCs (n = 6). Scale bar: 25 μm.
Table 1.
Summary of Salivary Gland Tumors Developed from Three Salivary Gland–Specific Knockout Mouse Models*
| GEMMs | No. | SGT Types |
|---|---|---|
| PtenSG-KO | 10 | |
| 7 | Salivary pleomorphic adenomas | |
| 3 | Normal | |
| Smad4SG-KO | 12 | |
| 5 | Salivary pleomorphic adenomas | |
| 7 | Normal | |
| PtenSmad4SG-KO | 11 | |
| 6 | Salivary adenoid cystic carcinomas | |
| 3 | Salivary duct carcinomas | |
| 2 | Salivary adenosquamous cell carcinomas | |
| Control mice | 15 | |
| K5CrePR1 | 6 | No tumors |
| Pten-flox | 4 | No tumors |
| Smad4-flox | 5 | No tumors |
All mice were treated with RU486 at 1 month old and harvested from 8 to 10 months after RU486 application.
Activation of mTOR and Increased TGFβ1 Ligand in Murine SGTs
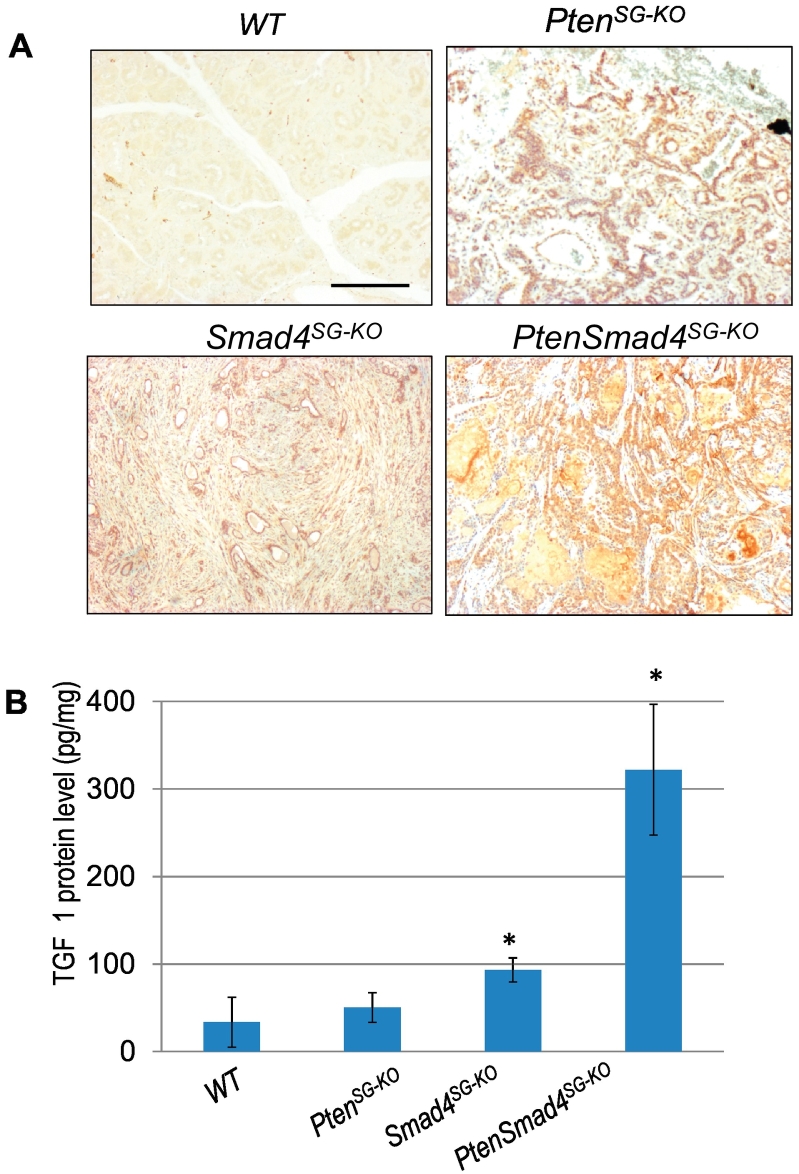
Pten is known as a negative regulator for the PI3K/mTOR pathway [11]. Deletion of Smad4 has also been shown to activate this pathway [40]. To investigate if the downstream mTOR activation is one of the potential mechanisms for mediating Pten or Smad4 loss in the SGTs from our mouse models, we examined p-mTOR by immunohistochemistry. As shown in Figure 5A, the mTOR is ubiquitously activated in both murine SPAs and SACCs from our mouse models. We have shown previously that increased TGFβ1 ligand occurs in HNSCC after Smad4 is deleted in oral epithelia, and promotes HNSCC progression by modulating tumor microenvironment [25]. We then examined the TGFβ1 ligand level by ELISA and found that TGFβ1 is significantly higher in SACCs developed from the PtenSmad4SG-KO mice compared to the SPAs developed from either PtenSG-KO or Smad4SG-KO mice (Figure 5B).
Figure 5.
Activation of mTOR and increased TGFβ1 ligand in murine SGTs.
(A) Immunohistochemistry staining of p-mTOR (brown) in SPAs developed from PtenSG-KO mice (n = 7) or Smad4SG-KO mice (n = 5), and SACCs developed from Pten.Smad4SG-KO mice (n = 6). WT: wild-type. Scale bar: 50 μm.
(B) ELISA measurement of TGFβ1 levels in SPAs developed from PtenSG-KO mice or Smad4SG-KO mice, and SACCs developed from Pten.Smad4SG-KO mice. n = 3 of each group. *P < .01.
Reduced Expression of Pten and Smad4 in Human SGTs
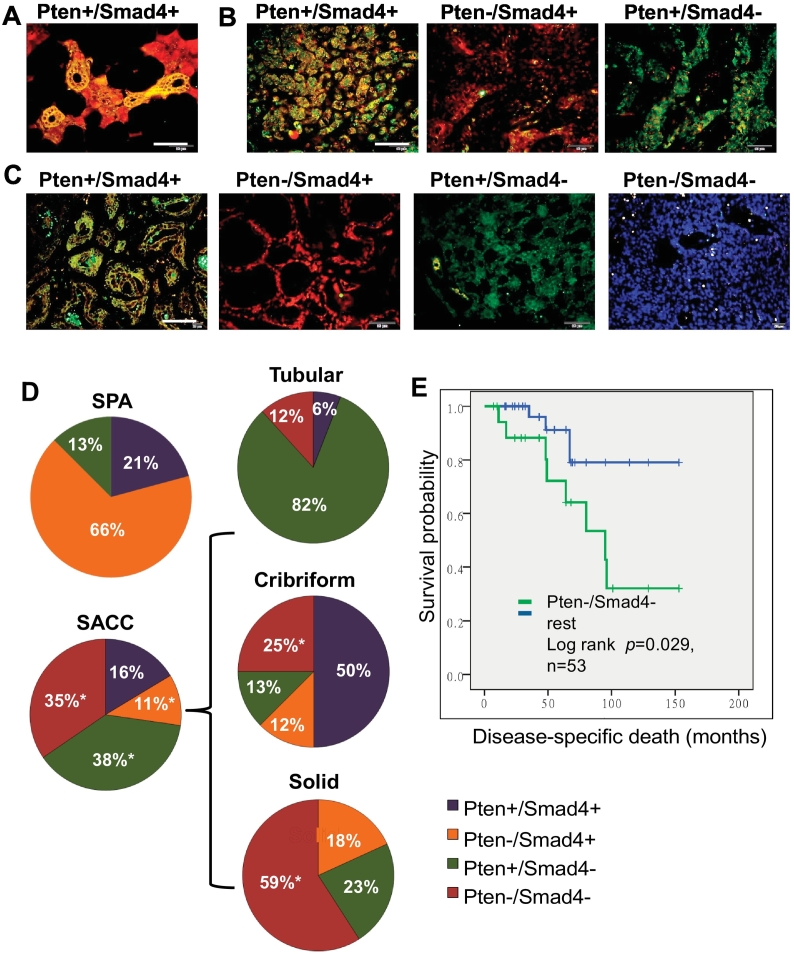
We and others recently showed that reduced Pten expression is a common event in human SACC patients and is correlated with poorer prognosis [12], [18], [19]. In this study, we further examined Smad4 expression and its association with Pten expression by double IF staining in the same cohort of human SGT patients. We first examined the expression pattern of Pten and Smad4 by double IF in normal human salivary glands. Individual IF for generating the double IF and their corresponding DAPI were also included in Supplementary Figure 6. As shown in Figure 6A and Supplementary Figure 6A, a strong cytoplasmic expression of both Smad4 and Pten was observed in ductal cells (yellow). Smad4, but not Pten, was also found to express in seromucous cells (red). We then examined Pten and Smad4 expression by double IF in 24 human SPA cases. Pten loss (Figure 6B and Supplementary Figure 6B, middle panels) was observed in 16 cases (66%, Figure 6D, SPA pie), and Smad4 loss (Figure 6B and Supplementary Figure 6B, right panels) was observed in 3 cases (13%, Figure 6D, SPA pie). There were no cases exhibiting both Pten and Smad4 loss, and there were five cases (21%) exhibiting normal expression of both Pten and Smad4 (Figure 6B and Supplementary Figure 6B, left panels and Figure 6D, SPA pie). Next, we examined Pten and Smad4 expression in 55 human SACC cases. Loss of Pten alone (Figure 6C and Supplementary Figure 6B, second panels) was observed in 6 cases (11%, Figure 6D, SACC pie), and loss of Smad4 alone (Figure 6C and Supplementary Figure 6C, third panels) was observed in 21 cases (38%, Figure 6D, SACC pie). In addition, loss of both Pten and Smad4 expression (Figure 6C and Supplementary Figure 6C, fourth panels) was observed in 19 cases (35%, Figure 6D, SACC pie), and normal expression of both Pten and Smad4 (Figure 6C and Supplementary Figure 6C, first panels) was observed in 9 cases (16%, Figure 6D, SACC pie). We further analyzed the expression of Pten and Smad4 in three forms of human SACC: tubular, cribriform, and solid forms. We found that loss of Pten and Smad4 was most frequent in solid-form SACC (59%) and less so in cribriform (25%) or tubular-form SACC (12%) (Figure 6D, SACC pie). Finally, we performed Kaplan-Meier survival analysis on a total of 53 human SACC cases stratified according to Pten and Smad4 status. We found that SACC patients with loss of both Pten and Smad4 had significantly shorter overall survival compared to all other forms combined (Figure 6E). Taken together, these data suggest that both Pten and Smad4 are required for suppression of human SACC formation and progression and support the human clinical relevance of our SGT mouse models.
Figure 6.
Reduced expression of Pten and/or Smad4 in human salivary gland tumors.
(A) Double IF staining of Pten (green) and Smad4 (red) in normal human salivary glands. Double positive staining of Pten and Smad4 was seen in the ductal cells (yellow); Smad4-positive staining was seen in seromucous cells (red). Scale bar: 50 μm.
(B) Double IF staining of Pten (green) and Smad4 (red) in human SPAs. Examples of double positive staining (yellow) of Pten and Smad4 (Pten+/Smad4+) is shown in the left panel, Pten-negative staining alone (red, Pten−/Smad4+) is shown in the middle panel, and Smad4-negative staining alone (green, Pten+/Smad4−) is shown in the right panel. Scale bar: 50 μm.
(C) Double IF staining of Pten (green) and Smad4 (red) in human SACCs. Examples of double positive staining (yellow) of Pten and Smad4 (Pten+/Smad4+) is shown in the first panel, Pten-negative staining alone (red, Pten−/Smad4+) is shown in the second panel, Smad4-negative staining alone (green, Pten+/Smad4−) is shown in the third panel, and double negative staining (blue, Pten−/Smad4−) is shown in the fourth panel. Scale bar: 50 μm.
(D) Pie chart of summary results of Pten and Smad4 double IF staining in human SPAs and SACCs. Double IF results from three forms of SACC, i.e., tubular, cribriform, and solid, are further shown in individual pie chart. Positive cases from each category are shown as percentage along each pie chart. *P < .01.
(E) Kaplan-Meier survival analysis in a total of 53 human SACC cases based on their Pten and Smad4 status.
Discussion
In this study, we reported a salivary gland–specific genetically engineered mouse model (GEMM), which developed the most common benign SGT, i.e., SPA, or several malignancies of SGTs, with the SACC the most commonly seen. One question is that if there are any influences of background SGTs reported previously by Sundberg et al. on the SGTs developed from our GEMMs [41]. We think that is unlikely the case for the following reasons: 1) There were no SGTs developed in the control mice of our study. 2) The incidence of background mouse SGT in the Sundberg's paper was 16/100,000, which is way lower than that in SGTs developed in our GEMMs. 3) The genetic background for developing background mouse SGTs in the Sundberg's paper was either AJ or BALB/C (Th2 type) mice. In contrast, the genetic background of our mice is C57BL6 (Th1 type).
Currently, most SGT mouse models are established by orthotopic injection of SGT cells, which cannot faithfully recapitulate the real situation of SGT formation and progression in humans [42]. SGT-GEMMs that faithfully recapitulate the natural course of formation and progression of human SGT have not been well established. Occasionally, SGTs develop in MMTV-driven mammary cancer mouse models [43], [44], [45]. In these instances, SGT was found to occur as a result of leaky expression in the secretory cells of murine salivary glands. This is further exemplified in a recent report of acinic cell carcinoma developing through a combination of Apc and Pten deletion [46]. Although their data demonstrated the in vivo role of oncogenes and tumor suppressor genes in SGT pathogenesis, these mice often develop a higher incidence of breast cancers since the MMTV promoter targets genes predominantly in mammary glands rather than salivary glands. Similarly, Raimondi et al. observed development of a few SGTs in addition to the expected squamous cell carcinomas in a Kras model driven by the K5 promoter. Although this study addressed SGTs originating from K5+ cells, the majority of SCCs developed in skin and other squamous epithelial cells due to ubiquitous expression of K5-positive cells in these organs [34]. Recently, Fu et al. reported an interesting model which developed a rare SDC variant sarcomatoid type when expressing a Kras mutant driven by an elastase I promoter [47]. The tumor developed and progressed very rapidly. However, the SGT developed from this model is extremely rare, the median survival is only 28 days, and the model also developed other systemic diseases such as pancreatic fibrosis; limit the applications of this model to study more common SGTs, such SACC, and testing therapies on this short-lived model. Development of SGTs was also seen in Justy mutant mouse strain. This stain bears a mutation in the Gon4l gene, which is a nuclear factor implicated in transcriptional regulation [48]. While the spontaneous development of SGTs under these genetic changes significantly suggests the role of these genes in the SGT pathogenesis and reflects a genetic heterogeneity of molecular alterations in SGT development, the human relevance of these alterations in human SGTs and underlying mechanistic studies would further define the function of each gene in SGT pathogenesis and translate into personalized target(s) for therapy based on their genetic alterations. The salivary gland–specific GEMMs we have developed overcome these problems. By application of RU486 directly into salivary glands, deletion of the gene(s) of interest occurs specifically in the salivary glands, which significantly improves the penetrance of developing SGTs and avoids generation of tumors in other organs. This novel salivary gland–specific knockout mouse model will allow us to evaluate the role of a clinically relevant tumor suppressor and to study the full spectrum of tumor formation and progression in salivary gland.
One question is why there was no tumor formation in Pten deletion driven by MMTV [46], but SPA formation by K5 in this study. This difference may be related to the cell origin in development of each type of SGTs. MMTV promoter targets mostly the secretory cells, while the K5 promoter predominantly targets basal layer of ductal cells of salivary glands. Thus, the difference of tumor developed in Pten deletion between K5 promoter versus MMTV promoter is more likely due to different cell type in which the Pten deletion is targeted. Another interesting question is why the same combination of gene knockout, i.e., deletion of Pten and Smad4, causes three distinct histopathological forms of SGTs, i.e., SACC, salivary ductal carcinoma, and salivary adenosquamous cell carcinoma, but not other common SGTs, such as mucoepidermiod carcinoma or acinic cell carcinoma. One possibility may be due to the tissue-specific promoter of K5, targeting gene deletion specifically in the K5+ cells, which are most expressed in the basal ductal but not acinar cells. Besides, K5-positive cells in salivary gland possess progenitor property, which suggests that SACC, salivary ductal carcinoma, and salivary adenosquamous carcinoma may develop from the same, such as K5+ cell origin, while mucoepidermoid carcinoma or acinic cell carcinoma may come from non-K5 progenitor cells [49]. The other possibility is that these three forms of SGTs may have similar molecular alterations in both Pten and Smad4 in humans, while other SGTs may not. Further molecular characterization of Pten and Smad4 at both genetic and expression levels in these SGTs is warranted.
Application of next-generation sequencing on human SGT samples has shown that the most common molecular alterations are p53, followed by Ras, PIK3CA, and Smad4 [8]. It would be interesting to generate p53 mutant mice either alone or in combination with other genetic alterations using the salivary gland–specific knockout system described in this manuscript to better understand the role of p53 mutation in SGT development and progression. However, although p53 mutation is the most common, the frequency of it in salivary gland cancers is around 30%. One of the most prominent finding from the Grunewald's study is the genetic heterogeneity revealed by sequencing, suggesting that personalized targets for therapy according to each mutation may be more effective [8]. In this scenario, molecular alterations of Pten or Smad4 have been shown in several publications including the Grunewald's study [8], our previous publication [12], and human data in this manuscript. These human relevant data suggest that the tumor formation driven by Pten and/Smad4 loss contributes to at least a part of SGT pathogenesis. K5 promoter targets gene of interest in the basal layer of ductal cells of salivary gland. It is the gene(s) of interest that exerts its function, but not K5 itself. In addition, the K5.CrePR1 alone, without Pten and/or Smad4 deletion, did not have tumor formation (Table 1), further emphasizing the role of Pten and/or Smad4, but not K5, in SGT pathogenesis.
An obvious and critical question to be answered now is: How are our SGT-GEMMs resulting from deletion of Pten and/or Smad4 relevant to human SGT patients? Molecular characterization on SGT samples revealed that, in addition to commonly seen chromosome translocations, molecular alterations of Pten and/or Smad4 occur in human SGTs. Three independent studies reported Pten mutations in human SGTs, particularly in SACC, SDC, and CxPA [8], [9], [10]. In addition, genetic loss or SGT-reduced Pten expression has been reported in human SGTs. We and others have shown that loss or reduced expression of Pten is common (~50%) in several human subtypes [12], [16], [17]. Similarly, Smad4 mutations have been found in human SGTs [8]. To further validate these findings and compare results from our mouse studies with human SGT patients, we further analyzed the expression of Pten and Smad4 in SPA and three forms of human ACC: tubular, cribriform, and solid forms. We found that loss of both Pten and Smad4 is more frequent in human SACCs, particularly in solid form, and is significantly associated with shorter overall survival. These data support the relevance of our mouse models to human SGTs and provide the unique SGT-GEMMs we have generated to determine the functional roles and molecular mechanisms of Pten and Smad4 in suppressing SGT formation and progression.
Given the poor understanding of the molecular mechanisms of SGT formation and progression, it is not surprising that there are no established diagnostic or prognostic biomarkers for SGTs. Human SGTs are extremely heterogeneous both pathologically and genetically. Different environmental exposure of human patients makes such analysis even more difficult and would require a restrictively large sample size. To this point, the SGTs developed from our salivary gland–specific mouse models provide innovative resources for identifying such biomarkers since these tumors occur naturally in a homogenous genetic background and closely mimic human SGT. Application of advanced genome-wide profiling of these murine SGTs will identify novel candidate genes capable of being translated into innovative diagnostic or prognostic markers for human SGT patients.
Current therapeutic options for human SGT are still very limited, in part, because the molecular mechanisms that contribute to SGT pathogenesis are largely unknown. Depending on their location, some SGTs are difficult to remove completely. Radiation therapy is controversial and not recommended for clinical treatment. The effects of current chemotherapies prove to be disappointing [2], [50]. Thus, the development of novel, effective therapies to treat patients with SGTs is critical for improving current therapeutic protocol. Here, we developed the capacity to assess the involvement of Pten and Smad4 in SGT pathogenesis through development of SGT-GEMMs in which Pten and Smad4 are specifically deleted in salivary gland. Characterization of SGT showed mTOR activation and TGFβ1 increase upon Pten and Smad4 deletion in our study. This mouse model positions us to dissect the roles of Pten and Smad4 in suppressing SGT development and will provide an ideal preclinical platform to test the effect of mTOR and TGFβ pathway inhibition, both of which are actively being developed and tested in clinical trials on multiple human cancers [51], [52].
Acknowledgments
Acknowledgements
This work is supported by National Institutes of Health grant R01DE021788 (to S.L. Lu), University of Colorado Academic Enrichment Fund (to S.L. Lu), Cancer League of Colorado (to S.L. Lu), and the National Natural Science Foundation of China 81272431 (to S.L. Lu). S.L. Lu is an investigator of THANC foundation. The authors thank Dr. Linda Johnson, a veterinary pathologist at the University of Colorado, for helping to answer one of the reviewer's question and the University of Colorado Skin Disease Research Center Morphology Phenotyping Core (P30 AR057212) for assisting with histological work.
Consent for Publication
All authors have agreed for this publication.
Conflict of Interests
The authors declare that they have no conflict interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2018.05.009.
Contributor Information
Jing Xiao, Email: xiaoj@dlmedu.edu.cn.
Shi-Long Lu, Email: shi-long.lu@ucdenver.edu.
Appendix A. Supplementary Data
Figure S1. Schematic of the head and neck inducible knockout mouse model.
Figure S2. Pictures showing murine salivary gland–specific delivery of RU486.
Left: Picture of RU486 injection into murine salivary gland (arrows).
Right: Picture of Evan blue dye to show injection into murine salivary gland. NM: neck muscle.
Figure S3. Validation of Pten or Smad4 deletion in the salivary glands from either the K5CrePR1.Ptenf/f mice or the K5CrePR1.Smad4f/f mice by deletion specific recombinant PCRs. VE: vehicle, RU: RU486
Figure S4. MRI images of murine salivary glands in control mice (upper) and PtenSG-KO mice (lower). N=3 of each group. Arrows: murine salivary glands.
Figure S5. Development of malignant salivary gland tumors in the Pten.Smad4SG-KO mice. (A) H&E staining of a murine salivary duct carcinoma developed in a Pten.Smad4SG-KO mouse. Scale bar: 100 μm.
(B) H&E staining of a murine salivary adenosquamous cell carcinoma developed in a Pten.Smad4SG-KO mouse. Scale bar: 100 μm.
Figure S6. Double IF (Ptenb&bSmad4, yellow), single IF of Pten (Green) or Smad4 (Red), and the corresponding DAPI staining (blue) in normal salivary glands (A), pleomorphic adenomas (B), and salivary adenoid cystic carcinomas (C).
References
- 1.Dunn LA, Ho AL, Laurie SA, Pfister DG. Unmet needs for patients with salivary gland cancer. Oral Oncol. 2016;60:142–145. doi: 10.1016/j.oraloncology.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Bell D, Hanna EY. Salivary gland cancers: biology and molecular targets for therapy. Curr Oncol Rep. 2012;14:166–174. doi: 10.1007/s11912-012-0220-5. [DOI] [PubMed] [Google Scholar]
- 3.Seethala RR. Salivary gland tumors: current concepts and controversies. Surg Pathol Clin. 2017;10:155–176. doi: 10.1016/j.path.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Seethala RR, Stenman G. Update from the 4th edition of the World Health Organization classification of head and neck tumours: tumors of the salivary gland. Head Neck Pathol. 2017;11:55–67. doi: 10.1007/s12105-017-0795-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coca-Pelaz A, Rodrigo JP, Bradley PJ, Vander Poorten V, Triantafyllou A, Hunt JL, Strojan P, Rinaldo A, Haigentz M, Jr., Takes RP. Adenoid cystic carcinoma of the head and neck—an update. Oral Oncol. 2015;51:652–661. doi: 10.1016/j.oraloncology.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Keller G, Steinmann D, Quaas A, Grunwald V, Janssen S, Hussein K. New concepts of personalized therapy in salivary gland carcinomas. Oral Oncol. 2017;68:103–113. doi: 10.1016/j.oraloncology.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 7.Yin LX, Ha PK. Genetic alterations in salivary gland cancers. Cancer. 2016;122:1822–1831. doi: 10.1002/cncr.29890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grunewald I, Vollbrecht C, Meinrath J, Meyer MF, Heukamp LC, Drebber U, Quaas A, Beutner D, Huttenbrink KB, Wardelmann E. Targeted next generation sequencing of parotid gland cancer uncovers genetic heterogeneity. Oncotarget. 2015;6:18224–18237. doi: 10.18632/oncotarget.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N, Ramaswami D, Walsh LA, Eng S, Huse JT. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;45:791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Russell JS, McDermott JD, Elvin JA, Khaira D, Johnson A, Jennings TA, Ali SM, Murray M, Marshall C. Profiling of 149 salivary duct carcinomas, carcinoma ex pleomorphic adenomas, and adenocarcinomas, not otherwise specified reveals actionable genomic alterations. Clin Cancer Res. 2016;22:6061–6068. doi: 10.1158/1078-0432.CCR-15-2568. [DOI] [PubMed] [Google Scholar]
- 11.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Du L, Wang R, Wei C, Liu B, Zhu L, Liu P, Liu Q, Li J, Lu SL. High frequency of loss of PTEN expression in human solid salivary adenoid cystic carcinoma and its implication for targeted therapy. Oncotarget. 2015;6:11477–11491. doi: 10.18632/oncotarget.3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D, Zhang B, Kang J, Ma X, Lu Y, Gong L. Expression and clinical significance of FAK, ILK, and PTEN in salivary adenoid cystic carcinoma. Acta Otolaryngol. 2013;133:203–208. doi: 10.3109/00016489.2012.728295. [DOI] [PubMed] [Google Scholar]
- 14.Kleist B, Poetsch M, Breitsprecher C, Dusterbehn G, Donath K, Lorenz G. Epithelial-myoepithelial carcinoma of the parotid gland—evidence of contrasting DNA patterns in two different histological parts. Virchows Arch. 2003;442:585–590. doi: 10.1007/s00428-003-0800-8. [DOI] [PubMed] [Google Scholar]
- 15.Villeneuve H, Tremblay S, Galiatsatos P, Hamel N, Guertin L, Morency R, Tischkowitz M. Acinic cell carcinoma of the retromolar trigone region: expanding the tumor phenotype in Cowden syndrome? Familial Cancer. 2011;10:691–694. doi: 10.1007/s10689-011-9472-8. [DOI] [PubMed] [Google Scholar]
- 16.Ettl T, Baader K, Stiegler C, Muller M, Agaimy A, Zenk J, Kuhnel T, Gosau M, Zeitler K, Schwarz S. Loss of PTEN is associated with elevated EGFR and HER2 expression and worse prognosis in salivary gland cancer. Br J Cancer. 2012;106:719–726. doi: 10.1038/bjc.2011.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffith CC, Seethala RR, Luvison A, Miller M, Chiosea SI. PIK3CA mutations and PTEN loss in salivary duct carcinomas. Am J Surg Pathol. 2013;37:1201–1207. doi: 10.1097/PAS.0b013e3182880d5a. [DOI] [PubMed] [Google Scholar]
- 18.Ach T, Zeitler K, Schwarz-Furlan S, Baader K, Agaimy A, Rohrmeier C, Zenk J, Gosau M, Reichert TE, Brockhoff G. Aberrations of MET are associated with copy number gain of EGFR and loss of PTEN and predict poor outcome in patients with salivary gland cancer. Virchows Arch. 2013;462:65–72. doi: 10.1007/s00428-012-1358-0. [DOI] [PubMed] [Google Scholar]
- 19.Locati LD, Perrone F, Cortelazzi B, Lo Vullo S, Bossi P, Dagrada G, Quattrone P, Bergamini C, Potepan P, Civelli E. Clinical activity of androgen deprivation therapy in patients with metastatic/relapsed androgen receptor-positive salivary gland cancers. Head Neck. 2016;38:724–731. doi: 10.1002/hed.23940. [DOI] [PubMed] [Google Scholar]
- 20.Malkoski SP, Wang XJ. Two sides of the story? Smad4 loss in pancreatic cancer versus head-and-neck cancer. FEBS Lett. 2012;586:1984–1992. doi: 10.1016/j.febslet.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lourenco SV, Uyekita SH, Lima DM, Soares FA. Developing human minor salivary glands: morphological parallel relation between the expression of TGF-beta isoforms and cytoskeletal markers of glandular maturation. Virchows Arch. 2008;452:427–434. doi: 10.1007/s00428-007-0552-y. [DOI] [PubMed] [Google Scholar]
- 22.Hall BE, Zheng C, Swaim WD, Cho A, Nagineni CN, Eckhaus MA, Flanders KC, Ambudkar IS, Baum BJ, Kulkarni AB. Conditional overexpression of TGF-beta1 disrupts mouse salivary gland development and function. Lab Investig. 2010;90:543–555. doi: 10.1038/labinvest.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuse C, Miguita L, Rosa AC, Soares AB, Martinez EF, Altemani A, de Araujo VC. Study of growth factors and receptors in carcinoma ex pleomorphic adenoma. J Oral Pathol Med. 2010;39:540–547. doi: 10.1111/j.1600-0714.2009.00858.x. [DOI] [PubMed] [Google Scholar]
- 24.Dong L, Wang YX, Li SL, Yu GY, Gan YH, Li D, Wang CY. TGF-beta1 promotes migration and invasion of salivary adenoid cystic carcinoma. J Dent Res. 2011;90:804–809. doi: 10.1177/0022034511401407. [DOI] [PubMed] [Google Scholar]
- 25.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, Reh D, Andersen P, Gross N, Olson S. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119:3408–3419. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Z, Wang D, Wang XJ, Roop DR. In utero activation of K5.CrePR1 induces gene deletion. Genesis. 2002;32:191–192. doi: 10.1002/gene.10064. [DOI] [PubMed] [Google Scholar]
- 27.Lesche R, Groszer M, Gao J, Wang Y, Messing A, Sun H, Liu X, Wu H. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis. 2002;32:80–81. doi: 10.1002/gene.10029. [DOI] [PubMed] [Google Scholar]
- 29.Lu SL, Herrington H, Reh D, Weber S, Bornstein S, Wang D, Li AG, Tang CF, Siddiqui Y, Nord J. Loss of transforming growth factor-beta type II receptor promotes metastatic head-and-neck squamous cell carcinoma. Genes Dev. 2006;20:1331–1342. doi: 10.1101/gad.1413306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Troiani T, Serkova NJ, Gustafson DL, Henthorn TK, Lockerbie O, Merz A, Long M, Morrow M, Ciardiello F, Eckhardt SG. Investigation of two dosing schedules of vandetanib (ZD6474), an inhibitor of vascular endothelial growth factor receptor and epidermal growth factor receptor signaling, in combination with irinotecan in a human colon cancer xenograft model. Clin Cancer Res. 2007;13:6450–6458. doi: 10.1158/1078-0432.CCR-07-1094. [DOI] [PubMed] [Google Scholar]
- 31.Lu SL, Reh D, Li AG, Woods J, Corless CL, Kulesz-Martin M, Wang XJ. Overexpression of transforming growth factor beta1 in head and neck epithelia results in inflammation, angiogenesis, and epithelial hyperproliferation. Cancer Res. 2004;64:4405–4410. doi: 10.1158/0008-5472.CAN-04-1032. [DOI] [PubMed] [Google Scholar]
- 32.Weber SM, Bornstein S, Li Y, Malkoski SP, Wang D, Rustgi AK, Kulesz-Martin MF, Wang XJ, Lu SL. Tobacco-specific carcinogen nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone induces AKT activation in head and neck epithelia. Int J Oncol. 2011;39:1193–1198. doi: 10.3892/ijo.2011.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu SL, Herrington H, Wang XJ. Mouse models for human head and neck squamous cell carcinomas. Head Neck. 2006;28:945–954. doi: 10.1002/hed.20397. [DOI] [PubMed] [Google Scholar]
- 34.Raimondi AR, Vitale-Cross L, Amornphimoltham P, Gutkind JS, Molinolo A. Rapid development of salivary gland carcinomas upon conditional expression of K-ras driven by the cytokeratin 5 promoter. Am J Pathol. 2006;168:1654–1665. doi: 10.2353/ajpath.2006.050847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, Raza S, Rosenbaum S, Van den Veyver I, Milosavljevic A. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X, Ehdaie B, Ohara N, Yoshino T, Deng CX. Synergistic action of Smad4 and Pten in suppressing pancreatic ductal adenocarcinoma formation in mice. Oncogene. 2010;29:674–686. doi: 10.1038/onc.2009.375. [DOI] [PubMed] [Google Scholar]
- 38.Teng Y, Sun AN, Pan XC, Yang G, Yang LL, Wang MR, Yang X. Synergistic function of Smad4 and PTEN in suppressing forestomach squamous cell carcinoma in the mouse. Cancer Res. 2006;66:6972–6981. doi: 10.1158/0008-5472.CAN-06-0507. [DOI] [PubMed] [Google Scholar]
- 39.Xu X, Kobayashi S, Qiao W, Li C, Xiao C, Radaeva S, Stiles B, Wang RH, Ohara N, Yoshino T. Induction of intrahepatic cholangiocellular carcinoma by liver-specific disruption of Smad4 and Pten in mice. J Clin Invest. 2006;116:1843–1852. doi: 10.1172/JCI27282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiao W, Li AG, Owens P, Xu X, Wang XJ, Deng CX. Hair follicle defects and squamous cell carcinoma formation in Smad4 conditional knockout mouse skin. Oncogene. 2006;25:207–217. doi: 10.1038/sj.onc.1209029. [DOI] [PubMed] [Google Scholar]
- 41.Sundberg JP, Hanson CA, Roop DR, Brown KS, Bedigian HG. Myoepitheliomas in inbred laboratory mice. Vet Pathol. 1991;28:313–323. doi: 10.1177/030098589102800408. [DOI] [PubMed] [Google Scholar]
- 42.El-Naggar AK, Kaye FJ, Shirazi Y, Gutkind JS, Forastiere AA. Meeting report—the NIDCR 2(nd) Salivary Gland Tumor Meeting, November 2008. Head Neck. 2009;31:1542–1543. doi: 10.1002/hed.21307. [DOI] [PubMed] [Google Scholar]
- 43.Adnane J, Jackson RJ, Nicosia SV, Cantor AB, Pledger WJ, Sebti SM. Loss of p21WAF1/CIP1 accelerates Ras oncogenesis in a transgenic/knockout mammary cancer model. Oncogene. 2000;19:5338–5347. doi: 10.1038/sj.onc.1203956. [DOI] [PubMed] [Google Scholar]
- 44.Brodie SG, Xu X, Li C, Kuo A, Leder P, Deng CX. Inactivation of p53 tumor suppressor gene acts synergistically with c-neu oncogene in salivary gland tumorigenesis. Oncogene. 2001;20:1445–1454. doi: 10.1038/sj.onc.1204222. [DOI] [PubMed] [Google Scholar]
- 45.Declercq J, Van Dyck F, Braem CV, Van Valckenborgh IC, Voz M, Wassef M, Schoonjans L, Van Damme B, Fiette L, Van de Ven WJ. Salivary gland tumors in transgenic mice with targeted PLAG1 proto-oncogene overexpression. Cancer Res. 2005;65:4544–4553. doi: 10.1158/0008-5472.CAN-04-4041. [DOI] [PubMed] [Google Scholar]
- 46.Diegel CR, Cho KR, El-Naggar AK, Williams BO, Lindvall C. Mammalian target of rapamycin-dependent acinar cell neoplasia after inactivation of Apc and Pten in the mouse salivary gland: implications for human acinic cell carcinoma. Cancer Res. 2010;70:9143–9152. doi: 10.1158/0008-5472.CAN-10-1758. [DOI] [PubMed] [Google Scholar]
- 47.Fu Y, Cruz-Monserrate Z, Helen Lin H, Chung Y, Ji B, Lin SM, Vonderfecht S, Logsdon CD, Li CF, Ann DK. Ductal activation of oncogenic KRAS alone induces sarcomatoid phenotype. Sci Rep. 2015;5 doi: 10.1038/srep13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simons AL, Lu P, Gibson-Corley KN, Robinson RA, Meyerholz DK, Colgan JD. The Justy mutant mouse strain produces a spontaneous murine model of salivary gland cancer with myoepithelial and basal cell differentiation. Lab Investig. 2013;93:711–719. doi: 10.1038/labinvest.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rugel-Stahl A, Elliott ME, Ovitt CE. Ascl3 marks adult progenitor cells of the mouse salivary gland. Stem Cell Res. 2012;8:379–387. doi: 10.1016/j.scr.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milano A, Longo F, Basile M, Iaffaioli RV, Caponigro F. Recent advances in the treatment of salivary gland cancers: emphasis on molecular targeted therapy. Oral Oncol. 2007;43:729–734. doi: 10.1016/j.oraloncology.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen SA, Walker D, Gillespie MB, Gutkind JS, Day TA. mTOR inhibitors and its role in the treatment of head and neck squamous cell carcinoma. Curr Treat Options in Oncol. 2012;13:71–81. doi: 10.1007/s11864-011-0180-2. [DOI] [PubMed] [Google Scholar]
- 52.Korpal M., Kang Y. Targeting the transforming growth factor-beta signalling pathway in metastatic cancer. Eur J Cancer. 2015;46:1232–1240. doi: 10.1016/j.ejca.2010.02.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Schematic of the head and neck inducible knockout mouse model.
Figure S2. Pictures showing murine salivary gland–specific delivery of RU486.
Left: Picture of RU486 injection into murine salivary gland (arrows).
Right: Picture of Evan blue dye to show injection into murine salivary gland. NM: neck muscle.
Figure S3. Validation of Pten or Smad4 deletion in the salivary glands from either the K5CrePR1.Ptenf/f mice or the K5CrePR1.Smad4f/f mice by deletion specific recombinant PCRs. VE: vehicle, RU: RU486
Figure S4. MRI images of murine salivary glands in control mice (upper) and PtenSG-KO mice (lower). N=3 of each group. Arrows: murine salivary glands.
Figure S5. Development of malignant salivary gland tumors in the Pten.Smad4SG-KO mice. (A) H&E staining of a murine salivary duct carcinoma developed in a Pten.Smad4SG-KO mouse. Scale bar: 100 μm.
(B) H&E staining of a murine salivary adenosquamous cell carcinoma developed in a Pten.Smad4SG-KO mouse. Scale bar: 100 μm.
Figure S6. Double IF (Ptenb&bSmad4, yellow), single IF of Pten (Green) or Smad4 (Red), and the corresponding DAPI staining (blue) in normal salivary glands (A), pleomorphic adenomas (B), and salivary adenoid cystic carcinomas (C).