Abstract
Growth differentiation factor 15 (GDF15), a member of the TGF-β superfamily of cytokines, has been reported to exert very heterogeneous functions in various tumors. However, its expression and roles in mediating non-small-cell lung cancer (NSCLC) progression remain unknown. In this study, we found that GDF15 is downregulated in paired NSCLC tissues and correlated with poor clinical outcomes in NSCLC. A functional experiment demonstrated that overexpression of GDF15 significantly repressed NSCLC proliferation both in vitro and in vivo. Mechanistic studies reveal that inhibition of EZH2 expression prevented its binding to the GDF15 promoter region and reduced the trimethylation modification pattern of H3K27. Together, our data uncover that GDF15 is a direct target of EZH2 and, as a regulator of proliferation, might serve as a candidate prognostic biomarker and target for new therapies in human NSCLC.
Keywords: EZH2, epigenetic, GDF15, cell proliferation, NSCLC
Introduction
Lung cancer is one of the leading causes of cancer-related mortality worldwide. Non-small-cell lung cancer (NSCLC), which is the dominant form of lung cancer, is an early asymptomatic disease that constitutes a major global health problem. Despite the development of progressive therapeutic strategies in lung cancer research, the 5-year survival rate among NSCLC patients remains around 15% of diagnosed cases.1, 2 Therefore, a better understanding of the molecular mechanisms underlying NSCLC development and progression is needed to develop more effective therapeutic options.
Growth differentiation factor 15 (GDF15), also known as macrophage inhibitory cytokine-1 (MIC-1), NSAID-activated gene (NAG-1), placental transformation growth factor-β (PTGFβ), and placental bone morphogenetic protein (PLAB), is a secreted protein with homology to members of the transforming growth factor β (TGF-β) superfamily.3 GDF15 is highly expressed in the placenta and adult prostate and at lower levels in fetal brain, liver, kidney, and pancreas.4, 5, 6, 7 Previous investigations revealed that GDF15 is an important regulator of intestinal adenoma growth and may act as a tumor suppressor gene.8, 9 Additionally, some nonsteroidal anti-inflammatory drugs (NSAIDs) induce apoptosis in colon cancer cells, which is possibly mediated by induction of GDF15.3, 10, 11 GDF15 has also been reported to reduce cell adhesion and induce caspase-dependent apoptosis in prostate cancer.12 In malignant gliomas, GDF-15 can promote cell proliferation and protects malignant glioma cells from natural killer (NK) and T cell-mediated cytotoxicity.13 Similarly, GDF15 could suppress macrophage surveillance, which is regulated by nuclear factor κB (NF-κB) during early human pancreatic adenocarcinoma development.14 These findings suggest that the tissue-specific GDF15 exerts diverse biological functions in tumors that remain largely obscure. To date, the role and detailed mechanisms of GDF15 in NSCLC tumorigenesis have not been elucidated.
In the present study, we aimed to explore whether GDF15 could be a key tumor suppressor gene which epigenetically repression mediated by EZH2 in NSCLC, and our findings provide new insights into the mechanisms of NSCLC tumorigenesis and support the potential of GDF15 as a therapeutic target in NSCLC treatment.
Results
GDF15 Expression Was Downregulated in NSCLC Tissues and Associated with a Poor Prognosis
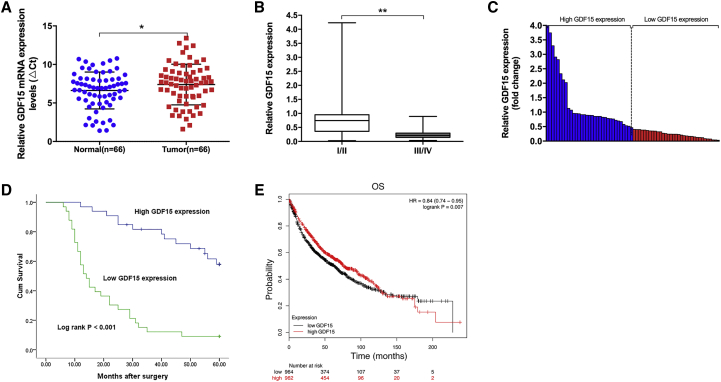
To initiate our investigation of GDF15, we first determined the GDF15 expression levels in 66 paired NSCLC samples and normal lung tissues by qRT-PCR, normalizing to GAPDH expression. The ρCt value was determined by subtracting the GAPDH Ct value from the GDF15 Ct value. Smaller ρCt values indicate higher expression. As shown in Figure 1A, GDF15 expression was significantly downregulated in 84.8% (56 of 66) cancerous tissues compared with normal counterparts. A paired t test was further performed, demonstrating a significant difference between NSCLC cancer tissues and normal counterparts (0.7749 ± 0.90148, p = 0.047). Next we explored the correlation between GDF15 expression and the clinicopathological factors of patients with NSCLC. In general, GDF15 expression was associated with TNM stage and tumor size. Specifically, patients with an advanced TNM stage (III/IV) were associated with lower GDF15 expression, whereas patients with a local TNM stage (I/II) were associated with a higher GDF15 level (0.9886 ± 0.99641 versus 0.2836 ± 0.25307, p < 0.001) (Figure 1B).
Figure 1.
Relative GDF15 Expression in NSCLC Tissues and Its Clinical Significance
(A) Relative expression levels of GDF15 in NSCLC tissues (n = 66) compared with corresponding non-tumor tissues (n = 66). GDF15 expression was examined using qPCR and was normalized to GAPDH expression. The ρCt value was determined by subtracting the GAPDH Ct value from the GDF15 Ct value (relative to a single reference value). Smaller ρCt values indicate higher expression. (B) GDF15 expression was significantly lower in patients with higher pathological stages (III and IV) than in those with lower pathological stages (I and II). (C) GDF15 expression was classified into two groups according to the median GDF15 expression level value in NSCLC tissue samples. (D) Kaplan-Meier overall survival (OS) curves according to GDF15 expression levels. (E) Kaplan-Meier survival plots demonstrating that low GDF15 expression levels correlated with worse OS in lung cancer patients (n = 1,926). *p < 0.05 and **p < 0.01.
According to the median value of GDF15 expression levels, we categorized the samples into a high group (above the median, n = 33) and a low group (below the median, n = 33) (Figure 1C). A chi-square test was performed to evaluate clinic-pathological factors between the two groups, and the clinicopathological characteristics of the NSCLC patients are summarized in Table 1. As shown in Table 2, GDF15 levels were also correlated to age (p = 0.014), TNM stage (p < 0.001) and tumor size (p = 0.017). Other clinicopathological features, such as gender and histological grade, were not statistically significant prognosis factors. Kaplan-Meier analysis and log rank test were used to evaluate the effects of GDF15 expression and the clinicopathological characteristics on overall survival (OS). Remarkably, down-expression of GDF15 was associated with a poor prognosis in patients with NSCLC (p < 0.001) (Figure 1D). In addition, analysis of Kaplan-Meier plots of 1,926 lung cancer patients (http://www.kmplot.com) also indicated a positive relationship between GDF15 and OS (Figure 1E).
Table 1.
The Clinicopathological Factors of 66 NSCLC Patients
| Clinical Factors | Number of Cases | Patients (%) |
|---|---|---|
| Sex | ||
| Male | 42 | 63.6 |
| Female | 24 | 36.4 |
| Age (Years) | ||
| ≤60 | 34 | 51.5 |
| >60 | 32 | 48.5 |
| Histological Grade | ||
| High | 7 | 10.6 |
| Middle | 23 | 34.8 |
| Middle to low | 16 | 24.2 |
| Low | 19 | 28.8 |
| Other | 1 | 1.5 |
| Histological Classification | ||
| Squamous cell carcinoma (SCC) | 40 | 60.6 |
| Adenocarcinoma (AD) | 22 | 33.3 |
| Other | 4 | 6.1 |
| Tumor Stage | ||
| I | 17 | 25.8 |
| II | 29 | 43.9 |
| III | 18 | 27.3 |
| IV | 2 | 3.0 |
| Tumor (T) | ||
| T1 | 21 | 31.8 |
| T2 | 27 | 40.9 |
| T3 | 13 | 19.7 |
| T4 | 5 | 7.6 |
| Lymph Node Metastasis (N) | ||
| N0 | 27 | 40.9 |
| N1 | 30 | 45.5 |
| N2 | 8 | 12.1 |
| N3 | 1 | 1.5 |
| History of Smoking | ||
| Ever | 39 | 59.1 |
| Never | 27 | 40.9 |
Table 2.
Correlation between GDF15 Expression and Clinicopathological Characteristics of 66 NSCLC Patients
| Characteristics | Expression of GDF15 |
p Value | |
|---|---|---|---|
| Low (n = 33) | High (n = 33) | ||
| Sex | |||
| Male | 19 | 23 | 0.306 |
| Female | 14 | 10 | |
| Age (Years) | |||
| ≤60 | 22 | 12 | 0.014∗ |
| >60 | 11 | 21 | |
| Histological Grade | |||
| Middle to low | 30 | 28 | 0.451 |
| High | 3 | 5 | |
| Histological Classification | |||
| SCC | 18 | 22 | 0.314 |
| AD or other | 15 | 11 | |
| TNM Stage | |||
| I and II | 16 | 30 | <0.001** |
| III and IV | 3 | 17 | |
| Lymph Node Metastasis | |||
| Negative | 11 | 16 | 0.211 |
| Positive | 22 | 17 | |
| Tumor Size | |||
| ≤3 cm | 6 | 15 | 0.017∗ |
| >3 cm | 27 | 18 | |
| History of Smoking | |||
| Ever | 20 | 19 | 0.802 |
| Never | 13 | 14 | |
p < 0.05 and **p < 0.01.
Univariate analysis identified TNM stage (I/II, III/IV) and GDF15 expression level as prognostic factors. Multivariate analysis further confirmed that a low GDF15 expression level was an independent predictor of poor survival for NSCLC (p = 0.036) as well as TNM stage (p < 0.001) (Table 3).
Table 3.
Univariate and Multivariate Analysis of Clinicopathologic Factors for Overall Survival in 66 Patients with NSCLC
| Risk Factors | Univariate Analysis |
Multivariate Analysis |
||||
|---|---|---|---|---|---|---|
| HR | p Value | 95% CI | HR | p Value | 95% CI | |
| GDF15 expression | 0.28 | 0.002* | 0.124∼0.630 | 0.477 | 0.036** | 0.239∼0.953 |
| TNM stage (I/II, III/IV) | 7.408 | <0.001* | 3.518∼15.596 | 4.964 | <0.001* | 2.255∼10.927 |
| Tumor size (≤3 cm, >3 cm) | 1.807 | 0.091 | 0.909∼3.592 | – | – | – |
| Histological grade (middle or low, high) | 0.97 | 0.945 | 0.409∼2.301 | – | – | – |
| Histological classification (SCC, AD, or other) | 1.51 | 0.181 | 0.825∼2.762 | – | – | – |
| Age (≤60, >60) | 0.599 | 0.097 | 0.327∼1.097 | – | – | – |
| N (negative, positive) | 1.661 | 0.114 | 0.885∼3.120 | – | – | – |
| History of smoking (ever, never) | 1.374 | 0.301 | 0.752∼2.509 | – | – | – |
| Sex (male, female) | 1.334 | 0.388 | 0.693∼2.567 | – | – | – |
HR, hazard ratio. *p < 0.01 and **p < 0.05.
GDF15 Inhibits NSCLC Cell Proliferation and Induces Apoptosis
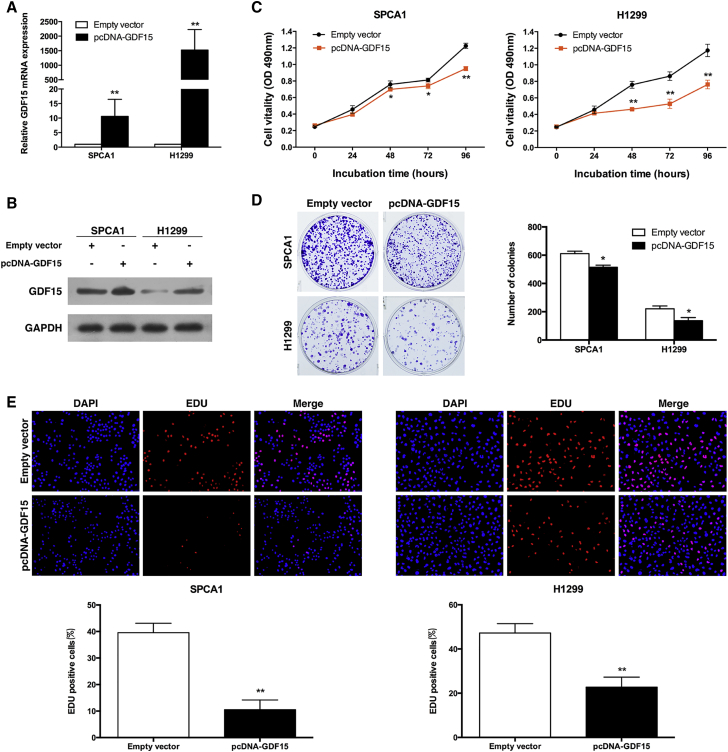
To evaluate the biological role of GDF15 in NSCLC, we first chose the human NSCLC cell lines SPCA1 and H1299 for further functional investigation. Then plasmid-mediated overexpression was used for exogenously manipulating the expression of GDF15 in both the SPCA1 and H1299 cell lines (Figure 2A). The results of the western blot showed that GDF15 expression was significantly upregulated in SPCA1 and H1299 cells transfected with pcDNA-GDF15 compared with control cells (Figure 2B). MTT assays showed that overexpression of GDF15 inhibited the proliferation of SPCA1 and H1299 cells compared with control cells (Figure 2C). Consistently, colony formation assay results revealed that clonogenic survival was inhibited after overexpression of GDF15 in SPCA1 and H1299 cells (Figure 2D). Similarly, ethynyldeoxyuridine (EdU) staining assays also indicated that overexpression of GDF15 decreased NSCLC cell proliferation (Figure 2E).
Figure 2.
Effects of GDF15 on NSCLC Cell Proliferation In Vitro
(A and B) qRT-PCR and western blot analysis of the GDF15 expression level in SPCA1 (A) and H1299 (B) cells transfected with the pcDNA-GDF15 vector. GAPDH protein expression was used as an internal control. (C) MTT assays were used to determine the viability of pcDNA-GDF15-transfected SPCA1 and H1299 cells. (D) Colony formation assays were performed to determine the proliferation of pcDNA-GDF15-transfected SPCA1 and H1299 cells. Colonies were counted and captured. (E) EdU (red) or DAPI (blue) assays were conducted 24 hr after transfection to determine the viability of the transfected SPCA1 and H1299 cells. The data represent the mean ± SD from three independent experiments. *p < 0.05 and **p < 0.01.
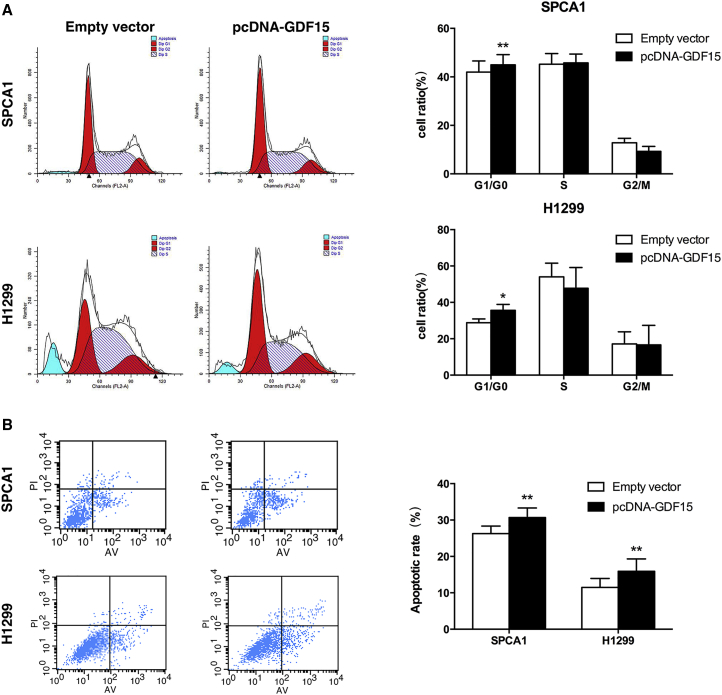
Next, flow cytometric analysis was performed to further examine whether GDF15 affected the proliferation of NSCLC cells by altering cell cycle progression. Results revealed that SPCA1 and H1299 cells transfected with pcDNA-GDF15 had increased cell cycle arrest in G0/G1 phase (Figure 3A). To determine whether NSCLC cell proliferation was influenced by cell apoptosis, we performed a flow cytometry analysis. The results showed that NSCLC cells transfected with pcDNA-GDF15 promoted apoptosis in comparison with control cells (Figure 3B). These results indicated that GDF15 could restrain NSCLC development.
Figure 3.
Effect of GDF15 on the NSCLC Cell Cycle and Cell Apoptosis In Vitro
(A) Forty-eight hours after transfection, the cell cycle stages of SPCA1 and H1299 cells were analyzed by flow cytometry. The bar chart represents the percentages of cells in G1/G0, S, or G2/M phases as indicated. (B) SPCA1 and H1299 cells were stained and analyzed by flow cytometry 48 hr after transfection. LR, early apoptotic cells; UR, terminal apoptotic cells. All experiments were conducted in biological triplicates with three technical replicates. Data are presented as the mean ± SD. *p < 0.05 and **p < 0.01.
EZH2 Repressed GDF15 Expression via H3K27me3 Modification
We explored the potential mechanism of decreased GDF15 expression in NSCLC and found that it was associated with DNA methylation.15 Previous studies have uncovered that histone methylation usually cooperates with DNA methylation in silencing gene activity.16, 17 As an important component of methylation, histone methylation is considered an important factor in transcriptional activation and repression.18 Enhancer of zeste homolog 2 (EZH2) is a methyltransferase and the core catalytic subunit of polycomb repressive complex 2 (PRC2), which catalyzes the tri-methylation of histone3 lysine 27 (H3K27me3) to induce chromatin compaction and transcriptional silencing of target genes that are involved in fundamental cellular processes, such as cell proliferation, cell cycle regulation, cell differentiation, and cancer.19, 20, 21 Recently accumulated evidence indicates that EZH2 is highly expressed in a wide range of cancer types, including NSCLC.22, 23, 24, 25 In addition, several genes with a growth-inhibitory function have recently been found to be targeted by EZH2 in cancer cells.26, 27, 28 Therefore, we examined whether GDF15 could also be a key tumor suppressor gene targeted for repression by EZH2 in NSCLC.
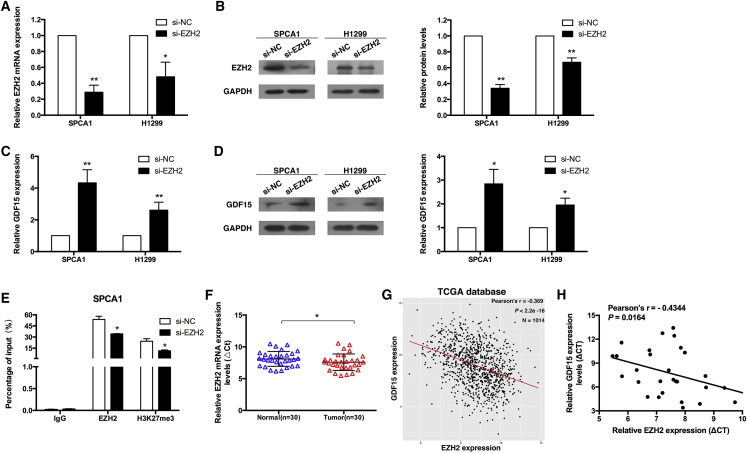
To assess the putative role of EZH2 in GDF15 repression, we first transiently depleted the expression of EZH2 in SPCA1 and H1299 cells (Figures 4A and 4B). To avoid off-target effects, we used an interference target sequence against EZH2, as studied in a previous article.29 We observed that the loss of EZH2 was associated with upregulation of GDF15 at the mRNA and protein levels (Figures 4C and 4D).
Figure 4.
EZH2 Is Involved in GDF15 Downregulation
(A and B) qPCR and western blot analysis of EZH2 expression levels following SPCA1 (A) and H1299 (B) cell treatment with si-EZH2. (C and D) qPCR and western blot assays were used to detect the levels of GDF15 in SPCA1 (C) and H1299 (D) cells after transfection of si-EZH2. (E) ChIP-qPCR of EZH2 and H3K27me3 of the promoter region of the GDF15 locus after siRNA treatment targeting si-NC or si-EZH2 in SPCA1 cells. Antibody enrichment was quantified relative to the amount of input DNA. An antibody directed against IgG was used as a negative control. (F) Relative expression levels of EZH2 in NSCLC tissues (n = 30) compared with corresponding non-tumor tissues (n = 30). EZH2 expression was examined using qPCR and was normalized to GAPDH expression. The ρCt value was determined by subtracting the GAPDH Ct value from the EZH2 Ct value (relative to a single reference value). Smaller ρCt values indicate higher expression. (G) Correlation analysis revealed that GDF15 expression levels were inversely correlated with EZH2 expression levels in 1,014 TCGA database NSCLC tissues. (H) Analysis of the relationship between GDF15 expression (ρCt value) and EZH2 expression (ρCt value) in 30 NSCLC tissues. *p < 0.05 and **p < 0.01.
To further verify the epigenetic silencing of GDF15 by EZH2, we performed chromatin immunoprecipitation (ChIP) assays to examine whether EZH2 and H3K27me3 occupy the GDF15 promoter in NSCLC cells transfected with si-EZH2 or non-specific small interfering RNA (si-NC). A nonspecific immunoglobulin G (IgG) antibody was used as a negative control. The qPCR results showed that the GDF15 promoter was enriched with endogenous EZH2 and H3K27me3, which was supported by the fact that knockdown of EZH2 expression reduced EZH2 binding and H3K27me3 modification (Figure 4E). To further validate the role of EZH2 in NSCLC, we detected the expression level of EZH2 in 30 pairs of NSCLC tissues and adjacent normal tissues. As shown in Figure 4F, EZH2 was obviously upregulated in NSCLC tissues. Finally, we verified that there was a negative correlation between the levels of GDF15 and EZH2 expression in NSCLC tissues from The Cancer Genome Atlas (TCGA) database (Figure 4G). In addition, the EZH2 mRNA expression level was negatively correlated with GDF15 in our 30 paired NSCLC tissues (Figure 4H). These data indicated that EZH2-mediated H3K27me3 modification has a key role in the repression of GDF15 expression.
GDF15 Inhibits NSCLC Cell Tumorigenesis In Vivo
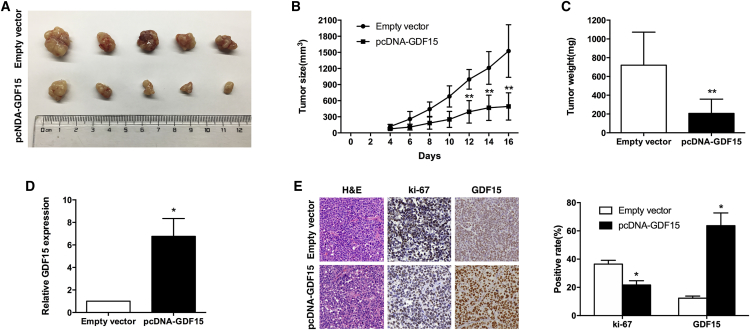
To explore whether the level of GDF15 expression could affect tumorigenesis, SPCA1 cells stably transfected with pcDNA-GDF15 or an empty vector were inoculated into male nude mice. Transplantable tumors derived from pcDNA-GDF15-transfected SPCA1 cells showed reduced tumorigenicity in athymic nude mice compared with vector-transfected SPCA1 cells. Sixteen days after the injection, the tumors formed in the pcDNA-GDF15 group were substantially smaller than those in the control group (Figures 5A and 5B). The average tumor weight in the pcDNA-GDF15 group was markedly lower than in the empty vector group (Figure 5C). In addition, qRT-PCR analysis found that the expression levels of GDF15 in tumor tissues formed from pcDNA-GDF15 cells were higher than in tumors formed in the control group (Figure 5D). Tumors formed from pcDNA-GDF15-transfected SPCA1 cells exhibited decreased positive for Ki67 and increased positive for GDF15 compared with those formed from control cells (Figure 5E). These data indicate that overexpression of GDF15 inhibits tumor growth in vivo.
Figure 5.
GDF15 Inhibits the Tumorigenesis of NSCLC Cells In Vivo
(A) Empty vector or pcDNA-GDF15 was transfected into SPCA1 cells, which were then injected individually into nude mice (n = 5). (B) The tumor volumes were calculated every 2 days after injection. The mean tumor volumes are indicated by points, and the bars indicate SD (n = 5). (C) The tumor weights are represented as the mean tumor weights ± SD. (D) qRT-PCR analysis was performed to detect the average expression levels of GDF15 in xenograft tumors (n = 5). (E) The tumor sections were examined using H&E staining and IHC staining with antibodies against Ki-67 and GDF15. Left: representative images. Right: statistical analysis. The error bars indicate the mean ± standard error. *p < 0.05 and **p < 0.01.
Discussion
NSCLC is one of the leading causes of cancer-related death worldwide. Because of late-stage diagnosis and frequency of metastasis, the 5-year survival rate for NSCLC remains poor (18.2%).30 In our present study, we found that expression of GDF15 in NSCLC tissues was frequently lower than in corresponding non-tumor tissues. The low expression of GDF15 in NSCLC patients was negatively correlated with advanced TNM stage and large tumor size. Importantly, decreased expression of GDF15 in NSCLC tissues was associated with a poor prognosis and could be an independent prognostic indicator. These results demonstrated that GDF15 might have an important role in NSCLC progression.
As a member of the TGF-β superfamily, GDF15 has been shown to have important roles in diverse cellular processes, such as proliferation, apoptosis, metabolism, inflammation, and immune escape.3, 12, 13, 31, 32 Some important clues have been obtained from studies of GDF15 gene expression and functional studies in breast cancer, glioma, prostate cancer, and intestinal adenoma.9, 12, 13, 33 However, very little is known about the biological significance and molecular mechanisms of GDF15 in NSCLC carcinogenesis. To this end, we used gain-of-function assays to explore whether GDF15 plays a critical role in NSCLC cell proliferation. In our study, we found that overexpression of GDF15 could significantly repress NSCLC cell proliferation and induce apoptosis. These findings suggest that GDF15 functions as a tumor suppressor in NSCLC.
Tumor suppressor genes are usually inactivated by epigenetic alterations in cancer cells. Previous studies have reported that epigenetic regulatory factors such as histone modification or DNA methylation can manipulate the expression of tumor suppressors. For instance, epigenetic silencing of KLF2 occurs in cancer cells through direct transcriptional repression mediated by the Polycomb group protein EZH2.28 In several glioblastoma cell lines, DNA methylation in the GDF15 promoter was strongly associated with low GDF15 expression.34 In addition, studies have shown that histone methylation usually cooperates with DNA methylation in silencing gene activity.16, 17
EZH2, a methyltransferase that is a key catalytic subunit of PRC2, functions as a histone methyltransferase that specifically induces transcriptionally incompetent H3K27me3 to the targeted genes.35 Increased EZH2 expression has been found in diverse malignancies and is associated with tumor cell cycle regulation and cell proliferation.36, 37 Here we found that the mRNA levels of the epigenetic factor EZH2 were upregulated in NSCLC tissue compared with normal tissue. Knockdown of EZH2 expression significantly upregulated GDF15 expression, whereas ChIP assays showed that inhibition of EZH2 expression prevented its binding to the GDF15 promoter region and reduced H3K27me3 modification, inducing GDF15 expression. Furthermore, the EZH2 mRNA expression level was inversely correlated with GDF15 in TCGA NSCLC samples. Our previous study also found that EZH2 could epigenetically repress the expression of SPRY4 in esophageal squamous cell carcinoma.38 Overall, our data indicate that the expression of GDF15 was significantly decreased in NSCLC tissues, which was at least partially mediated by the histone methyltransferase EZH2.
In summary, this study identified that the expression of GDF15 was markedly decreased in NSCLC tissues and that its downregulation may be a negative prognostic factor for NSCLC patients for the first time. Overexpression of GDF15 repressed cell proliferation, facilitated cell apoptosis in vitro, and inhibited tumorigenesis in vivo. Furthermore, GDF15 undergoes transcriptional silencing in human tumorigenesis by the direct repression of an oncogenic Polycomb protein, the histone methyltransferase EZH2. EZH2-mediated inactivation of GDF15 blocks the tumor suppressor features of GDF15. Our findings elucidate a potential mechanism underlying the tumor suppressor GDF15 in NSCLC and indicate that GDF15 could be a useful marker in NSCLC.
Materials and Methods
Tissue Collection and Ethics Statement
A total of 66 patients analyzed in this study underwent resection of NSCLC at the First Affiliated Hospital of Nanjing Medical University. The study was approved by the Research Ethics Committee of Nanjing Medical University (Nanjing, Jiangsu, China), and written informed consent was obtained from all patients. The clinicopathological characteristics of the NSCLC patients are summarized in Table 1.
Cell Cultures
The NSCLC cell lines SPCA1 and NCI-H1299 were purchased from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI 1640 medium or DMEM (Gibco-BRL) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin in incubators at 37°C with 5% CO2.
RNA Extraction and qRT-PCR Analyses
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For qRT-PCR, RNA was reverse-transcribed to cDNA by using a reverse transcription kit (Takara, Dalian, China). Real-time PCR analyses were performed with SYBR Green (Takara, Dalian China). Results were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The gene-specific primers were as follows: GDF15 forward, 5′-CTCCAGATTCCGAGAGTTGC-3′ and reverse, 5′-AGAGATACGCAGGTGCAGGT-3′; GAPDH forward, 5′-AGCCACATCGCTCAGACAC-3′ and reverse, 5′-GCCCAATACGACCAAATCC-3′; EZH2 forward, 5′-TGCACATCCTGACTTCTGTG-3′ and reverse, 5′-AAGGGCATTCACCAACTCC-3′. qRT-PCR and data collection were carried out on an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The relative expression of GDF15 was calculated and normalized using the 2−ΔΔCt method relative to GAPDH.
Transfection of Cell Lines
The GDF15 sequence was synthesized according to the full-length GDF15 sequence and then subcloned into a pcDNA3.1 vector (Invitrogen, China). The empty pcDNA3.1 vector was used as the control. The plasmid was transfected by X-treme GENE HP DNA transfection reagent (Roche, Basel, Switzerland) according to the manufacturer’s instructions. The nucleotide sequences of small interfering RNAs (siRNAs) for EZH2 were as follows: sense, 5′-GAGGUUCAGACGAGCUGAUUU-3′; antisense 5′-AAAUCAGCUCGUCUGAACCUC-3′.29 si-NC was purchased from Invitrogen. Typically, cells were seeded in six-well plates and then transfected the next day with specific siRNA (100 nM) and control siRNA (100 nM) using Lipofectamine RNAi MAX according to the manufacturer’s protocol (Invitrogen). 48 hr post-transfection, cells were harvested and processed for the following experiments.
Flow Cytometric Analysis
Cells were harvested after transfection by trypsinization. For cell cycle analysis, cells were stained with propidium oxide using the Cycle Test Plus DNA Reagent Kit (BD Biosciences) following the manufacturer’s protocol and then analyzed by flow cytometry (FACScan, BD Biosciences). The percentage of cells in G0/G1, S, and G2/M phase were counted and compared. Double staining with fluorescein isothiocyanate (FITC)-Annexin V and propidium iodide was done using the FITC Annexin V Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s recommendations. The cells were analyzed with a flow cytometer (FACScan, BD Biosciences) equipped with Cell Quest software (BD Biosciences). Cells were discriminated into viable cells, dead cells, early apoptotic cells, and apoptotic cells, and then the relative ratio of early apoptotic cells was compared with control transfection from each experiment. All of the samples were assayed in triplicate.
EdU Analysis
An EdU labeling or detection kit (Ribobio, Guangzhou, China) was used to assess cell proliferation. The cells were cultured in 96-well plates at 5 × 103 cells/well. Forty-eight hours after transfection, 50 μM EdU labeling medium was added to the 96-well plates, and they were incubated for 2 hr at 37°C under 5% CO2. After treatment with 4% paraformaldehyde and 0.5% Triton X-100, the cells were stained with anti-EdU working solution. DAPI was used to label the cell nuclei. The percentage of EdU-positive cells was calculated after fluorescence microscopy analysis. Five fields of view were randomly assessed for each treatment group.
ChIP Assays
ChIP assays were performed using EZ-CHIPKIT according to the manufacturer’s instructions (Millipore, USA). The EZH2 antibody was obtained from Millipore. H3 trimethyl Lys 27 antibody was purchased from Abcam. Quantification of immunoprecipitated DNA was performed using qPCR with SYBR Green Mix (Takara). The ChIP primer sequences (forward 5′- GTATTCTCATGCAGATTCCGTT-3′ and reverse 5′- CCACTTCCTTGTGACCGCA-3′) were used to amplify the promoter region of GDF15. ChIP data were calculated as a percentage relative to the input DNA by the following equation: 2[Input Ct-Target Ct] × 0.1 × 100.
Tumor Formation Assay in a Nude Mouse Model
Five-week-old male athymic BALB/c mice were maintained under specific pathogen-free conditions and manipulated according to protocols approved by the Shanghai Medical Experimental Animal Care Commission. SPCA1 cells transfected with the pcDNA3.1 vector or pcDNA-GDF15 were harvested at a concentration of 2 × 107 cells/mL. Of the suspended cells, 0.1 mL was subcutaneously injected into either side of the posterior flanks of the nude mouse. Tumor volumes and weights were examined every 2 days in mice from the control (seven mice) or pcDNA-GDF15 (seven mice) group; tumor volumes were measured (length × width2 × 0.5). Sixteen days after injection, the mice were killed, and tumor weights were measured and used for further analysis. The primary tumors were excised, and tumor tissues were used to perform qRT-PCR analysis of GDF15 levels and immunostaining analysis of Ki- 67 and GDF15 protein expression.
Statistical Analysis
Statistical analysis was performed using SPSS version 22 software (Chicago, IL). Chi-square and t tests were performed to explore the associations between the GDF15 expression level and the clinical characteristics. The survival curves were estimated using the Kaplan-Meier method. A log rank test was used to estimate the significance of the differences between the survival curves. A Cox proportional hazards analysis was performed to calculate the hazard ratio (HR) and the 95% confidence interval (CI) to evaluate the association between GDF15 expression and OS. A multivariate Cox regression was performed to adjust for other covariates. p values less than 0.05 were considered to be statistically significant.
Author Contributions
X. Lu, X.H., J.S., and J.W. contributed to designing and organizing the experiments, carrying out data analysis, and writing the manuscript. X. Liu, K.X., and W.D. contributed to laboratory measurements and data analysis. Y.E.S., R.G., and E.Z. contributed to conceiving the ideas, supervising the study, and writing the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81702266, 81230054, 91029739, and 81172217), the National Key Technology R&D Program for the 12th Five-Year Plan of China (2013BAI01B06), the State Key Laboratory of Reproductive Medicine, Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Natural Science Foundation of Jiangsu Province (BK20161593), the Medical Important Talent Foundation of Jiangsu Province (RC201157), and the Wu Jie-ping Foundation (320.6799.15032). This work was also supported by the China Postdoctoral Science Foundation (2017M610339), Jiangsu Planned Projects for Postdoctoral Research Funds (1701041A), and the Scientific Foundation of Wuxi City of Jiangsu (Q201728).
Contributor Information
Erbao Zhang, Email: erbaozhang@njmu.edu.cn.
Renhua Guo, Email: rhguonjmu@sina.com.
Yuenian Eric Shi, Email: daweieric@163.com.
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Baek S.J., Kim K.S., Nixon J.B., Wilson L.C., Eling T.E. Cyclooxygenase inhibitors regulate the expression of a TGF-beta superfamily member that has proapoptotic and antitumorigenic activities. Mol. Pharmacol. 2001;59:901–908. [PubMed] [Google Scholar]
- 4.Yokoyama-Kobayashi M., Saeki M., Sekine S., Kato S. Human cDNA encoding a novel TGF-beta superfamily protein highly expressed in placenta. J. Biochem. 1997;122:622–626. doi: 10.1093/oxfordjournals.jbchem.a021798. [DOI] [PubMed] [Google Scholar]
- 5.Böttner M., Suter-Crazzolara C., Schober A., Unsicker K. Expression of a novel member of the TGF-beta superfamily, growth/differentiation factor-15/macrophage-inhibiting cytokine-1 (GDF-15/MIC-1) in adult rat tissues. Cell Tissue Res. 1999;297:103–110. doi: 10.1007/s004410051337. [DOI] [PubMed] [Google Scholar]
- 6.Strelau J., Böttner M., Lingor P., Suter-Crazzolara C., Galter D., Jaszai J., Sullivan A., Schober A., Krieglstein K., Unsicker K. GDF-15/MIC-1 a novel member of the TGF-beta superfamily. J. Neural Transm. Suppl. 2000;60:273–276. doi: 10.1007/978-3-7091-6301-6_18. [DOI] [PubMed] [Google Scholar]
- 7.Paralkar V.M., Vail A.L., Grasser W.A., Brown T.A., Xu H., Vukicevic S., Ke H.Z., Qi H., Owen T.A., Thompson D.D. Cloning and characterization of a novel member of the transforming growth factor-beta/bone morphogenetic protein family. J. Biol. Chem. 1998;273:13760–13767. doi: 10.1074/jbc.273.22.13760. [DOI] [PubMed] [Google Scholar]
- 8.Kim K.S., Baek S.J., Flake G.P., Loftin C.D., Calvo B.F., Eling T.E. Expression and regulation of nonsteroidal anti-inflammatory drug-activated gene (NAG-1) in human and mouse tissue. Gastroenterology. 2002;122:1388–1398. doi: 10.1053/gast.2002.32972. [DOI] [PubMed] [Google Scholar]
- 9.Baek S.J., Okazaki R., Lee S.H., Martinez J., Kim J.S., Yamaguchi K., Mishina Y., Martin D.W., Shoieb A., McEntee M.F., Eling T.E. Nonsteroidal anti-inflammatory drug-activated gene-1 over expression in transgenic mice suppresses intestinal neoplasia. Gastroenterology. 2006;131:1553–1560. doi: 10.1053/j.gastro.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Baek S.J., Kim J.S., Moore S.M., Lee S.H., Martinez J., Eling T.E. Cyclooxygenase inhibitors induce the expression of the tumor suppressor gene EGR-1, which results in the up-regulation of NAG-1, an antitumorigenic protein. Mol. Pharmacol. 2005;67:356–364. doi: 10.1124/mol.104.005108. [DOI] [PubMed] [Google Scholar]
- 11.Baek S.J., Wilson L.C., Lee C.H., Eling T.E. Dual function of nonsteroidal anti-inflammatory drugs (NSAIDs): inhibition of cyclooxygenase and induction of NSAID-activated gene. J. Pharmacol. Exp. Ther. 2002;301:1126–1131. doi: 10.1124/jpet.301.3.1126. [DOI] [PubMed] [Google Scholar]
- 12.Liu T., Bauskin A.R., Zaunders J., Brown D.A., Pankhurst S., Russell P.J., Breit S.N. Macrophage inhibitory cytokine 1 reduces cell adhesion and induces apoptosis in prostate cancer cells. Cancer Res. 2003;63:5034–5040. [PubMed] [Google Scholar]
- 13.Roth P., Junker M., Tritschler I., Mittelbronn M., Dombrowski Y., Breit S.N., Tabatabai G., Wick W., Weller M., Wischhusen J. GDF-15 contributes to proliferation and immune escape of malignant gliomas. Clin. Cancer Res. 2010;16:3851–3859. doi: 10.1158/1078-0432.CCR-10-0705. [DOI] [PubMed] [Google Scholar]
- 14.Ratnam N.M., Peterson J.M., Talbert E.E., Ladner K.J., Rajasekera P.V., Schmidt C.R., Dillhoff M.E., Swanson B.J., Haverick E., Kladney R.D. NF-κB regulates GDF-15 to suppress macrophage surveillance during early tumor development. J. Clin. Invest. 2017;127:3796–3809. doi: 10.1172/JCI91561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monteiro-Reis S., Leça L., Almeida M., Antunes L., Monteiro P., Dias P.C., Morais A., Oliveira J., Henrique R., Jerónimo C. Accurate detection of upper tract urothelial carcinoma in tissue and urine by means of quantitative GDF15, TMEFF2 and VIM promoter methylation. Eur. J. Cancer. 2014;50:226–233. doi: 10.1016/j.ejca.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 16.Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 17.Fahrner J.A., Eguchi S., Herman J.G., Baylin S.B. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- 18.McGrath J., Trojer P. Targeting histone lysine methylation in cancer. Pharmacol. Ther. 2015;150:1–22. doi: 10.1016/j.pharmthera.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Kim K.H., Roberts C.W. Targeting EZH2 in cancer. Nat. Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Croce L., Helin K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 21.Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behrens C., Solis L.M., Lin H., Yuan P., Tang X., Kadara H., Riquelme E., Galindo H., Moran C.A., Kalhor N. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin. Cancer Res. 2013;19:6556–6565. doi: 10.1158/1078-0432.CCR-12-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleer C.G., Cao Q., Varambally S., Shen R., Ota I., Tomlins S.A., Ghosh D., Sewalt R.G., Otte A.P., Hayes D.F. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsukawa Y., Semba S., Kato H., Ito A., Yanagihara K., Yokozaki H. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006;97:484–491. doi: 10.1111/j.1349-7006.2006.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu C., Han H.D., Mangala L.S., Ali-Fehmi R., Newton C.S., Ozbun L., Armaiz-Pena G.N., Hu W., Stone R.L., Munkarah A. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beke L., Nuytten M., Van Eynde A., Beullens M., Bollen M. The gene encoding the prostatic tumor suppressor PSP94 is a target for repression by the Polycomb group protein EZH2. Oncogene. 2007;26:4590–4595. doi: 10.1038/sj.onc.1210248. [DOI] [PubMed] [Google Scholar]
- 27.Yu J., Cao Q., Mehra R., Laxman B., Yu J., Tomlins S.A., Creighton C.J., Dhanasekaran S.M., Shen R., Chen G. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12:419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi H., Jacinto F.V., Villanueva A., Fernandez A.F., Yamamoto H., Carmona F.J., Puertas S., Marquez V.E., Shinomura Y., Imai K., Esteller M. Silencing of Kruppel-like factor 2 by the histone methyltransferase EZH2 in human cancer. Oncogene. 2012;31:1988–1994. doi: 10.1038/onc.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prensner J.R., Iyer M.K., Balbin O.A., Dhanasekaran S.M., Cao Q., Brenner J.C., Laxman B., Asangani I.A., Grasso C.S., Kominsky H.D. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeSantis C.E., Lin C.C., Mariotto A.B., Siegel R.L., Stein K.D., Kramer J.L., Alteri R., Robbins A.S., Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J. Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 31.Lee S.E., Kang S.G., Choi M.J., Jung S.B., Ryu M.J., Chung H.K., Chang J.Y., Kim Y.K., Lee J.H., Kim K.S. Growth Differentiation Factor 15 Mediates Systemic Glucose Regulatory Action of T-Helper Type 2 Cytokines. Diabetes. 2017;66:2774–2788. doi: 10.2337/db17-0333. [DOI] [PubMed] [Google Scholar]
- 32.Li P.X., Wong J., Ayed A., Ngo D., Brade A.M., Arrowsmith C., Austin R.C., Klamut H.J. Placental transforming growth factor-beta is a downstream mediator of the growth arrest and apoptotic response of tumor cells to DNA damage and p53 overexpression. J. Biol. Chem. 2000;275:20127–20135. doi: 10.1074/jbc.M909580199. [DOI] [PubMed] [Google Scholar]
- 33.Peake B.F., Eze S.M., Yang L., Castellino R.C., Nahta R. Growth differentiation factor 15 mediates epithelial mesenchymal transition and invasion of breast cancers through IGF-1R-FoxM1 signaling. Oncotarget. 2017;8:94393–94406. doi: 10.18632/oncotarget.21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadowaki M., Yoshioka H., Kamitani H., Watanabe T., Wade P.A., Eling T.E. DNA methylation-mediated silencing of nonsteroidal anti-inflammatory drug-activated gene (NAG-1/GDF15) in glioma cell lines. Int. J. Cancer. 2012;130:267–277. doi: 10.1002/ijc.26082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 36.Chang C.J., Hung M.C. The role of EZH2 in tumour progression. Br. J. Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chase A., Cross N.C. Aberrations of EZH2 in cancer. Clin. Cancer Res. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 38.Zhang E., Han L., Yin D., He X., Hong L., Si X., Qiu M., Xu T., De W., Xu L. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:3086–3101. doi: 10.1093/nar/gkw1247. [DOI] [PMC free article] [PubMed] [Google Scholar]