Abstract
Progresses in the past two decades have greatly expanded our understanding of inflammatory bowel disease (IBD), an incurable disease with multifaceted and challenging clinical manifestations. The pathogenesis of IBD involves multiple processes on the cellular level, which include the stress response signaling such as endoplasmic reticulum (ER) stress, oxidative stress, and hypoxia. Under physiological conditions, the stress responses play key roles in cell survival, mucosal barrier integrity, and immunomodulation. However, they can also cause energy depletion, trigger cell death and tissue injury, promote inflammatory response, and drive the progression of clinical disease. In recent years, gut microflora has emerged as an essential pathogenic factor and therapeutic target for IBD. Altered compositional and metabolic profiles of gut microbiota, termed dysbiosis, are associated with IBD. Recent studies, although limited, have shed light on how ER stress, oxidative stress, and hypoxic stress interact with gut microorganisms, a potential source of stress in the microenvironment of gastrointestinal tract. Our knowledge of cellular stress responses in intestinal homeostasis as well as their cross-talks with gut microbiome will further our understanding of the pathogenesis of inflammatory bowel disease and probably open avenues for new therapies.
1. Introduction
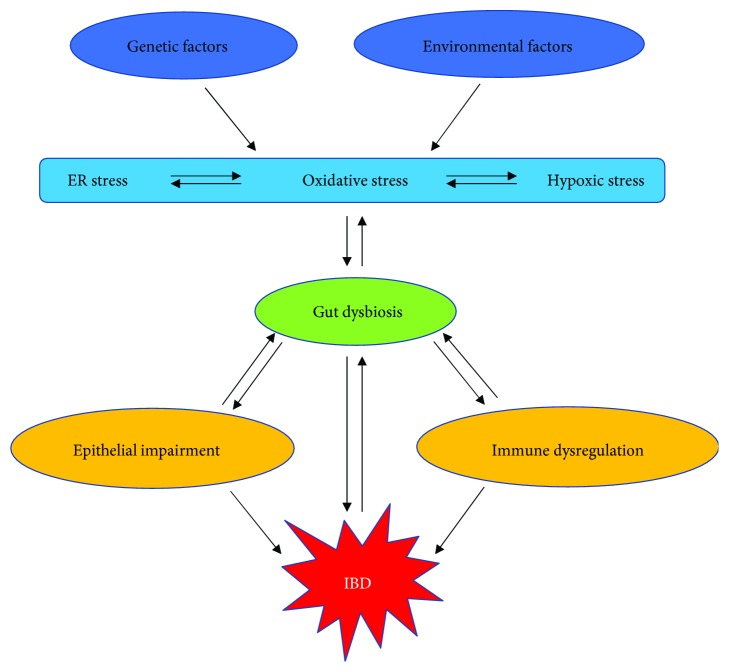
Inflammatory bowel disease (IBD) affects more than 3.1 million people in the United States and 2.5 million in Europe and is increasing in incidence worldwide especially in regions such as Eastern and South Asia [1, 2]. IBD including Crohn's disease (CD) and ulcerative colitis (UC) are characterized by chronic intestinal inflammation as well as extraintestinal manifestations, which are likely triggered by a combination of genetic predispositions and environmental factors such as diet, infection, and medication use [3]. The complex pathophysiology of IBD involves multiple cell populations in the gastrointestinal tract and numerous signaling pathways from energy homeostasis to innate immune response. Cellular stress signaling, including endoplasmic reticulum (ER) stress, oxidative stress, and hypoxic stress, has been implicated in multiple human pathologies such as metabolic disease, neurodegenerative disease, cardiovascular disease, cancer, and autoimmune disease. Recent evidence suggests that stress signaling plays a key role in mucosal homeostasis in the gut. As an integral component of the gastrointestinal environment, the gut microbiome has been shown to promote intestinal health as a physiological stressor. Meanwhile, through complex interactions with the stress pathways in the host cells, these microorganisms may contribute to the pathogenesis of IBD by inducing cell death and differentiation, epithelial barrier breakdown, and inflammatory response. Recently, targeting cellular stress signaling and the gut microbiota have been proposed as new therapies for UC and CD. The understanding of both sides and the bridge between the two will likely spur the progresses in this arena.
2. Cellular Stress Signaling in Gastrointestinal Tract
2.1. ER Stress
Approximately one-third of all proteins synthesized in the cell travel across the ER where they are folded into their proper three-dimensional structures with posttranslational modifications including disulfide bond formation, glycosylation, hydroxylation, and lipidation. Protein folding in the ER is subject to numerous insults such as increased mRNA translation, elevated demand of protein secretion, genetic mutations of client proteins, deficiency of ER chaperones or foldases, ER redox or calcium perturbations, bacterial and viral infection, ATP depletion, nutrient deficiency, and pharmacological and toxicological insults. The accumulation of unfolded or misfolded proteins in the ER lumen activates the unfolded protein response (UPR), which is signaled through three ER transmembrane proteins: inositol-requiring enzyme 1α (IRE1α), dsRNA-activated protein kinase-related ER protein kinase (PERK), and activating transcription factor 6α (ATF6α). All three transmembrane sensors are maintained in an inactive state through interaction between their ER luminal domains and the protein chaperone immunoglobulin heavy chain-binding protein (BiP). A recent study showed that the ER luminal cochaperone ERdj4/DNAJB9 is required for the complex formation between BiP and the luminal domain of IRE1a, thereby maintaining IRE1a in a monomeric, inactive state in the absence of ER stress [4]. Upon ER stress, accumulated unfolded/misfolded proteins in the ER lumen bind and sequester BiP, thereby triggering dissociation of BiP from IRE1α, PERK, and ATF6α [5–7]. All three branches of the UPR have been involved in intestinal mucosal homeostasis and the pathogenesis of IBD.
The most evolutionarily conserved ER stress transducer is IRE1α, which contains both endoribonuclease and kinase domains within its cytosolic region. During ER stress, IRE1α activates via dimerization and auto-transphosphorylation, which then leads to removal of a 26-nucleotide intron from the mRNA encoding an active and stable transcription factor XBP1s. The activity of XBP1s has been linked to cellular proteostasis by inducing the expression of various genes involved in ER protein translocation, protein folding and modification, quality-control mechanisms, biogenesis of the ER and Golgi compartments, and ER-associated protein degradation (ERAD) [5, 6]. In addition to the cleavage of Xbp1 mRNA, IRE1α regulates the UPR, cell death and differentiation, and inflammation via regulated IRE1a-dependent decay (RIDD) of various mRNAs and miRNAs, which was recently shown to orchestrate the survival of conventional dendritic cell in the gut [8]. Furthermore, IRE1α also cross-talks with a number of other stress-related signaling pathways including autophagy, nuclear factor-κB (NF-κB), mitogen-activated protein (MAP) kinases, and the c-Jun N-terminal kinase (JNK) pathways [6].
The IRE1α-XBP1 pathway has been well known to play an essential role in immunity and inflammation. XBP1 is required for the differentiation of plasma cells, dendritic cells, CD8+ T cells, and eosinophils, as well as the production of several cytokines in macrophages [9, 10]. XBP1 is one of the first UPR effectors to associate with IBD. Findings from both animal and human studies suggested an essential role of XBP1-mediated adaptive UPR signaling in the intestinal homeostasis. Loss of Xbp1 in murine intestinal epithelial cells (IECs) hyperactivates IRE1α, which results in a NF-κB and JNK-dependent inflammatory response. A recent study showed that the selective autophagy receptor optineurin may interact with and target IRE1α for degradation, thereby containing IRE1a aggregation and hyperactivation in IECs [11]. XBP1 deficiency leads to Paneth cell death, spontaneous small bowel inflammation, and susceptibility to bacterial infection [12]. Interestingly, ablation of Xbp1 in Paneth cells alone (using Defa6-Cre) is sufficient to produce similar phenotype of spontaneous ileitis as observed in the previous model [13]. In addition to Paneth cells, the goblet cells in the large bowel were also affected by Xbp1 deletion as shown by impaired mucin secretion. These animals were more susceptible to dextran sodium sulfate- (DSS-) induced colitis, which is probably caused by multiple mechanisms including the absence of XBP1-mediated adaptive UPR, proapoptotic effectors such as CCAAT-enhancer-binding protein homologous protein (CHOP), cellular inflammatory response in the settings of IRE1α activation, and its downstream signaling [14]. In colonic mucosal tissues from mice and humans, Xbp1 was targeted by a colitis-induced miRNA, miR665, which was shown to promote apoptosis and DSS-induced colitis in mice [15]. A recent study suggested that loss of Xbp1 in the IECs leads to increased expression of natural killer group 2 member D ligand (NKG2DL) mouse UL16-binding protein-like transcript 1 via CHOP, which likely contributes to the recruitment of NKG2D-expressing group 1 innate lymphoid cells and mucosal inflammation [16]. In addition, a human deep-sequencing study identified hypomorphic, rare variants of XBP1 that associate with both CD and UC [12]. Despite that hyperactivation of IRE1α seems to contribute to mucosal inflammation, the deletion of Ire1α in murine IECs resulted in the loss of colonic goblet cells, impaired barrier function, and subsequent spontaneous colitis and rectal bleeding. There was a decreased Xbp1 mRNA splicing, as expected, and an increased CHOP in the colonocytes of the mutant animals [14]. Although the IRE1α and XBP1 deficiencies in IECs cause mucosal inflammation from different mechanisms, it seems that both models involve proapoptotic UPR signaling such as CHOP. It would be interesting to test whether the ablation of Chop in IECs rescues the phenotypes (see more discussions below). It is also worthwhile to notice that Xbp1 deletion produced more dramatic abnormalities in the small intestine, while the loss of IRE1α generated phenotype mainly in the large bowel. It is unclear whether this is due to the differential roles of IRE1α versus XBP1 in different IECs (Paneth cells versus goblet cells) or it is related to the microflora in specific animal facilities [17–19].
PERK is an ER transmembrane protein with a serine/threonine kinase domain on its cytosolic side. The activated PERK directly phosphorylates the eukaryotic mRNA translation initiation factor eIF2α, which dampens global protein synthesis and therefore relieving protein folding overload in the ER [5, 20, 21]. Meanwhile, the phosphorylated eIF2α undergoes selective translation of some mRNAs including that of activating transcription factor 4 (ATF4), which induces various genes involved in ER protein translocation and folding, antioxidative response, autophagy, and amino acid metabolism. ATF4 also engages apoptotic and inflammatory pathways through the transactivation of Chop and Bcl-2 family members, as well as Mcp1, Il6, and Tnfα [5, 22]. The role of eIF2α phosphorylation in murine IECs was studied using mice with IEC-specific expression of nonphosphorylatable eIF2α [23]. Electron microscopy of Paneth cells in the mutant mice revealed a decreased number of secretory granules, a fragmented ER, and damaged mitochondria, which resembled the morphology of the Paneth cells deficient of autophagy mediator ATG16L1 or ATG5 [24]. The mutant mice exhibited a diminished secretion of lysozyme and cryptdins in their small bowel, which were likely responsible for the increased susceptibility to Salmonella infection. Interestingly, the ER-associated translation of Lysozyme and Cryptdin mRNAs was specifically impaired, which is associated with reduced expression of ER protein translocation machinery in the absence of eIF2a phosphorylation [5, 23]. A recent study showed that lysozyme is transported via secretory autophagy in Paneth cells, a process requiring bacteria-induced PERK-eIF2α activation [25]. In addition, eIF2α phosphorylation is linked to mucosal homeostasis in the large bowel. The unaffected mucosal tissue of UC patients exhibited decreased eIF2α phosphorylation, which may indicate a defective integrated stress response in colonic epithelial cells [26]. Most data so far support a protective role of eIF2α phosphorylation in IECs for intestinal homeostasis. In contrast, the overexpression of transcription factor Chop exacerbated DSS-induced colitis, which seems due to impaired epithelial cell proliferation rather than increased apoptosis [27]. Consistently, a recent study showed that the absence of CHOP alleviates bile duct ligation-induced loss of stemness in intestinal stem cells, although the underlying mechanism is unclear [28]. Besides ER stress, eIF2α phosphorylation serves as the regulatory hub of the so-called integrated stress response, which can also be initiated by three other serine/threonine eIF2α kinases in response to distinct stimuli including viral infection (via the double-stranded RNA-activated protein kinase, a.k.a. PKR), amino acid deprivation (via the general control nonderepressible 2, a.k.a. GCN2), and heme deficiency (via the heme-regulated eIF2α kinase, a.k.a. HRI) [6, 29]. PKR has been implicated in a broad spectrum of infectious and inflammatory processes in addition to its well-known antiviral response [30–33]. PKR is activated in murine colonic epithelial cells during DSS-induced colitis. The deletion of Pkr exacerbated epithelial cell death and colonic inflammation, which are likely due to reduced ER chaperones and other prosurvival pathways such as Akt/PKB and the signal transducer and activator of transcription 3 [34, 35]. A recent study linked GCN2 to intestinal inflammation. The genetic ablation of Gcn2 in either IECs or CD11c+ antigen-presenting cells leads to Th17 cell activation and intestinal inflammation, which are likely due to impaired autophagy, elevated reactive oxygen species, and subsequent inflammasome activation [36].
Unlike IRE1α and PERK, ATF6α contains a basic leucine zipper transcription factor domain within its cytosolic region. Upon dissociation of BiP during ER stress, ATF6α translocates to the Golgi apparatus, where it is cleaved by endopeptidases S1P and S2P. The released ATF6α p50 fragment then travels to the nucleus and induces genes that are involved in protein folding and ERAD [7]. The Atf6a−/− mice reconstituted with wild-type bone marrow cells are more vulnerable to DSS-induced colitis, likely due to diminished expression of ER chaperones including BiP and P58IPK and increased proapoptotic UPR signaling including CHOP in the colonocytes [37].
2.2. Oxidative Stress
Oxygen metabolism produces reactive oxygen species (ROS) including superoxide, hydroxyl radicals, hydrogen peroxide, hypochlorous acid, and lipid hydroperoxides [38]. The electron transport chain in mitochondria generates a large proportion of ROS [39, 40]. Other sources of endogenous ROS in mammals include the ER, peroxisomes, nucleus as well as the cytosol, and extracellular matrix. Under physiological conditions, the cell is protected from ROS by its antioxidant capacity. However, an excessive ROS production or insufficient antioxidative response can cause lipid and protein modifications, DNA damage, altered membrane permeability, inflammatory response, and cell death, all of which can contribute to the pathogenesis of IBD [41–44]
Multiple cell types in the gastrointestinal tract can be both perpetrators and victims of oxidative stress. Induced by inflammatory cytokines, the IECs produce superoxide and nitric oxide that can damage cytoskeleton proteins and alter tight junctions, subsequently leading to barrier disruption and exacerbation of mucosal inflammation [45, 46]. Upon activation, neutrophils and macrophages in the intestine generate more ROS, which forms a vicious circle of mucosal injury and inflammation [47, 48]. Furthermore, oxidative stress can be initiated by environmental factors, such as luminal antigens, cigarette smoking, alcohol, drugs, xenobiotics, radiation, and chemotherapy, all of which contribute to IBD [3, 49]. Tobacco smoking is one of best known environmental risk factors for CD, whereas it is considered as an alleviating factor of UC. How smoking affects CD risks is not fully understood. Several mechanisms have been proposed. First of all, the gas and tar of cigarette smoke contain a large amount of ROS. Second, cigarette smoke can affect the ROS production in the cell. For example, metal ions from tobacco smoke promote the generation of highly reactive hydroxyl radical from hydrogen peroxide [50]. The activity of inducible nitric oxide synthase is elevated in the small intestine after nicotine administration, although the underlying mechanism remains unclear. Third, tobacco smoke can impair the antioxidative response via multiple mechanisms including the inactivation of superoxide dismutase [51–53]. In recent years, the genetic variations of several antioxidant enzyme genes have been associated with IBD. Some examples include the genes encoding superoxide dismutase 2, glutathione S-transferase, NAD(P)H:quinone oxidoreductase 1, paraoxonase 1, and nuclear factor- (erythroid-derived 2) like 2 [54–61]. Interestingly, the loss of peroxiredoxin 6, an important antioxidative protein, partially protected mice against both acute and chronic DSS-induced colitis, likely by promoting a compensatory elevation of other antioxidative enzymes in the gastrointestinal tract [62]. However, it is unclear whether peroxiredoxin 6 has similar function in different cell populations (e.g. IECs versus immune cells) during intestinal inflammation. This data suggests that the network of antioxidative mechanisms in the gut may be more complex than we previously appreciated.
2.3. Hypoxia
The association between mucosal hypoxia and intestinal inflammation has been demonstrated by hypoxic staining of mucosal tissues in animal models of colitis as well as elevated hypoxia-induced transcription factors in inflamed intestinal samples from individuals with IBD [63–68]. There is also an interesting finding that travelling to altitude > 2000 meters or taking flights increased the risk of IBD exacerbations within 4 weeks of travel, suggesting the role of hypoxia in the course of IBD [69]. Under normal conditions, there is a steep oxygen gradient from the intestinal lumen towards the submucosa in mammalian gastrointestinal tract [68, 70]. Intestinal mucosa becomes more hypoxic during inflammation due to increased oxygen consumption by infiltrated immune cells as well as decreased oxygen supply due to local microvascular occlusion and thrombosis [71, 72]. Mammalian cells have developed evolutionarily conserved mechanisms, including the prolyl hydroxylase domain protein- (PHD-) hypoxia-inducible factors (HIFs) pathway and NF-κB, to code with hypoxic stress [73].
The degree of hypoxia in the GI tract was measured with different measures. The level of intestinal hypoxia under normal conditions, termed physiologic hypoxia, is indispensable for intestinal homeostasis [74]. Low-grade hypoxia promotes intestinal barrier function by inducing the expression of genes such as trefoil factor 3, mucin-3, multidrug resistance protein 1, CD39, CD73, and creatine kinases in IECs [75–78]. In addition to signal transduction via HIFs, PHD3 has been shown to stabilize the tight junction protein occludin in IECs by preventing its interaction with the E3 ligase Itch in a hydroxylase-independent manner [79]. Using a zebrafish model, intestinal hif1ab was shown to bind to a hypoxia-inducible response element of mammalian anterior gradient 2 (AGR2) that encodes an ER protein disulfide-isomerase [80]. AGR2 is associated with IBD and required for the differentiation and maturation of intestinal goblet cells in mice [81–84]. Furthermore, physiological level hypoxia in IECs is important for the constitutive expression of β defensin, an antimicrobial peptide, and netrin-1 that impedes neutrophil infiltration into intestinal mucosa [85, 86]. In contrast, severe hypoxia can elicit an inflammatory response in the gut by targeting multiple cell types. IECs produce higher level of inflammatory cytokines such as TNFα and even enter apoptosis when the local oxygen drops below the physiological level [87, 88]. Mucosal immune cells are also affected by hypoxia during inflammation. Similar to IECs, pathologic hypoxia induces expression of proinflammatory cytokines in macrophages and dendritic cells [89–91]. Importantly, hypoxic macrophages were shown to downregulate autophagy in IECs, a protective signaling against mucosal inflammation. Upon hypoxia, HIF-1 induces Wnt1 expression in macrophages, which suppressed autophagy in cocultured epithelial cells via the activation of β-catenin and mTOR [92]. Knockout of the gene encoding PHD1, an inhibitor of the HIF signaling, drives macrophages towards an anti-inflammatory M2 phenotype and attenuates IL-1b production in dendritic cells in response to lipopolysaccharide [93]. In addition, dendritic cell-specific deletion of Hif1α in mice worsened DSS-induced colitis. This is likely due to impaired activation of Foxp3+ regulatory T cells in the settings of decreased thymic stromal lymphopoietin receptor and IL-10 in Hif1a−/− dendritic cells [94]. Interestingly, hypoxia-challenged neutrophils may actually survive longer, thereby delaying the resolution of inflammation [95, 96]. Hypoxia also elicits the expression of β2 integrin that promotes leukocyte adhesion and extravasation [97]. Upon severe hypoxia, endothelial cells release proinflammatory cytokines, prostaglandins, platelet-activating factor, and P-selectin, all of which contribute to neutrophil recruitment and mucosal inflammation [98, 99].
3. Cellular Stress Responses Interact with Microbiome in Gastrointestinal Tract
A role of the gut microbiota in the pathogenesis of IBD has long been studied. In particular, both UC and DC are associated with dysbiosis, defined as a decrease in gut bacterial diversity due to a shift from commensal to potentially pathogenic species [100–102]. For example, the Firmicutes phylum usually decreases in the fecal samples of patients with CD compared to healthy individuals, while the Proteobacteria phylum including Escherichia coli is often increased in proportional abundance in patients with UC or CD [101, 103–109]. Studies of pediatric cohorts have revealed substantial differences in gut bacterial populations between diseased and healthy children [110–112]. Importantly, dysbiosis has also been observed in the stool and mucosal samples from newly diagnosed, treatment-naive children with CD [113], suggesting that dysbiotic alternations might precede the onset of clinical disease. The gut microbiota also includes viruses and fungi, whose roles in IBD have been increasingly appreciated. Bacteriophages are the predominant species of the gut virome as shown by metagenomic analyses of viral particles from human stool samples [114, 115]. Altered bacteriophage composition has been associated with IBD. In particular, expansion of Caudovirales bacteriophages was observed in patients with CD [116, 117]. A recent study suggests that some phages have immunomodulatory functions on human peripheral mononuclear cells by downregulating CD14, TLR4 while upregulating IL-10, IL-1R antagonist, and suppressor of cytokine signaling 3 [118]. However, whether bacteriophages in the gut have similar properties is unclear. There are very limited data about the role of eukaryotic viruses in IBD [119]. Similarly, changes in fungal composition in the gut have been reported in both fecal and mucosa samples. Animal studies suggest that fungi may affect intestinal homeostasis by regulating host metabolism, mucosal immune response, and other microorganisms in the gut [120], although a causal relationship between specific fungal species and IBD remains undetermined.
3.1. ER Stress and Gut Microbiota
The interactions between bacteria and the ER in mammalian cells have long been recognized and investigated (Figure 1). As a biosynthetic factory of the cell, the ER is a nutrient-rich environment presumably free of antimicrobial or hydrolytic enzymes, making it a safe haven for the survival and proliferation of many intracellular bacteria. There are few studies so far that directly examined how gut commensals interact with the ER. However, several gastrointestinal pathogens have been shown to induce ER stress and the UPR both in vitro and in vivo. For example, Helicobacter pylori can elicit ER stress in gastric epithelial cells with its vacuolating cytotoxin VacA, which activates the PERK-eIF2α-CHOP pathway and causes mitochondrial damage and subsequent apoptosis [120]. ER stress induction is not restricted to ER-dwelling microorganisms. Secreted toxins enable extracellular bacteria to disrupt ER homeostasis without entering the cell. Listeria monocytogenes can activate all three branches of the UPR and downstream apoptotic signaling via secretion of cytolysin listeriolysin O (LLO), likely by disturbing ER calcium homeostasis through its pore-forming activity [121, 122]. Other bacteria produce and deliver proteins with enzymatic activities specifically targeting the UPR pathway. Shiga-toxigenic Escherichia coli secretes subtilase cytotoxin that cleaves the ER chaperone BiP, triggering the UPR activation and subsequent cell cycle arrest via translation attenuation and degradation of cyclin D1 [123–125]. Several gastrointestinal pathogens are known to exploit ER stress and the UPR for its survival and/or replication. ER stress may play a key role in the mucosal inflammatory response to microorganisms and their secreted molecules. For example, cholera toxin and Shiga toxin undergo retro-translocation to the ER, where they fold appropriately before arriving at their final destinations in the IECs. In the ER lumen, the A subunits of cholera toxin (CTA) and Shiga toxin (Stx1A) activate NF-κB through direct binding to IRE1α and subsequent activation of RIDD [126]. The UPR activation by pathogens and their molecules also involves the pattern recognition receptors including Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain- (NOD-) like receptors (NLRs). In murine and human macrophages, ligands of TLR2/4 specifically activate the IRE1α-XBP1 pathway that requires recruitment of TNF receptor-associated factor 6 (TRAF6) to the TLR and subsequent ROS production by NADPH oxidase NOX2. The spliced XBP1 then induces the expression of Tnfα and Il6 by binding to their promoter regions [127]. IRE1α is known to activate the JNK-dependent inflammatory signaling through the recruitment of TRAF2 [128]. A recent study showed that the interaction between TRAF2 and NOD1/NOD2 is indispensable in this process [129]. This is of particular interest because some genetic variants of both NOD2 and XBP1 have been implicated in Crohn's disease [10]. So far, most studies have focused on the interactions between pathogenic bacteria and the ER. There is lack of data on how commensal bacteria affect the ER function and homeostasis.
Figure 1.
Cellular stress signaling interacts with the gut microbiome in the pathogenesis of IBD.
Viral invasion and replication can active the UPR in host cells. At least 36 eukaryotic viruses have been shown to trigger ER stress and induce downstream pathways including inflammatory and immune responses [130]. However, little is known about how gut virome affects the ER homeostasis in any cell type in the gastrointestinal tract. For the last decade, the findings that helminth infection may improve IBD have drawn our attention to the role of parasites in intestinal health [131, 132]. A recent study showed that infection of Schistosoma japonicum mitigated DSS-induced colitis in mice. Alleviated ER stress in the colonic tissues was shown to be partly responsible for the improved inflammation and mucosal cell apoptosis in infected mice, although the underlying mechanisms are unclear [133].
3.2. Oxidative Stress and Gut Microbiota
Oxidative stress in the gastrointestinal tract plays a multifactorial role in the pathogenesis of IBD. Previous studies have associated elevated ROS with dysbiosis in the gut. Given the radial oxygen gradient in the intestinal tract, bacteria residing on the colonic mucosa have higher oxygen tolerance and catalase expression relative to their luminal or stool-associated counterparts [134]. As inflammation usually drives an oxidative state, it might favor the outgrowth of aerotolerant phyla such as Actinobacteria and Proteobacteria in the gut, one example is the murine pathogen Citrobacter rodentium [135]. Furthermore, intestinal inflammation has been shown to upregulate the production of small molecules that function as terminal electron acceptors for facultative anaerobes such as Enterobacteriaceae [136, 137], thus contributing to dysbiosis in the intestine.
During intestinal inflammation, the gut microbiota can contribute to ROS production directly and/or indirectly through the mucosal cells [43] (Figure 1). One example is H. pylori that generates ROS both by itself and by inducing neutrophils to produce ROS [138]. Some bacteria in the gut can enhance the production of nitric oxide and nitrous oxide by activating macrophages in the settings of inflammation-induced DNA damage [139, 140]. Importantly, the host DNA repair mechanisms can be altered by bacterial modulation. Some strains of enteropathogenic E. coli have been shown to interfere with the DNA mismatch repair by secreted toxins, leading to apoptotic cell death and carcinogenesis [141, 142].
3.3. Hypoxic Stress and Gut Microbiota
The gut microbiota influence intestinal health in a number of ways, one of which is the production of short-chain fatty acids (SCFA) including butyrate, propionate, and acetate [143]. In addition to being absorbed into the circulation, SCFAs serve as energy sources of colonocytes and are important for intestinal barrier function. Butyrate augments regulatory T cell function by activating G-protein coupled receptors and leads to epigenetic modification through the inhibition of histone deacetylases [97]. In addition, metabolism of butyrate has recently been shown to boost the oxygen consumption and to stabilize HIFs in murine IECs. Consistently, the level of hypoxia is lower with decreased expression of HIFs in germ-free mice [143]. Depletion of butyrate-producing Clostridia using antibiotics facilitated the expansion of Salmonella in the more aerobic (or less hypoxic) mucosal microenvironment [144]. These data indicate that the gut microbiota composition dramatically impacts intestinal oxygenation versus hypoxia status, which orchestrates the composition of aerobes versus anaerobes and gut dysbiosis. A recent study linked the peroxisome proliferator-activated receptor-γ (PPARγ) to the bioavailability of oxygen in the microbial composition and intestinal homeostasis. Butyrate in the colon activates the nuclear receptor PPARγ, which drives the β-oxidation of butyrate in the colonocytes. This process is highly oxygen-consuming and thereby generates a hypoxic environment that limits the expansion of facultative anaerobic bacteria including the Enterobacteriaceae [145]. The ileal mucosa of patients with CD is predominantly colonized by adherent-invasive Escherichia coli (AIEC) [146], which requires the interaction between type 1 pili on AIEC and CEACAM6 receptors on the IECs [147]. Another study revealed potentially methylated dinucleotide CpGs within the hypoxia-responsive elements (HREs) of CEACAM6 promoter and the inverse correlation between the methylation and CEACAM6 expression in the IECs [148]. This data highlight a role of HRE methylation and HIF-1 in the regulation of AIEC colonization and ileal mucosal inflammation. Furthermore, hypoxia is a known regulator of autophagy and NLR signaling, two key innate immune mechanisms associated with IBD. A recent study showed that hypoxia suppresses NF-κB and NLRP3 signaling via hypoxia-induced autophagy in IECs, thereby relieving colitis in mice [149]. This data suggests that hypoxia is an important regulator of the microbial-epithelial interaction and innate immunity in the gut (Figure 1).
Importantly, the luminal oxygen level diminishes significantly from the small intestine to large intestine, together with other factors such as antimicrobial peptides and pH, creating distinct habitats for commensal bacteria. Fast-growing facultative anaerobes that effectively utilize simple carbohydrates while tolerating oxidative stress, bile acids, and antimicrobial peptides dominate in the small intestine [150]. For example, certain Clostridium spp. and some members of the phylum Proteobacteria are enriched in ileostomy samples from humans [151]. In contrast, the cecum and colon cultivate a more diverse community of bacteria. Fermentative polysaccharide-degrading anaerobes, notably the Bacteroidaceae and Clostridia, flourish in the large bowel thanks to its more hypoxic environment, slower transit time, a lack of simple carbon sources, and lower levels of antimicrobials [152]. The longitudinal gradient of oxygenation plays an important role in the intestinal homeostasis, either dependent or independent of the microbiota. It remains unclear whether the hypoxia gradient along the GI tract contributes to the longitudinal distribution of disease in UC versus CD.
4. Discussions
The complex pathophysiology of IBD is linked to the tremendous complexity of interactions between the host cells and microorganisms in the gastrointestinal tract. Gut microflora is a key component of the mammalian digestive system and has been shown to play a critical role in intestinal health in humans. Although the causative relationship between dysbiosis and the pathogenesis of IBD has not yet been established, data from both animal and human studies suggest that the altered composition and metabolic profiles are not merely a byproduct of intestinal inflammation. It is evident that gut microbiome can influence intestinal homeostasis by interacting with multiple types of cells in the gut through numerous signaling pathways. During evolution, cellular stress responses have developed as important players in the cross-talks with gut microorganisms, an essential component of environmental stress one faces on a daily basis. Both in vitro and in vivo studies have shown how gut microorganisms and their molecules activate cellular stress pathways, which can produce distinct effects on cell fate and tissue homeostasis. Recent studies found that the ER stress/UPR, antioxidative stress response, and hypoxic stress response may contribute to IBD via multiple mechanisms such as changes in cellular metabolism, altered differentiation and survival of cells, epithelial barrier impairment, proinflammatory response, and fibrosis and wound healing. The picture is further complicated by the fact that ER stress, oxidative stress, and hypoxia interact extensively during intestinal inflammation [10, 153, 154]. Future studies should focus on the network of cellular stress responses in the interaction with gut microbiome using tools such as systems biology.
There is evidence from animal studies that deletion of a single gene associated with the stress signaling (e.g., Xbp1) or transfer of colitogenic bacteria to wild-type mice is sufficient to cause inflammation [11, 155, 156]. However, such examples are sparse in humans and the vast majority of IBD cases are probably multifactorial involving both host genetics and environmental factors. It is possible that an altered stress response from either genetic or epigenetic defects can initiate clinical disease of IBD, when the individuals encounter certain microorganisms/molecules in the gut that target these pathways and/or their compensatory pathways directly or indirectly. One example of this susceptibility gene-microbial interaction in mice is that a combination of impaired autophagy (from a mutated Atg16L1) and colonization of murine norovirus results in Paneth cell dysfunction, resembling the abnormalities seen in some patients with Crohn's ileitis [157]. Microbiome GWAS (mGWAS) has shed light on how host genetics may interact with gut microbiota in IBD pathogenesis by combining human genome sequencing and 16s rRNA or metagenome sequencing of microbiome [158]. Using these tools, the human lactase gene LCT was associated with the abundance of Bifidobacterium in a small mGWAS and validated in subsequent cohorts [159]. Future mGWAS with larger sample sizes are likely to achieve higher statistical power [154]. Longitudinal studies that start tracking genetically susceptible individuals before the onset of clinical disease may shed light on the causality between certain microorganism populations and IBD. Following this strategy, a recently published study showed an alternation of intestinal virome that occurred before the development of type-1 diabetes-associated serum auto-antibodies in genetically susceptible children [160].
Bacteria are by far the most studied microorganisms in the gut. Some gastrointestinal pathogens are known to disturb ER function and modulate the UPR either by physically invading the ER lumen or via their secreted molecules. However, how specific gut commensal populations influence host's stress signaling remains elusive. It is likely that the interactions between the host and the commensal microorganisms follow rules different from those between the host and pathogens. Given the findings that certain bacterial species can trigger intestinal inflammation upon transfer to wild-type mice, it would be worthwhile to explore how these microorganisms affect cellular stress responses in the gastrointestinal tract. In addition, some bacterial metabolites can affect mucosal homeostasis via interactions with IECs and immune cells. Future studies should explore how these molecules cross-talk with the stress signaling pathways in these cells. While many mutations in the stress pathways alone do not cause clinical disease in mice, these models provide unique opportunities to study the interactions between susceptibility genes and specific populations of the microbiome in intestinal inflammation. While many human viruses have been shown to compromise ER function during their invasion and/or replication, the relevance of gut virome to ER stress or other stress pathways is unknown. Similarly, the potential interactions between the stress pathways and fungal species in the gut remain to be determined. The exploration into these puzzles will not only further our understanding of IBD but also spur the efforts in developing novel therapies for this disease.
Acknowledgments
The author thanks Dr. Randal Kaufman at Sanford Burnham Prebys Medical Discovery Institute, Dr. Nicholas Davidson at Washington University in St. Louis, and Dr. Averil Ma at the University of California, San Francisco, for discussions.
Conflicts of Interest
The author has no conflicts of interest to disclose.
References
- 1.Dahlhamer J. M., Zammitti E. P., Ward B. W., Wheaton A. G., Croft J. B. Prevalence of inflammatory bowel disease among adults aged ≥18 years — United States, 2015. MMWR. Morbidity and Mortality Weekly Report. 2016;65(42):1166–1169. doi: 10.15585/mmwr.mm6542a3. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan G. G. The global burden of IBD: from 2015 to 2025. Nature Reviews Gastroenterology & Hepatology. 2015;12(12):720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 3.Ananthakrishnan A. N., Bernstein C. N., Iliopoulos D., et al. Environmental triggers in IBD: a review of progress and evidence. Nature Reviews Gastroenterology & Hepatology. 2017;15(1):39–49. doi: 10.1038/nrgastro.2017.136. [DOI] [PubMed] [Google Scholar]
- 4.Amin-Wetzel N., Saunders R. A., Kamphuis M. J., et al. A J-protein co-chaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell. 2017;171(7):1625–1637.e13. doi: 10.1016/j.cell.2017.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han J., Kaufman R. J. Physiological/pathological ramifications of transcription factors in the unfolded protein response. Genes & Development. 2017;31(14):1417–1438. doi: 10.1101/gad.297374.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hetz C., Papa F. R. The unfolded protein response and cell fate control. Molecular Cell. 2018;69(2):169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Cao S. S., Kaufman R. J. Unfolded protein response. Current Biology. 2012;22(16):R622–R626. doi: 10.1016/j.cub.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Tavernier S. J., Osorio F., Vandersarren L., et al. Regulated IRE1-dependent mRNA decay sets the threshold for dendritic cell survival. Nature Cell Biology. 2017;19(6):698–710. doi: 10.1038/ncb3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M., Kaufman R. J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529(7586):326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 10.Grootjans J., Kaser A., Kaufman R. J., Blumberg R. S. The unfolded protein response in immunity and inflammation. Nature Reviews Immunology. 2016;16(8):469–484. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tschurtschenthaler M., Adolph T. E., Ashcroft J. W., et al. Defective ATG16L1-mediated removal of IRE1α drives Crohn’s disease–like ileitis. The Journal of Experimental Medicine. 2017;214(2):401–422. doi: 10.1084/jem.20160791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaser A., Lee A.-H., Franke A., et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134(5):743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adolph T. E., Tomczak M. F., Niederreiter L., et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503(7475):272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H.-S., Chen Y., Fan L., et al. The endoplasmic reticulum stress sensor IRE1α in intestinal epithelial cells is essential for protecting against colitis. Journal of Biological Chemistry. 2015;290(24):15327–15336. doi: 10.1074/jbc.M114.633560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M., Zhang S., Qiu Y., et al. Upregulation of miR-665 promotes apoptosis and colitis in inflammatory bowel disease by repressing the endoplasmic reticulum stress components XBP1 and ORMDL3. Cell Death & Disease. 2017;8(3, article e2699) doi: 10.1038/cddis.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosomi S., Grootjans J., Tschurtschenthaler M., et al. Intestinal epithelial cell endoplasmic reticulum stress promotes MULT1 up-regulation and NKG2D-mediated inflammation. Journal of Experimental Medicine. 2017;214(10):2985–2997. doi: 10.1084/jem.20162041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao S. S. Endoplasmic reticulum stress and unfolded protein response in inflammatory bowel disease. Inflammatory Bowel Diseases. 2015;21(3):636–644. doi: 10.1097/MIB.0000000000000238. [DOI] [PubMed] [Google Scholar]
- 18.Cao S. S. Epithelial ER stress in Crohn’s disease and ulcerative colitis. Inflammatory Bowel Diseases. 2016;22(4):984–993. doi: 10.1097/MIB.0000000000000660. [DOI] [PubMed] [Google Scholar]
- 19.Cao S. S., Luo K. L., Shi L. Endoplasmic reticulum stress interacts with inflammation in human diseases. Journal of Cellular Physiology. 2016;231(2):288–294. doi: 10.1002/jcp.25098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman R. J., Davies M. V., Pathak V. K., Hershey J. W. The phosphorylation state of eucaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Molecular and Cellular Biology. 1989;9(3):946–958. doi: 10.1128/MCB.9.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murtha-Riel P., Davies M. V., Scherer B. J., Choi S. Y., Hershey J. W., Kaufman R. J. Expression of a phosphorylation-resistant eukaryotic initiation factor 2 alpha-subunit mitigates heat shock inhibition of protein synthesis. Journal of Biological Chemistry. 1993;268(17):12946–12951. [PubMed] [Google Scholar]
- 22.Han J., Back S. H., Hur J., et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature Cell Biology. 2013;15(5):481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao S. S., Wang M., Harrington J. C., Chuang B.-M., Eckmann L., Kaufman R. J. Phosphorylation of eIF2α is dispensable for differentiation but required at a posttranscriptional level for paneth cell function and intestinal homeostasis in mice. Inflammatory Bowel Diseases. 2014;20(4):712–722. doi: 10.1097/MIB.0000000000000010. [DOI] [PubMed] [Google Scholar]
- 24.Cadwell K., Liu J. Y., Brown S. L., et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456(7219):259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bel S., Pendse M., Wang Y., et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357(6355):1047–1052. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tréton X., Pédruzzi E., Cazals–Hatem D., et al. Altered endoplasmic reticulum stress affects translation in inactive colon tissue from patients with ulcerative colitis. Gastroenterology. 2011;141(3):1024–1035. doi: 10.1053/j.gastro.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 27.Waldschmitt N., Berger E., Rath E., et al. C/EBP homologous protein inhibits tissue repair in response to gut injury and is inversely regulated with chronic inflammation. Mucosal Immunology. 2014;7(6):1452–1466. doi: 10.1038/mi.2014.34. [DOI] [PubMed] [Google Scholar]
- 28.Liu R., Li X., Huang Z., et al. CHOP-induced loss of intestinal epithelial stemness contributes to bile duct ligation-induced cholestatic liver injury. Hepatology (Baltimore, Md.) 2017;67(4):1441–1457. doi: 10.1002/hep.29540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taniuchi S., Miyake M., Tsugawa K., Oyadomari M., Oyadomari S. Integrated stress response of vertebrates is regulated by four eIF2α kinases. Scientific Reports. 2016;6(1, article 32886) doi: 10.1038/srep32886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalet A., Gatti E., Pierre P. Integration of PKR-dependent translation inhibition with innate immunity is required for a coordinated anti-viral response. FEBS Letters. 2015;589(14):1539–1545. doi: 10.1016/j.febslet.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Dhar A. The role of PKR as a potential target for treating cardiovascular diseases. Current Cardiology Reviews. 2017;13(1):28–31. doi: 10.2174/1573403X12666160526122600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hull C. M., Bevilacqua P. C. Discriminating self and non-self by RNA: roles for RNA structure, misfolding, and modification in regulating the innate immune sensor PKR. Accounts of Chemical Research. 2016;49(6):1242–1249. doi: 10.1021/acs.accounts.6b00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sud N., Rutledge A. C., Pan K., Su Q. Activation of the dsRNA-activated protein kinase PKR in mitochondrial dysfunction and inflammatory stress in metabolic syndrome. Current Pharmaceutical Design. 2016;22(18):2697–2703. doi: 10.2174/1381612822666160202141845. [DOI] [PubMed] [Google Scholar]
- 34.Cao S. S., Song B., Kaufman R. J. PKR protects colonic epithelium against colitis through the unfolded protein response and prosurvival signaling. Inflammatory Bowel Diseases. 2012;18(9):1735–1742. doi: 10.1002/ibd.22878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao S. S., Kaufman R. J. PKR in DSS-induced colitis: a matter of genetic background and maternal microflora? Inflammatory Bowel Diseases. 2013;19(3):E49–E50. doi: 10.1002/ibd.22978. [DOI] [PubMed] [Google Scholar]
- 36.Ravindran R., Loebbermann J., Nakaya H. I., et al. The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature. 2016;531(7595):523–527. doi: 10.1038/nature17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao S. S., Zimmermann E. M., Chuang B.–. M., et al. The unfolded protein response and chemical chaperones reduce protein misfolding and colitis in mice. Gastroenterology. 2013;144(5):989–1000.e6. doi: 10.1053/j.gastro.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bedard K., Krause K.-H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiological Reviews. 2007;87(1):245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 39.Novak E. A., Mollen K. P. Mitochondrial dysfunction in inflammatory bowel disease. Frontiers in Cell and Development Biology. 2015;3 doi: 10.3389/fcell.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poyton R. O., Castello P. R., Ball K. A., Woo D. K., Pan N. Mitochondria and hypoxic signaling: a new view. Annals of the New York Academy of Sciences. 2009;1177(1):48–56. doi: 10.1111/j.1749-6632.2009.05046.x. [DOI] [PubMed] [Google Scholar]
- 41.Ridnour L. A., Isenberg J. S., Espey M. G., Thomas D. D., Roberts D. D., Wink D. A. Nitric oxide regulates angiogenesis through a functional switch involving thrombospondin-1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(37):13147–13152. doi: 10.1073/pnas.0502979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valko M., Leibfritz D., Moncol J., Cronin M. T. D., Mazur M., Telser J. Free radicals and antioxidants in normal physiological functions and human disease. The International Journal of Biochemistry & Cell Biology. 2007;39(1):44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Tian T., Wang Z., Zhang J. Pathomechanisms of oxidative stress in inflammatory bowel disease and potential antioxidant therapies. Oxidative Medicine and Cellular Longevity. 2017;2017:18. doi: 10.1155/2017/4535194.4535194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereira C., Coelho R., Grácio D., et al. DNA damage and oxidative DNA damage in inflammatory bowel disease. Journal of Crohn's & Colitis. 2016;10(11):1316–1323. doi: 10.1093/ecco-jcc/jjw088. [DOI] [PubMed] [Google Scholar]
- 45.Banan A., Choudhary S., Zhang Y., Fields J. Z., Keshavarzian A. Ethanol-induced barrier dysfunction and Its prevention by growth factors in human intestinal monolayers: evidence for oxidative and cytoskeletal mechanisms. The Journal of Pharmacology and Experimental Therapeutics. 1999;291(3):1075–1085. [PubMed] [Google Scholar]
- 46.Rao R., Baker R. D., Baker S. S. Inhibition of oxidant-induced barrier disruption and protein tyrosine phosphorylation in Caco-2 cell monolayers by epidermal growth factor. Biochemical Pharmacology. 1999;57(6):685–695. doi: 10.1016/S0006-2952(98)00333-5. [DOI] [PubMed] [Google Scholar]
- 47.Knutson C. G., Mangerich A., Zeng Y., et al. Chemical and cytokine features of innate immunity characterize serum and tissue profiles in inflammatory bowel disease. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(26):E2332–E2341. doi: 10.1073/pnas.1222669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pacher P., Beckman J. S., Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiological Reviews. 2007;87(1):315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danese S., Sans M., Fiocchi C. Inflammatory bowel disease: the role of environmental factors. Autoimmunity Reviews. 2004;3(5):394–400. doi: 10.1016/j.autrev.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Wang H., Yu M., Ochani M., et al. Nicotinic acetylcholine receptor α7 subunit is an essential regulator of inflammation. Nature. 2003;421(6921):384–388. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- 51.Eliakim R., Karmeli F., Cohen P., Heyman S. N., Rachmilewitz D. Dual effect of chronic nicotine administration: augmentation of jejunitis and amelioration of colitis induced by iodoacetamide in rats. International Journal of Colorectal Disease. 2001;16(1):14–21. doi: 10.1007/s003840000262. [DOI] [PubMed] [Google Scholar]
- 52.Guo X., Ko J. K. S., Mei Q.-B., Cho C. H. Aggravating effect of cigarette smoke exposure on experimental colitis is associated with leukotriene B4 and reactive oxygen metabolites. Digestion. 2001;63(3):180–187. doi: 10.1159/000051887. [DOI] [PubMed] [Google Scholar]
- 53.Guo X., Wang W. P., Ko J. K. S., Cho C. H. Involvement of neutrophils and free radicals in the potentiating effects of passive cigarette smoking on inflammatory bowel disease in rats. Gastroenterology. 1999;117(4):884–892. doi: 10.1016/S0016-5085(99)70347-1. [DOI] [PubMed] [Google Scholar]
- 54.Mittal R. D., Manchanda P. K., Bid H. K., Ghoshal U. C. Analysis of polymorphisms of tumor necrosis factor-α and polymorphic xenobiotic metabolizing enzymes in inflammatory bowel disease: study from northern India. Journal of Gastroenterology and Hepatology. 2007;22(6):920–924. doi: 10.1111/j.1440-1746.2006.04538.x. [DOI] [PubMed] [Google Scholar]
- 55.Kosaka T., Yoshino J., Inui K., et al. Involvement of NAD(P)H:quinone oxidoreductase 1 and superoxide dismutase polymorphisms in ulcerative colitis. DNA and Cell Biology. 2009;28(12):625–631. doi: 10.1089/dna.2009.0877. [DOI] [PubMed] [Google Scholar]
- 56.Karban A., Hartman C., Eliakim R., et al. Paraoxonase (PON)1 192R allele carriage is associated with reduced risk of inflammatory bowel disease. Digestive Diseases and Sciences. 2007;52(10):2707–2715. doi: 10.1007/s10620-006-9700-5. [DOI] [PubMed] [Google Scholar]
- 57.Arisawa T., Tahara T., Shibata T., et al. Nrf2 gene promoter polymorphism is associated with ulcerative colitis in a Japanese population. Hepato-Gastroenterology. 2008;55(82-83):394–397. [PubMed] [Google Scholar]
- 58.Karban A., Krivoy N., Elkin H., et al. Non-Jewish Israeli IBD patients have significantly higher glutathione S-transferase GSTT1-null frequency. Digestive Diseases and Sciences. 2011;56(7):2081–2087. doi: 10.1007/s10620-010-1543-4. [DOI] [PubMed] [Google Scholar]
- 59.Senhaji N., Kassogue Y., Fahimi M., Serbati N., Badre W., Nadifi S. Genetic polymorphisms of multidrug resistance gene-1 (MDR1/ABCB1) and glutathione S-transferase gene and the risk of inflammatory bowel disease among Moroccan patients. Mediators of Inflammation. 2015;2015:8. doi: 10.1155/2015/248060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varzari A., Deyneko I. V., Tudor E., Turcan S. Polymorphisms of glutathione S-transferase and methylenetetrahydrofolate reductase genes in Moldavian patients with ulcerative colitis: genotype-phenotype correlation. Meta Gene. 2016;7:76–82. doi: 10.1016/j.mgene.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mrowicki J., Mrowicka M., Majsterek I., Mik M., Dziki A., Dziki Ł. Evaluation of effect CAT -262C/T, SOD + 35A/C, GPx1 Pro197Leu polymorphisms in patients with IBD in the Polish population. Polish Journal of Surgery. 2016;88(6):321–327. doi: 10.1515/pjs-2016-0071. [DOI] [PubMed] [Google Scholar]
- 62.Melhem H., Spalinger M. R., Cosin-Roger J., et al. Prdx6 deficiency ameliorates DSS colitis: relevance of compensatory antioxidant mechanisms. Journal of Crohn's & Colitis. 2017;11(7):871–884. doi: 10.1093/ecco-jcc/jjx016. [DOI] [PubMed] [Google Scholar]
- 63.Karhausen J., Furuta G. T., Tomaszewski J. E., Johnson R. S., Colgan S. P., Haase V. H. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. Journal of Clinical Investigation. 2004;114(8):1098–1106. doi: 10.1172/JCI200421086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giatromanolaki A., Sivridis E., Maltezos E., et al. Hypoxia inducible factor 1α and 2α overexpression in inflammatory bowel disease. Journal of Clinical Pathology. 2003;56(3):209–213. doi: 10.1136/jcp.56.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu C., Dong W. Role of hypoxia-inducible factor-1α in pathogenesis and disease evaluation of ulcerative colitis. Experimental and Therapeutic Medicine. 2016;11(4):1330–1334. doi: 10.3892/etm.2016.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hindryckx P., Waeytens A., Laukens D., et al. Absence of placental growth factor blocks dextran sodium sulfate-induced colonic mucosal angiogenesis, increases mucosal hypoxia and aggravates acute colonic injury. Laboratory Investigation. 2010;90(4):566–576. doi: 10.1038/labinvest.2010.37. [DOI] [PubMed] [Google Scholar]
- 67.Schreiber S., Nikolaus S., Hampe J. Activation of nuclear factor κB in inflammatory bowel disease. Gut. 1998;42(4):477–484. doi: 10.1136/gut.42.4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown E., Taylor C. T. Hypoxia-sensitive pathways in intestinal inflammation. The Journal of Physiology. 2017 doi: 10.1113/JP274350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vavricka S. R., Rogler G., Maetzler S., et al. High altitude journeys and flights are associated with an increased risk of flares in inflammatory bowel disease patients. Journal of Crohn's & Colitis. 2014;8(3):191–199. doi: 10.1016/j.crohns.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 70.Fisher E. M., Khan M., Salisbury R., Kuppusamy P. Noninvasive monitoring of small intestinal oxygen in a rat model of chronic mesenteric ischemia. Cell Biochemistry and Biophysics. 2013;67(2):451–459. doi: 10.1007/s12013-013-9611-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hatoum O. A., Miura H., Binion D. G. The vascular contribution in the pathogenesis of inflammatory bowel disease. American Journal of Physiology-Heart and Circulatory Physiology. 2003;285(5):H1791–H1796. doi: 10.1152/ajpheart.00552.2003. [DOI] [PubMed] [Google Scholar]
- 72.Campbell E. L., Bruyninckx W. J., Kelly C. J., et al. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity. 2014;40(1):66–77. doi: 10.1016/j.immuni.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Welden S., Selfridge A. C., Hindryckx P. Intestinal hypoxia and hypoxia-induced signalling as therapeutic targets for IBD. Nature Reviews Gastroenterology & Hepatology. 2017;14:596–611. doi: 10.1038/nrgastro.2017.101. [DOI] [PubMed] [Google Scholar]
- 74.Colgan S. P., Dzus A. L., Parkos C. A. Epithelial exposure to hypoxia modulates neutrophil transepithelial migration. Journal of Experimental Medicine. 1996;184(3):1003–1015. doi: 10.1084/jem.184.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Synnestvedt K., Furuta G. T., Comerford K. M., et al. Ecto-5′-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. Journal of Clinical Investigation. 2002;110(7):993–1002. doi: 10.1172/JCI0215337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Furuta G. T., Turner J. R., Taylor C. T., et al. Hypoxia-inducible factor 1–dependent induction of intestinal trefoil factor protects barrier function during hypoxia. The Journal of Experimental Medicine. 2001;193(9):1027–1034. doi: 10.1084/jem.193.9.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Louis N. A., Hamilton K. E., Canny G., Shekels L. L., Ho S. B., Colgan S. P. Selective induction of mucin-3 by hypoxia in intestinal epithelia. Journal of Cellular Biochemistry. 2006;99(6):1616–1627. doi: 10.1002/jcb.20947. [DOI] [PubMed] [Google Scholar]
- 78.Comerford K. M., Wallace T. J., Karhausen J., Louis N. A., Montalto M. C., Colgan S. P. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Research. 2002;62(12):3387–3394. [PubMed] [Google Scholar]
- 79.Chen Y., Zhang H.-S., Fong G.-H., et al. PHD3 stabilizes the tight junction protein occludin and protects intestinal epithelial barrier function. Journal of Biological Chemistry. 2015;290(33):20580–20589. doi: 10.1074/jbc.M115.653584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lai Y.-R., Lu Y.-F., Lien H.-W., Huang C. J., Hwang S. P. L. Foxa2 and Hif1ab regulate maturation of intestinal goblet cells by modulating agr2 expression in zebrafish embryos. Biochemical Journal. 2016;473(14):2205–2218. doi: 10.1042/BCJ20160392. [DOI] [PubMed] [Google Scholar]
- 81.Zheng W., Rosenstiel P., Huse K., et al. Evaluation of AGR2 and AGR3 as candidate genes for inflammatory bowel disease. Genes & Immunity. 2006;7(1):11–18. doi: 10.1038/sj.gene.6364263. [DOI] [PubMed] [Google Scholar]
- 82.Chen Y.-C., Lu Y.-F., Li I.-C., Hwang S. P. L. Zebrafish Agr2 is required for terminal differentiation of intestinal goblet cells. PLoS One. 2012;7(4, article e34408) doi: 10.1371/journal.pone.0034408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao F., Edwards R., Dizon D., et al. Disruption of Paneth and goblet cell homeostasis and increased endoplasmic reticulum stress in Agr2−/− mice. Developmental Biology. 2010;338(2):270–279. doi: 10.1016/j.ydbio.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pelaseyed T., Bergström J. H., Gustafsson J. K., et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunological Reviews. 2014;260(1):8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kelly C. J., Glover L. E., Campbell E. L., et al. Fundamental role for HIF-1α in constitutive expression of human β defensin-1. Mucosal Immunology. 2013;6(6):1110–1118. doi: 10.1038/mi.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rosenberger P., Schwab J. M., Mirakaj V., et al. Hypoxia-inducible factor–dependent induction of netrin-1 dampens inflammation caused by hypoxia. Nature Immunology. 2009;10(2):195–202. doi: 10.1038/ni.1683. [DOI] [PubMed] [Google Scholar]
- 87.Taylor C. T., Dzus A. L., Colgan S. P. Autocrine regulation of epithelial permeability by hypoxia: role for polarized release of tumor necrosis factor α. Gastroenterology. 1998;114(4):657–668. doi: 10.1016/S0016-5085(98)70579-7. [DOI] [PubMed] [Google Scholar]
- 88.Taylor C. T., Fueki N., Agah A., Hershberg R. M., Colgan S. P. Critical role of cAMP response element binding protein expression in hypoxia-elicited induction of epithelial tumor necrosis factor-α. Journal of Biological Chemistry. 1999;274(27):19447–19454. doi: 10.1074/jbc.274.27.19447. [DOI] [PubMed] [Google Scholar]
- 89.Yun J. K., McCormick T. S., Villabona C., Judware R. R., Espinosa M. B., Lapetina E. G. Inflammatory mediators are perpetuated in macrophages resistant to apoptosis induced by hypoxia. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(25):13903–13908. doi: 10.1073/pnas.94.25.13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Köhler T., Reizis B., Johnson R. S., Weighardt H., Förster I. Influence of hypoxia-inducible factor 1α on dendritic cell differentiation and migration. European Journal of Immunology. 2012;42(5):1226–1236. doi: 10.1002/eji.201142053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blengio F., Raggi F., Pierobon D., et al. The hypoxic environment reprograms the cytokine/chemokine expression profile of human mature dendritic cells. Immunobiology. 2013;218(1):76–89. doi: 10.1016/j.imbio.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 92.Ortiz-Masiá D., Cosín-Roger J., Calatayud S., et al. Hypoxic macrophages impair autophagy in epithelial cells through Wnt1: relevance in IBD. Mucosal Immunology. 2014;7(4):929–938. doi: 10.1038/mi.2013.108. [DOI] [PubMed] [Google Scholar]
- 93.Sun M., He C., Wu W., et al. Hypoxia inducible factor-1α-induced interleukin-33 expression in intestinal epithelia contributes to mucosal homeostasis in inflammatory bowel disease. Clinical & Experimental Immunology. 2017;187(3):428–440. doi: 10.1111/cei.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Flück K., Breves G., Fandrey J., Winning S. Hypoxia-inducible factor 1 in dendritic cells is crucial for the activation of protective regulatory T cells in murine colitis. Mucosal Immunology. 2016;9(2):379–390. doi: 10.1038/mi.2015.67. [DOI] [PubMed] [Google Scholar]
- 95.Hannah S., Mecklenburgh K., Rahman I., et al. Hypoxia prolongs neutrophil survival in vitro. FEBS Letters. 1995;372(2-3):233–237. doi: 10.1016/0014-5793(95)00986-J. [DOI] [PubMed] [Google Scholar]
- 96.Mecklenburgh K. I., Walmsley S. R., Cowburn A. S., et al. Involvement of a ferroprotein sensor in hypoxia-mediated inhibition of neutrophil apoptosis. Blood. 2002;100(8):3008–3016. doi: 10.1182/blood-2002-02-0454. [DOI] [PubMed] [Google Scholar]
- 97.Kong L., Wang J., Han W., Cao Z. Modeling heterogeneity in direct infectious disease transmission in a compartmental model. International Journal of Environmental Research and Public Health. 2016;13(3):3–253. doi: 10.3390/ijerph13030253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Michiels C., Arnould T., Knott I., Dieu M., Remacle J. Stimulation of prostaglandin synthesis by human endothelial cells exposed to hypoxia. American Journal of Physiology-Cell Physiology. 1993;264(4):C866–C874. doi: 10.1152/ajpcell.1993.264.4.C866. [DOI] [PubMed] [Google Scholar]
- 99.Michiels C., Arnould T., Remacle J. Endothelial cell responses to hypoxia: initiation of a cascade of cellular interactions. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2000;1497(1):1–10. doi: 10.1016/S0167-4889(00)00041-0. [DOI] [PubMed] [Google Scholar]
- 100.Ott S. J., Musfeldt M., Wenderoth D. F., et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53(5):685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Manichanh C., Rigottier-Gois L., Bonnaud E., et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55(2):205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ni J., Wu G. D., Albenberg L., Tomov V. T. Gut microbiota and IBD: causation or correlation? Nature Reviews Gastroenterology & Hepatology. 2017;14(10):573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sokol H., Pigneur B., Watterlot L., et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Walker A. W., Sanderson J. D., Churcher C., et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiology. 2011;11(1):p. 7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gophna U., Sommerfeld K., Gophna S., Doolittle W. F., Veldhuyzen van Zanten S. J. O. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. Journal of Clinical Microbiology. 2006;44(11):4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Frank D. N., St. Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(34):13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martinez-Medina M., Aldeguer X., Gonzalez-Huix F., Acero D., Garcia-Gil J. L. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflammatory Bowel Diseases. 2006;12(12):1136–1145. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- 108.Rehman A., Rausch P., Wang J., et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut. 2016;65(2):238–248. doi: 10.1136/gutjnl-2014-308341. [DOI] [PubMed] [Google Scholar]
- 109.Baumgart M., Dogan B., Rishniw M., et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. The ISME Journal. 2007;1(5):403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 110.Assa A., Butcher J., Li J., et al. Mucosa-associated ileal microbiota in new-onset pediatric Crohn’s disease. Inflammatory Bowel Diseases. 2016;22(7):1533–1539. doi: 10.1097/MIB.0000000000000776. [DOI] [PubMed] [Google Scholar]
- 111.Kellermayer R., Mir S. A. V., Nagy-Szakal D., et al. Microbiota separation and C-reactive protein elevation in treatment-naïve pediatric granulomatous Crohn disease. Journal of Pediatric Gastroenterology and Nutrition. 2012;55(3):243–250. doi: 10.1097/MPG.0b013e3182617c16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shah R., Cope J. L., Nagy-Szakal D., et al. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes. 2016;7(5):384–396. doi: 10.1080/19490976.2016.1190073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gevers D., Kugathasan S., Denson L. A., et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host & Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reyes A., Haynes M., Hanson N., et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Minot S., Sinha R., Chen J., et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Research. 2011;21(10):1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wagner J., Maksimovic J., Farries G., et al. Bacteriophages in gut samples from pediatric Crohn’s disease patients: metagenomic analysis using 454 pyrosequencing. Inflammatory Bowel Diseases. 2013;19(8):1598–1608. doi: 10.1097/MIB.0b013e318292477c. [DOI] [PubMed] [Google Scholar]
- 117.Norman J. M., Handley S. A., Baldridge M. T., et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160(3):447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Van Belleghem J. D., Clement F., Merabishvili M., Lavigne R., Vaneechoutte M. Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Scientific Reports. 2017;7(1):p. 8004. doi: 10.1038/s41598-017-08336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Magin W. S., Van Kruiningen H. J., Colombel J.-F. Immunohistochemical search for viral and bacterial antigens in Crohn’s disease. Journal of Crohn's & Colitis. 2013;7(2):161–166. doi: 10.1016/j.crohns.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 120.Akazawa Y., Isomoto H., Matsushima K., et al. Endoplasmic reticulum stress contributes to Helicobacter pylori VacA-induced apoptosis. PLoS One. 2013;8(12, article e82322) doi: 10.1371/journal.pone.0082322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gekara N. O., Groebe L., Viegas N., Weiss S. Listeria monocytogenes desensitizes immune cells to subsequent Ca2+ signaling via listeriolysin O-induced depletion of intracellular Ca2+ stores. Infection and Immunity. 2008;76(2):857–862. doi: 10.1128/IAI.00622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pillich H., Loose M., Zimmer K.-P., Chakraborty T. Activation of the unfolded protein response by listeria monocytogenes. Cellular Microbiology. 2012;14(6):949–964. doi: 10.1111/j.1462-5822.2012.01769.x. [DOI] [PubMed] [Google Scholar]
- 123.Paton A. W., Beddoe T., Thorpe C. M., et al. AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature. 2006;443(7111):548–552. doi: 10.1038/nature05124. [DOI] [PubMed] [Google Scholar]
- 124.Wolfson J. J., May K. L., Thorpe C. M., Jandhyala D. M., Paton J. C., Paton A. W. Subtilase cytotoxin activates PERK, IRE1 and ATF6 endoplasmic reticulum stress-signalling pathways. Cellular Microbiology. 2008;10(9):1775–1786. doi: 10.1111/j.1462-5822.2008.01164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morinaga N., Yahiro K., Matsuura G., Moss J., Noda M. Subtilase cytotoxin, produced by Shiga-toxigenic Escherichia coli, transiently inhibits protein synthesis of Vero cells via degradation of BiP and induces cell cycle arrest at G1 by downregulation of cyclin D1. Cellular Microbiology. 2008;10(4):921–929. doi: 10.1111/j.1462-5822.2007.01094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cho J. A., Lee A.-H., Platzer B., et al. The unfolded protein response element IRE1α senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell Host & Microbe. 2013;13(5):558–569. doi: 10.1016/j.chom.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 127.Martinon F., Chen X., Lee A.-H., Glimcher L. H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nature Immunology. 2010;11(5):411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Urano F., Wang X., Bertolotti A., et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 129.Keestra-Gounder A. M., Byndloss M. X., Seyffert N., et al. NOD1 and NOD2 signalling links ER stress with inflammation. Nature. 2016;532(7599):394–397. doi: 10.1038/nature17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li S., Kong L., Yu X. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Critical Reviews in Microbiology. 2015;41(2):150–164. doi: 10.3109/1040841X.2013.813899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heylen M., Ruyssers N. E., Gielis E. M., et al. Of worms, mice and man: an overview of experimental and clinical helminth-based therapy for inflammatory bowel disease. Pharmacology & Therapeutics. 2014;143(2):153–167. doi: 10.1016/j.pharmthera.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 132.Cheifetz A. S., Gianotti R., Luber R., Gibson P. R. Complementary and alternative medicines used by patients with inflammatory bowel diseases. Gastroenterology. 2017;152(2):415–429.e15. doi: 10.1053/j.gastro.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 133.Liu Y., Ye Q., Liu Y.-L., Kang J., Chen Y., Dong W. G. Schistosoma japonicum attenuates dextran sodium sulfate-induced colitis in mice via reduction of endoplasmic reticulum stress. World Journal of Gastroenterology. 2017;23(31):5700–5712. doi: 10.3748/wjg.v23.i31.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Albenberg L., Esipova T. V., Judge C. P., et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147(5):1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lopez C. A., Miller B. M., Rivera-Chavez F., et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science. 2016;353(6305):1249–1253. doi: 10.1126/science.aag3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Winter S. E., Lopez C. A., Bäumler A. J. The dynamics of gut-associated microbial communities during inflammation. EMBO Reports. 2013;14(4):319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lewis J. D., Chen E. Z., Baldassano R. N., et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host & Microbe. 2017;22(2):p. 247. doi: 10.1016/j.chom.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 138.Handa O., Naito Y., Yoshikawa T. Helicobacter pylori: a ROS-inducing bacterial species in the stomach. Inflammation Research. 2010;59(12):997–1003. doi: 10.1007/s00011-010-0245-x. [DOI] [PubMed] [Google Scholar]
- 139.Irrazábal T., Belcheva A., Girardin S. E., Martin A., Philpott D. J. The multifaceted role of the intestinal microbiota in colon cancer. Molecular Cell. 2014;54(2):309–320. doi: 10.1016/j.molcel.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 140.Lundberg J. O., Weitzberg E., Cole J. A., Benjamin N. Nitrate, bacteria and human health. Nature Reviews Microbiology. 2004;2(7):593–602. doi: 10.1038/nrmicro929. [DOI] [PubMed] [Google Scholar]
- 141.Maddocks O. D. K., Short A. J., Donnenberg M. S., Bader S., Harrison D. J. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One. 2009;4(5, article e5517) doi: 10.1371/journal.pone.0005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Maddocks O. D. K., Scanlon K. M., Donnenberg M. S. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. mBio. 2013;4(3):e00152–e00113. doi: 10.1128/mBio.00152-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kelly C. J., Zheng L., Campbell E. L., et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host & Microbe. 2015;17(5):662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rivera-Chávez F., Zhang L. F., Faber F., et al. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host & Microbe. 2016;19(4):443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Byndloss M. X., Olsan E. E., Rivera-Chávez F., et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017;357(6351):570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Martin H. M., Campbell B. J., Hart C. A., et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127(1):80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 147.Carvalho F. A., Barnich N., Sivignon A., et al. Crohn’s disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. The Journal of Experimental Medicine. 2009;206(10):2179–2189. doi: 10.1084/jem.20090741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Denizot J., Desrichard A., Agus A., et al. Diet-induced hypoxia responsive element demethylation increases CEACAM6 expression, favouring Crohn’s disease-associated Escherichia coli colonisation. Gut. 2015;64(3):428–437. doi: 10.1136/gutjnl-2014-306944. [DOI] [PubMed] [Google Scholar]
- 149.Cosin-Roger J., Simmen S., Melhem H., et al. Hypoxia ameliorates intestinal inflammation through NLRP3/mTOR downregulation and autophagy activation. Nature Communications. 2017;8(1):p. 98. doi: 10.1038/s41467-017-00213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Islam K. B. M. S., Fukiya S., Hagio M., et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141(5):1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 151.Zoetendal E. G., Raes J., van den Bogert B., et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. The ISME Journal. 2012;6(7):1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Donaldson G. P., Lee S. M., Mazmanian S. K. Gut biogeography of the bacterial microbiota. Nature Reviews Microbiology. 2016;14(1):20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cao S. S., Kaufman R. J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxidants & Redox Signaling. 2014;21(3):396–413. doi: 10.1089/ars.2014.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chong W. C., Shastri M. D., Eri R. Endoplasmic reticulum stress and oxidative stress: a vicious nexus implicated in bowel disease pathophysiology. International Journal of Molecular Sciences. 2017;18(4) doi: 10.3390/ijms18040771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Garrett W. S., Lord G. M., Punit S., et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131(1):33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Palm N. W., de Zoete M. R., Cullen T. W., et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158(5):1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cadwell K., Patel K. K., Maloney N. S., et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141(7):1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hall A. B., Tolonen A. C., Xavier R. J. Human genetic variation and the gut microbiome in disease. Nature Reviews Genetics. 2017;18(11):690–699. doi: 10.1038/nrg.2017.63. [DOI] [PubMed] [Google Scholar]
- 159.Blekhman R., Goodrich J. K., Huang K., et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biology. 2015;16(1):p. 191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhao G., Vatanen T., Droit L., et al. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(30):E6166–E6175. doi: 10.1073/pnas.1706359114. [DOI] [PMC free article] [PubMed] [Google Scholar]