Abstract
Introducing new chemical reactivity into proteins in living cells would endow innovative covalent bonding ability to proteins for research and engineering in vivo. Latent bioreactive unnatural amino acids (Uaas) can be incorporated into proteins to react with target natural amino acid residues via proximity-enabled reactivity. To expand the diversity of proteins amenable to such reactivity in vivo, a chemical functionality that is biocompatible and able to react with multiple natural residues under physiological conditions is highly desirable. Here we report the genetic encoding of fluorosulfate-L-tyrosine (FSY), the first latent bioreactive Uaa that undergoes sulfur-fluoride exchange (SuFEx) on proteins in vivo. FSY was found nontoxic to E. coli and mammalian cells; after being incorporated into proteins, it selectively reacted with proximal lysine, histidine, and tyrosine via SuFEx, generating covalent intra-protein bridge and inter-protein crosslink of interacting proteins directly in living cells. The proximity-activatable reactivity, multi-targeting ability, and excellent biocompatibility of FSY will be invaluable for covalent manipulation of proteins in vivo. Moreover, genetically encoded FSY hereby empowers general proteins with the next generation of click chemistry, SuFEx, which will afford broad utilities in chemical biology, drug discovery, and biotherapeutics.
Graphical Abstract
The ability to endow proteins with new chemical reactivity will expand novel avenues for protein research and engineering, potentially impacting biological studies, biotherapeutics, and synthetic biology.1 The genetic code has recently been engineered to encode a new class of unnatural amino acids (Uaas), the latent bioreactive Uaas.2 These Uaas, after incorporation into proteins, react with a target natural amino acid residue via proximity-enabled reactivity, enabling selective formation of new covalent linkages within and between proteins. The new covalent bonding ability has been harnessed within proteins to enhance photostability,2 increase thermostability,3 staple proteins recombinantly,4 and build optical nano-switches,5 and between proteins to pinpoint ligand-receptor interaction,6 target native receptors irreversibly,4,7 generate covalent macromolecular inhibitors,8 and capture weak protein-protein interactions.9 Bioreactive Uaas have initially been developed to target Cys2 and subsequently to target Lys, His and Tyr.3–4,7,10 Reacting with Lys, His and Tyr often requires basic conditions that are infeasible in vivo.4,10b At this burgeoning stage, most genetically encoded bioreactive Uaas react with 1 to 2 natural residues only, have modest to severe cytotoxicity, and are primarily used for in vitro applications.
An ideal chemical functionality for latent bioreactive Uaas should be stable, nontoxic, and nonreactive inside cells, yet upon placed in proximity to target residues becomes reactive under cellular conditions. Aryl fluorosulfates are weak electrophiles attracting great attention for the new generation of click chemistry:11 they are relatively unreactive toward hydrolysis, thermolysis and reduction,11–12 and are unreactive toward free amino acids under physiological conditions,13 but form covalent bonds with Tyr, Lys and Ser through sulfur-fluoride exchange (SuFEx) reaction when used as war-heads in chemical probes specifically binding proteins,14 although a positively charged microenvironment is required for reactivity. Given the tremendous potential of aryl fluorosulfate for biocompatible reactivity emerging from recent small molecule studies, it is desirable to genetically install this functional group onto proteins so that its unique properties can be exploited in large biomolecules and in vivo.
Here we evolved a new tRNA-synthetase pair to genetically incorporate fluorosulfate-L-tyrosine (FSY, Fig. 1a) into proteins in E. coli and mammalian cells. We found that FSY was generally nontoxic to cells, and was able to react with Lys, His, and Tyr specifically via proximity-enabled SuFEx reaction within and between proteins under physiological conditions (Fig. 1b). We demonstrated the crosslinking of interacting proteins using FSY directly in vivo.
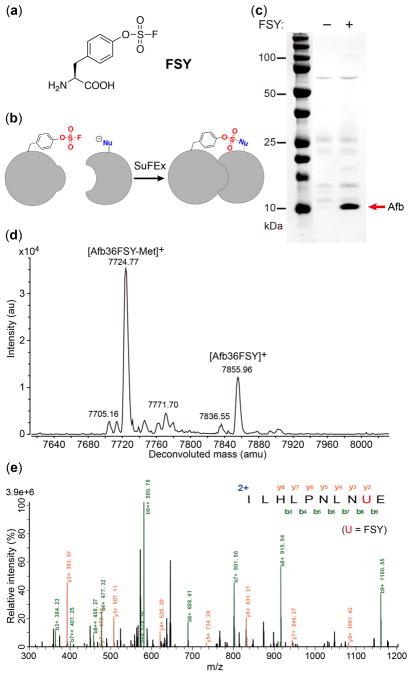
Figure 1.
Genetically encode FSY into proteins in E. coli. (a) Structure of FSY. (b) Scheme showing proximity-enabled SuFEx reaction between FSY and a natural nucleophilic residue. (c) SDS-PAGE showing FSY incorporation into Afb(36TAG) in E. coli. (d) ESI-TOF MS spectrum of intact Afb-36FSY. (e) Tandem MS spectrum of Z-24FSY.
FSY was synthesized using the SO2F2/borax method (88% yield, Supplementary methods).11,15 To genetically encode FSY, we evolved a mutant pyrrolysyl-tRNA synthetase (PylRS) specific for FSY. A PylRS mutant library was generated by mutating residues Ala302, Leu305, Tyr306, Leu309, Ile322, Asn346, Cys348, Tyr384, Val401, and Trp417 of the Methanosarcina mazei PylRS using the small-intelligent mutagenesis approach,16 and subjected to selection as described.17 Six hits showing FSY-dependent phenotype were identified; they all converged on the same amino acid sequence (302I/346T/348I/384L/417K), which we named as FSYRS.
We evaluated the incorporation specificity of FSY into proteins in E. coli. The Zspa affibody (Afb) gene containing a TAG codon at position 36 (Afb-36TAG) was co-expressed with the pair in E. coli. In the absence of FSY, no full-length Afb was detected; when 1 mM FSY was added in growth media, full-length Afb36FSY was produced with a yield of 1.6 mg/L (Fig. 1c). The purified Afb36FSY was analyzed by electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS) (Fig. 1d). A peak observed at 7855.96 Da corresponds to intact Afb containing FSY at site 36 (Afb36FSY: expected 7856.69 Da). A peak measured at 7724.77 Da corresponds to Afb36FSY lacking the initiating Met (Afb36FSY-Met: expected 7725.50 Da). Two minor peaks observed at 7836.55 and 7705.16 Da correspond to Afb36FSY lacking F (expected 7836.69 Da) and Afb36FSY-Met lacking F (expected 7705.49 Da), respectively, suggesting slight F elimination during MS measurement. Notably, no peaks corresponding to Afb containing other amino acids at position 36 were observed. We also incorporated FSY at position 24 of the Z protein and analyzed with tandem MS. A series of b and y ions unambiguously indicate that FSY was incorporated at the TAG-specified position 24 (Fig. 1e). The presence of 1 mM FSY did not affect E. coli growth (Fig. S1), suggesting no obvious cytotoxicity. These results indicate that the evolved pair was able to incorporate FSY with high efficiency and specificity in E. coli.
We next tested FSY incorporation into proteins in mammalian cells. We transfected HeLa-EGFP-182TAG reporter cells18 with plasmid pMP-FSYRS-3xtRNA, which expresses FSYRS and genes. Suppression of the 182TAG codon of the genome-integrated EGFP gene would produce full-length EGFP rendering cells fluorescent. Strong EGFP fluorescence was measured from cells by flow cytometry only when FSY was added (Fig. 2a). The fluorescence intensity increased with FSY concentration and incubation time (Fig. 2b). As a positive control, we incorporated p-azido-L-phenylalanine (AzF) into reporter cells in parallel using plasmid pIre-Azi3 (Fig. S2), which is the most efficient Uaa incorporation system in mammalian cells in our hands.6 FSY incorporation reached 76% of the AzF level. Notably, while cellular toxicity is often an issue with bioreactive Uaas, we did not observe obvious toxicity of FSY to HeLa or HEK293T cells (Fig. S3), a valuable property of FSY possibly owing to the extremely low background reactivity of aryl fluorosulfate inside cells.14a The above results were also confirmed by fluorescence confocal microscopy (Fig. 2c). In the presence of FSY, strong EGFP fluorescence was observed throughout the cells, and cell morphology remained normal. No fluorescence signal was detected when FSY was not added. We also incorporated FSY into proliferating cell nuclear antigen (PCNA) through transient transfection of PCNA-165TAG and genes into HEK293T cells. Full-length PCNA was produced in the presence of FSY, with incorporation efficiency reaching 41% of AzF (Fig. 2d). These results suggest that FSY was incorporated into proteins in mammalian cells with high efficiency and specificity without causing detrimental effects.
Figure 2.
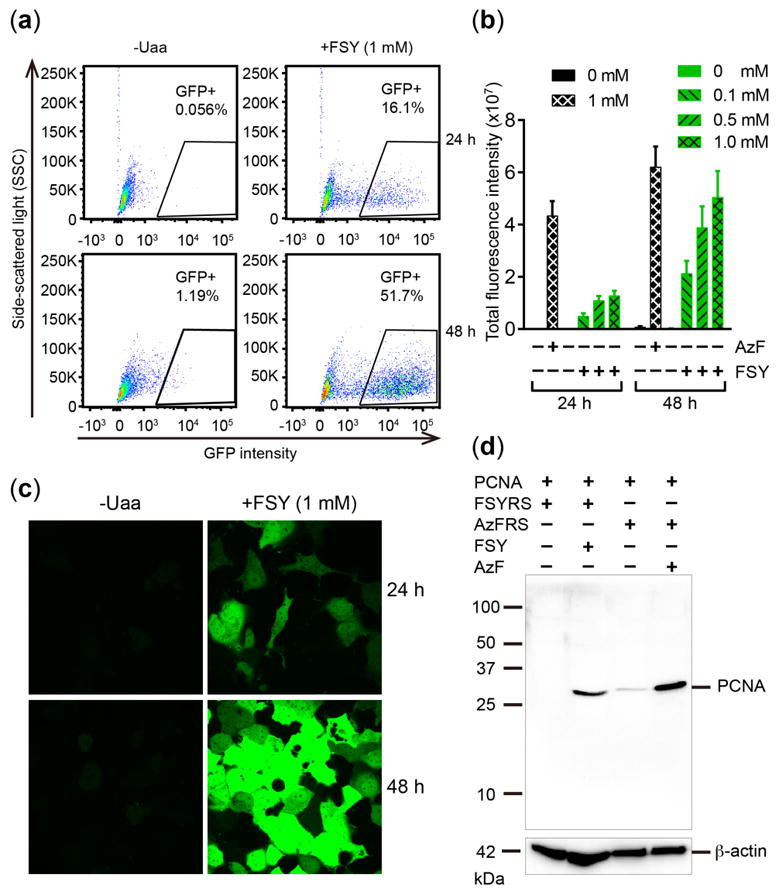
Genetically encode FSY into proteins in mammalian cells. (a) FACS analysis of FSY incorporation into EGFP-182TAG in HeLa cells. (b) Total EGFP fluorescence intensity measured from the same number of HeLa-EGFP-182TAG reporter cells. Error bar: s.e.m., n = 6. (c) Fluorescence images of HeLa-EGFP-182TAG reporter cells. (d) Western blot analysis of FSY and AzF incorporation into PCNA-165TAG in HEK293T cells. An anti-His antibody was used to detect the 6xHis-tag appended at the C-terminus of PCNA.
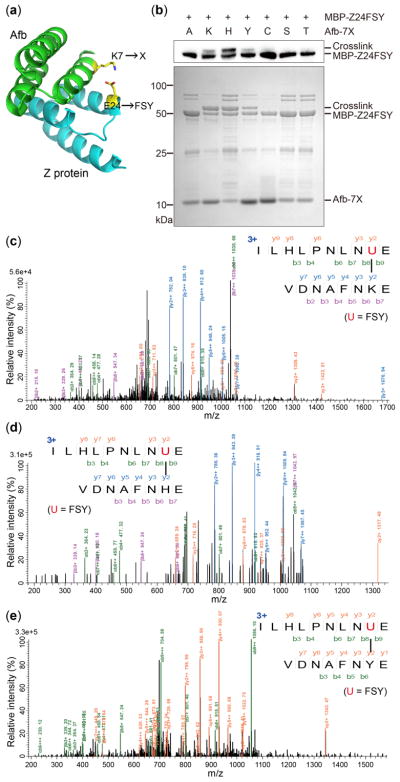
We then determined whether the incorporated FSY could react with natural amino acid residues via proximity-enabled reactivity directly in E. coli. Afb binds to its substrate Z protein with a moderate affinity, providing a suitable protein framework to study FSY crosslinking in vivo. In light of the crystal structure of Afb-Z complex,19 we introduced FSY at position 24 of Z protein and the target natural residue at position 7 of Afb, placing the two residues in close proximity upon Afb-Z binding (Fig. 3a). As aryl fluorosulfate is a weak electrophile, we decided to test FSY’s reactivity toward Lys, His, Tyr, Cys, Ser, and Thr using Ala as a negative control. To better separate the Afb and Z proteins of similar molecular weights, we fused maltose binding protein (MBP) to the N-terminus of Z (MBP-Z). MBP-Z and Afb were both appended with a 6xHis-tag at C-terminus. To determine whether chemical crosslinking could occur in living cells, we co-expressed MBP-Z24FSY and Afb-7X (X = target residue) in E. coli. After culturing at 37 °C for 6 h, the same number of cells were analyzed using Western blot under denatured conditions. From cells expressing Afb-7Lys, Afb-7His, or Afb-7Tyr, crosslinking bands were observed with molecular weight corresponding to MBP-Z24FSY and Afb adducts (Fig. 3b). We then purified 6xHis-tagged proteins from cells and analyzed with SDS-PAGE. Consistently, a protein band corresponding to the crosslinked MBP-Z with Afb was clearly observed for Afb-7Lys, Afb-7His, and Afb-7Tyr (Fig. 3b), with crosslinking efficiency (determined by band intensities) of 59%, 53% and 35%, respectively. In contrast, no cross-linking bands were observed when MBP-Z24FSY was co-expressed with Afb-7Cys, Afb-7Ser, Afb-7Thr, or Afb-7Ala. While aryl carbamate requires basic pH to crosslink Lys or Tyr at Afb/Z interface in vitro,10b FSY was able to crosslink Lys, His or Tyr directly in live E. coli cells.
Figure 3.
FSY crosslinks proximal Lys, His and Tyr via SuFEx directly in E. coli cells. (a) Structure of Afb-Z complex showing two proximal sites for FSY and target residue X incorporation. (b) Top: Western blot of E. coli cell lysates; Bottom: SDS-PAGE of proteins His-tag purified from E. coli. (c–e) Tandem MS spectrum of MBP-Z-24FSY/Afb-7Lys (c), MBP-Z-24FSY/Afb-7His (d) and MBP-Z-24FSY/Afb-7Tyr (e).
To further validate the in vivo chemical crosslinking ability of FSY, we analyzed the purified proteins using tandem MS. As expected, strong signals corresponding to the covalently-linked peptides of MBP-Z24FSY and Afb-7Lys were identified (Fig. 3c). A series of b and y fragmented ions clearly indicate that the incorporated FSY crosslinked exclusively with Lys 7 of Afb. Similar MS results were also obtained for MBP-Z24FSY co-expressed with Afb-7His or Afb-7Tyr, confirming FSY crosslinked with the target His7 (Fig. 3d) or Tyr7 (Fig. 3e), respectively. Meanwhile, consistent with Western and SDS-PAGE results, no crosslinked peptides of MBP-Z24FSY with Afb-7Ser, Afb-7Thr, Afb-7Cys, or Afb-7Ala were detected by tandem MS.
In addition, we incorporated FSY and Tyr into a single protein to test intramolecular crosslinking in vivo. We co-expressed in E. coli the pair with a mutant calmodulin gene (CaM-76TAG), which encoded 76TAG for FSY incorporation and Tyr at a nearby site 80 (Fig. 4a). This CaM protein was expressed in the presence of 1 mM FSY, purified (Fig. 4b), and analyzed with tandem MS (Fig. 4c). A series of b and y fragment ions unambiguously show that FSY formed a covalent linkage with Tyr80 via SuFEx.
Figure 4.
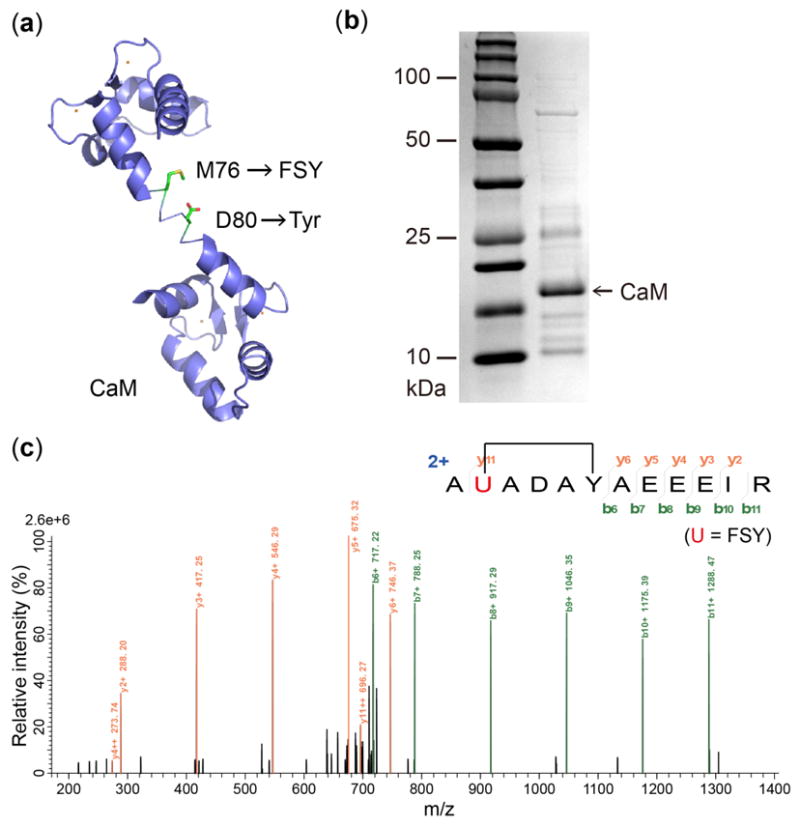
FSY crosslinks Tyr via SuFEx intramolecularly in E. coli cells. (a) Structure of CaM showing sites for FSY and target Tyr. (b) SDS-PAGE and (c) tandem MS spectrum of purified CaM-76FSY-80Tyr.
To demonstrate that FSY could crosslink interacting proteins through targeting a native Tyr residue, we tested whether FSY-armed thioredoxin (Trx) could covalently capture 3′-phosphoadenosine-5′-phosphosulfate (PAPS) reductase. Trx interacts with PAPS reductase to facilitate the reduction of adenylated sulfate to sulfite for de novo cysteine biosynthesis.20 On the basis of the complex structure of PAPS reductase with Trx1,20 we incorporated FSY into E. coli Trx1 at site 62 to target the proximal Tyr191 of PAPS reductase (Fig. 5a). We expressed and purified Trx1(62FSY) and WT PAPS reductase, and incubated them in Tris buffer at pH 7.4 or 8.0 for 12 h. SDS-PAGE showed clear bands corresponding to the covalent complex of Trx1 with PAPS reductase (Fig. 5b, Fig. S4). Tandem MS analysis of the sample unambiguously indicate that FSY of Trx1 covalently crosslinked with the target Tyr191 of PAPS reductase via SuFEx reaction (Fig. 5c). Taken together, the intramolecular CaM crosslinking and intermolecular Trx1-PAPS reductase crosslinking further corroborate that FSY reacted with Tyr in proximity via SuFEx reaction.
Figure 5.
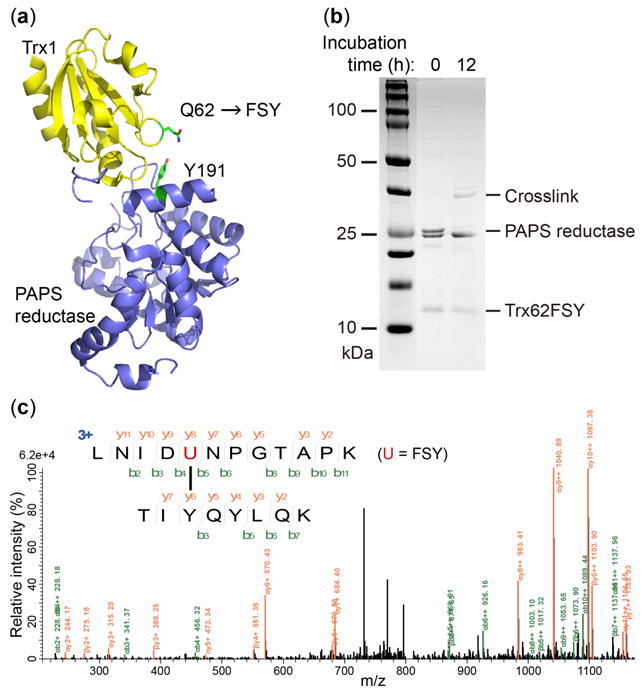
FSY crosslinks Tyr via SuFEx intermolecularly. (a) Structure of Trx1 in complex with PAPS reductase showing FSY site and the native Tyr191. (b) SDS-PAGE and (c) tandem MS spectrum of Trx1 crosslinked with PAPS reductase.
In summary, we genetically encoded the live-cell friendly FSY into proteins in E. coli and mammalian cells, which selectively reacted with proximal Lys, His and Tyr residues via SuFEx directly in live E. coli cells. Intermolecular crosslinking using bioreactive Uaas has been mainly limited to in vitro usage with few exceptions targeting Cys;6,9a FSY enables intermolecular crosslinking of interacting proteins in vivo and targeting three different residues. Since FSY’s target residues are often found at protein surface and interface, FSY will dramatically expand the diversity of proteins amenable to covalent bonding and enable creative in vivo applications to exploit protein covalent bonding ability. Moreover, genetically encoding FSY now empowers proteins with the new generation of click chemistry, SuFEx, which will find broad applications in chemical biology, drug discovery, and biotherapeutics.
Supplementary Material
Acknowledgments
L.W. acknowledges the support of the NIH (R01GM118384, RF1MH114079).
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information. The Supporting Information is available free of charge on the ACS Publications website. Experimental details, Figure S1–S3 (PDF)
References
- 1.Wang L. N Biotechnol. 2017;38:16–25. doi: 10.1016/j.nbt.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiang Z, Ren H, Hu YS, Coin I, Wei J, Cang H, Wang L. Nat Methods. 2013;10:885–888. doi: 10.1038/nmeth.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang Z, Lacey VK, Ren H, Xu J, Burban DJ, Jennings PA, Wang L. Angew Chem Int Ed Engl. 2014;53:2190–2193. doi: 10.1002/anie.201308794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen XH, Xiang Z, Hu YS, Lacey VK, Cang H, Wang L. ACS Chem Biol. 2014;9:1956–1961. doi: 10.1021/cb500453a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Hoppmann C, Lacey VK, Louie GV, Wei J, Noel JP, Wang L. Angew Chem Int Ed Engl. 2014;53:3932–3936. doi: 10.1002/anie.201400001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hoppmann C, Maslennikov I, Choe S, Wang L. J Am Chem Soc. 2015;137:11218–11221. doi: 10.1021/jacs.5b06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coin I, Katritch V, Sun T, Xiang Z, Siu FY, Beyermann M, Stevens RC, Wang L. Cell. 2013;155:1258–1269. doi: 10.1016/j.cell.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furman JL, Kang M, Choi S, Cao Y, Wold ED, Sun SB, Smider VV, Schultz PG, Kim CH. J Am Chem Soc. 2014;136:8411–8417. doi: 10.1021/ja502851h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoppmann C, Wang L. Chem Commun (Camb) 2016;52:5140–5143. doi: 10.1039/c6cc01226d. [DOI] [PubMed] [Google Scholar]
- 9.(a) Yang B, Tang S, Ma C, Li ST, Shao GC, Dang B, DeGrado WF, Dong MQ, Wang PG, Ding S, Wang L. Nat Commun. 2017;8:2240. doi: 10.1038/s41467-017-02409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cigler M, Muller TG, Horn-Ghetko D, von Wrisberg MK, Fottner M, Goody RS, Itzen A, Muller MP, Lang K. Angew Chem Int Ed Engl. 2017;56:15737–15741. doi: 10.1002/anie.201706927. [DOI] [PubMed] [Google Scholar]
- 10.(a) Xuan W, Li J, Luo X, Schultz PG. Angew Chem Int Ed Engl. 2016;55:10065–10068. doi: 10.1002/anie.201604891. [DOI] [PubMed] [Google Scholar]; (b) Xuan W, Shao S, Schultz PG. Angew Chem Int Ed Engl. 2017;56:5096–5100. doi: 10.1002/anie.201611841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong J, Krasnova L, Finn MG, Sharpless KB. Angew Chem Int Ed Engl. 2014;53:9430–9448. doi: 10.1002/anie.201309399. [DOI] [PubMed] [Google Scholar]
- 12.Narayanan A, Jones LH. Chem Sci. 2015;6:2650–2659. doi: 10.1039/c5sc00408j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee H, Debreczeni J, Breed J, Tentarelli S, Aquila B, Dowling JE, Whitty A, Grimster NP. Org Biomol Chem. 2017;15:9685–9695. doi: 10.1039/c7ob02028g. [DOI] [PubMed] [Google Scholar]
- 14.(a) Chen W, Dong J, Plate L, Mortenson DE, Brighty GJ, Li S, Liu Y, Galmozzi A, Lee PS, Hulce JJ, Cravatt BF, Saez E, Powers ET, Wilson IA, Sharpless KB, Kelly JW. J Am Chem Soc. 2016;138:7353–7364. doi: 10.1021/jacs.6b02960. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mortenson DE, Brighty GJ, Plate L, Bare G, Chen W, Li S, Wang H, Cravatt BF, Forli S, Powers ET, Sharpless KB, Wilson IA, Kelly JW. J Am Chem Soc. 2018;140:200–210. doi: 10.1021/jacs.7b08366. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Fadeyi OO, Hoth LR, Choi C, Feng X, Gopalsamy A, Hett EC, Kyne RE, Jr, Robinson RP, Jones LH. ACS Chem Biol. 2017;12:2015–2020. doi: 10.1021/acschembio.7b00403. [DOI] [PubMed] [Google Scholar]
- 15.Chen W, Dong J, Li S, Liu Y, Wang Y, Yoon L, Wu P, Sharpless KB, Kelly JW. Angew Chem Int Ed Engl. 2016;55:1835–1838. doi: 10.1002/anie.201509016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacey VK, Louie GV, Noel JP, Wang L. Chem Bio Chem. 2013;14:2100–2105. doi: 10.1002/cbic.201300400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Wang L, Schultz PG. Angew Chem Int Ed Engl. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]; (b) Takimoto JK, Dellas N, Noel JP, Wang L. ACS Chem Biol. 2011;6:733–743. doi: 10.1021/cb200057a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee KF, Slesinger PA, Wang L. Nat Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogbom M, Eklund M, Nygren PA, Nordlund P. Proc Natl Acad Sci U S A. 2003;100:3191–3196. doi: 10.1073/pnas.0436100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chartron J, Shiau C, Stout CD, Carroll KS. Biochemistry. 2007;46:3942–3951. doi: 10.1021/bi700130e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.