Abstract
The block copolymer VIPER (virus-inspired polymer for endosomal release) has been reported to be a promising novel delivery system of DNA plasmids both in vitro and in vivo. VIPER is comprised of a polycation segment for condensation of nucleic acids as well as a pH-sensitive segment that exposes the membrane lytic peptide melittin in acidic environments to facilitate endosomal escape. The objective of this study was to investigate VIPER/siRNA polyplex characteristics, and compare their in vitro and in vivo performance with commercially available transfection reagents and a control version of VIPER lacking melittin. VIPER/siRNA polyplexes were formulated and characterized at various charge ratios and shown to be efficiently internalized in cultured cells. Target mRNA knockdown was confirmed by both flow cytometry and qRT-PCR and the kinetics of knockdown was monitored by live cell spinning disk microscopy, revealing knockdown starting by 4 hours post-delivery. Intratracheal instillation of VIPER particles formulated with sequence specific siRNA to the lung of mice resulted in a significantly more efficient knockdown of GAPDH compared to treatment with VIPER particles formulated with scrambled sequence siRNA. We also demonstrated using pH-sensitive labels that VIPER particles experience less acidic environments compared to control polyplexes. In summary, VIPER/siRNA polyplexes efficiently deliver siRNA in vivo resulting in robust gene silencing (>75% knockdown) within the lung.
Keywords: siRNA delivery, pulmonary delivery, endosomal escape, pH-sensitive polymer, melittin
1. Introduction
The potential to treat a variety of diseases with nucleic acid-based drugs has remained high ever since the first gene transfer approaches were developed in the early 1980s for animal models. [1] In the last decade, several gene therapy products have been approved for clinical use around the world.[2–4] A critical component in these formulations is the delivery vector. While extremely efficient at gene transfer, viral systems are well known to have numerous safety concerns along with high manufacturing costs.[5] Non-viral synthetic polymer systems on the other hand, have enhanced safety profiles but suffer from lower gene transfer efficiencies.[6] Numerous studies have identified endosomal release to be one of the key factors contributing to successful transfection efficiency in cells treated with polymeric carriers of nucleic acid-based drugs.[7] In order to address the issue of endosomal entrapment, various mechanisms have been utilized to improve endosomal release of synthetic polymer carriers.[8] The use of polymers with buffering capabilities has been hypothesized to lead to the rupture of the endosomal membrane, but requires concentrations of polymer that are difficult to achieve in vivo. Conversely, while the use of membrane-active peptides to disrupt the endosomal membrane are promising, off-site toxicity may occur if the peptides are not properly shielded.
Recently, Pun et al. designed a membrane lytic peptide – polymer system that mimics the mechanism of efficient endosomal escape employed by adenovirus.[9] The polymer consists of a hydrophilic block for nucleic acid binding and a reversibly hydrophobic block that is selectively membrane disruptive in acidic environments. Upon cellular internalization and trafficking to acidic endosomes, the hydrophobic block switches to hydrophilic and exposes a membrane-lytic peptide, melittin.[10] Plasmid DNA delivered by the polymer, named VIPER (virus-inspired polymer for endosomal release), avoids endosomal entrapment, resulting in efficient delivery in vitro and in vivo.
In addition to plasmid DNA gene therapy, another promising gene therapy strategy to address various untreatable diseases involves the use of RNA interference (RNAi) through the delivery of small-interfering RNA (siRNA). siRNA has long been proposed as a highly selective approach of disease treatment due to its ability to potentially knock down any gene of interest involved in disease development or progression, [11] and is therefore an attractive alternative to conventional cancer therapy.[12] Although plasmid DNA and siRNA are both comprised of synthetic nucleic acids, the two molecules differ greatly.[13] Most importantly, plasmid DNA consists of a double stranded sequence that can range in size from a few hundred to thousands of base pairs whereas siRNA only contains 20-25 base pairs. Due to the differences between these therapeutic molecules, our study set out to investigate the ability of the VIPER system to package siRNA and evaluate its ability to mediate gene knockdown in vitro and in vivo.
2. Materials and Methods
2.1. Materials
Hyperbranched polyethylenimine (PEI, 25k Da) was obtained from BASF (Ludwigshafen, DE). Ham's F-12K (Kaighn's) medium and RPMI-1640 medium with HEPES was purchased from Gibco (Carlsbad, CA, US). Dulbecco’s phosphate buffered saline (PBS), heat-inactivated fetal bovine serum (FBS), D-(+)-glucose, sodium bicarbonate, sodium pyruvate, 2-mercaptoethanol, dimethyl sulfoxide (DMSO, ≥ 99.7%), ethylenediaminetetraacetic acid (EDTA, 99.4−100.06%) and trypan blue (0.4%, sterile filtered) were purchased from Sigma-Aldrich (St. Louis, MO, US). SYBR Gold dye and Lipofectamine 2000 were obtained from Life Technologies (Carlsbad, CA, US). Dicer substrate double-stranded siRNA (DsiRNA) targeting the enhanced green fluorescent protein gene (EGFP siRNA,25/27), human (hGAPDH) and mouse GAPDH (msGAPDH), scrambled nonspecific control DsiRNA as well as amine-modified and TYE™-563 labelled siRNA were purchased from Integrated DNA Technologies (IDT, Coralville, IA, US). Chloroquine diphosphate was bought from MP Biomedicals (Santa Ana, CA, US).
2.2. Synthesis of p(OEGMA-DMAEMA)-b-p(DIPAMA-PDSEMA)
The block copolymer p(OEGMA11-DMAEMA56)-b-p(DIPAMA33-PDSEMA1) was synthesized via reversible addition-fragmentation chain transfer (RAFT) polymerization as described previously and was characterized by 1H NMR, GPC, and UV spectroscopy. [9]
2.3. Conjugation of cys-melittin to p(OEGMA11-DMAEMA56)-b-p(DIPAMA33-PDSEMA1)
The block copolymer was further modified with cys-melittin through a disulfide exchange reaction which was described before.[14] The unmodified copolymer was used as a control polymer in all studies and is denoted as CP whereas the block copolymer that was modified by cys-melittin is denoted as VIPER.
2.4. Preparation of siRNA polyplexes
The polymer/siRNA polyplexes were assembled by dissolving each polymer in water to yield separate 0.5 mg/ml solutions which were then filtered through a 0.22 μm filter for sterilization. These stock solutions were further diluted to pre-calculated concentrations with sterile 5% glucose solution. An equal volume of each polymer solution was added to a defined amount of siRNA solution and incubated for 20 minutes to yield polyplexes at various N/P ratios (the molar ratio of nitrogen in amine to phosphate in siRNA). All calculations were based on previous experiments with plasmid DNA.[14]
2.5. SYBR gold binding assay
A SYBR® gold assay was used to investigate the ability of the unmodified block copolymer (CP) or melittin-conjugated block copolymer (VIPER) to successfully condense siRNA at various N/P ratios. Upon intercalation with uncomplexed double-stranded nucleic acids, the SYBR® Gold fluorescent dye (Life Technologies) undergoes a more than 1000-fold signal enhancement. However, this signal is reduced if the siRNA is condensed within polyplexes, thereby blocking the dye from intercalating with the nucleic acids. To test the polyplexes that were prepared using either CP, VIPER or PEI, 50 µl of siRNA in 5% glucose (1 pmol/µl) was distributed in each well of a white FluoroNunc 96-well plate (Thermo Fisher Scientific, Waltham, MA, US) followed by adding 50 µl of the diluted polymer solution to form polyplexes at increasing N/P ratios. Once mixed, the plate was incubated for 20 min at room temperature. Following incubation, 30 µl of a 4× SYBR® gold solution was added to each well and incubated for 10 min in the dark at room temperature. The fluorescence of each sample was quantified using a Synergy 2 multi-mode microplate reader (BioTek Instrument, Winooski, VT, US) at excitation wavelength of 485/20 nm and emission wavelength of 520/20 nm. Samples were normalized based on the following criteria. The fluorescence level of 100% free siRNA was calculated based on the fluorescence of uncondensed free siRNA incubated with SYBR® gold dye. The fluorescence of zero percent free siRNA was calculated with SYBR® gold dye in the absence of any siRNA. All measurements were performed in triplicates and the results are displayed as mean values.
2.6. Hydrodynamic size and Zeta (ζ) Potential Measurements of siRNA Polyplexes
Hydrodynamic diameter measurements of polyplexes were performed by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments Inc., Malvern, UK). Polyplexes were formed at various N/P ratios with scrambled siRNA in 5% glucose solution. A total volume of 50 µl of each sample was added into a disposal cuvette (Brand GmbH, Wertheim, Germany) and the 173° backscatter angle measurement was read in triplicates with each run consisting of 15 scans. Results are represented as average size (nm) ± standard deviation. The samples were then diluted to 1 mL with filtered Nanopure water and transferred to a folded capillary cell (Malvern Instruments Inc., Malvern, UK), and ζ-potential measurements were taken. ζ-Potential measurements were read in triplicates by laser Doppler anemometry (LDA), with each run consisting of 30-50 scans. Results are shown in mV ± standard deviation.
2.7. Transmission electron microscope (TEM)
siRNA (10 µL at 0.1 µg/µL) and either VIPER or CP were mixed at N/P 8 and incubated for 10 minutes for polyplex formation. Polyplexes were then deposited on 400-mesh formvar/copper grids (recently glow discharged for 45 s) and incubated at room temperature for 30 min. The grids were touched to a 50 μL drop of 4% (w/v) uranyl acetate for 10-15 s and then dragged across filter paper to create a negative staining gradient across the grid. Grids were dried in a desiccator overnight and imaged with a JEOL JEM 1400 Transmission Electron Microscope at the Fred Hutchinson Cancer Research Center.
2.8. Cell culture
NCI-H1299 cells are a human non-small cell lung carcinoma cell line and were obtained from ATTC (Manassas, VA, US). H1299/GFP cells are a H1299 cell line stably expressing the reporter gene EGFP. Both H1299 and H1299/GFP cells were cultured in RPMI-1640 medium supplemented with 10% heat inactivated fetal bovine serum, 1% penicillin/streptomycin, 2 mM L-Glutamine, 10 mM HEPES, 1.5 g/L NaHCO3, 4.5 g/L glucose and 1mM sodium pyruvate. Jurkat cells are a human T lymphocyte cell line and were a kind gift from Dr. Larry H. Matherly (Wayne State School of Medicine, Detroit, MI, US). Jurkat T lymphocytes were cultured at a concentration of 106 cells/ml in RPMI-1640 medium supplemented with 10 % heat inactivated fetal bovine serum, 1% penicillin/streptomycin, 2 mM L- Glutamine and 10 mM HEPES. All cells were maintained in a humidified atmosphere within 75 cm2 cell culture flasks (Thermo Fisher Scientific) and 5% CO2 at 37 °C.
2.9. In vitro cellular uptake
Amine modified scrambled sequence siRNA was labeled with succinimidyl ester (NHS) modified Alexa Fluor-488 dye (Life Technologies) as described previously. [15] For all uptake experiments, negative controls consisted of blank/untreated cells while positive control cells were transfected with Lipofectamine 2000 (Life Technologies) lipoplexes, which were prepared according to the manufacturer’s protocol. Briefly, every 10 pmol of AF488-siRNA were formulated with 0.5 µL Lipofectamine solution (LF). One day prior to transfection, 50,000 H1299 cells were seeded and incubated overnight at 37°C and 5% CO2 in 24-well plates (Corning Incorporated, Corning, NY, US). Unless otherwise stated, cells were transfected for 4 h in 37°C and 5% CO2 with 50 μL of polyplex/lipoplex solution containing 50 pmol siRNA within a total volume of 450 μL of serum containing cell culture media. Following transfection, cells were treated with a 0.4% trypan blue solution for 5 min. Cells were then washed with PBS, trypsinized and spun down at 350 g for 10 min. After centrifugation, the supernatant was decanted, and the cells were washed three times and resuspended in 400 µl PBS/2mM EDTA. Samples were analyzed via flow cytometry (Applied Biosystems Attune® Acoustic Focusing Cytometer, Life Technologies) and the median fluorescence intensity (MFI) was measured using 488 nm excitation and 530/30 nm bandpass emission filter set. Samples were run in triplicates, with each sample gated by morphology based on forward/sideward scattering for a minimum of 10,000 viable cells. Analysis and presentation of the data was performed by GraphPad Prism 5.0 software calculating mean values and standard deviation.
2.10. In vitro cellular uptake in suspension cells
To test the cellular uptake of polyplexes in hard-to-transfect suspension cells, the Jurkat human T lymphocyte cell line was used. One day prior to transfection, 400,000 Jurkat cells were seeded and incubated overnight at 37°C and 5% CO2 in 96-well plates (Corning Incorporated). Cells were transfected for 24 hr in 37°C and 5% CO2 with 50 μL of polyplex/lipoplex solution containing 50 pmol siRNA within a total volume of 200 μL of serum containing cell culture media. Following transfection, cells were treated with a 0.4% trypan blue solution. Cells were then washed with PBS, trypsinized and spun down at 350 g for 10 min. After centrifugation, the supernatant was decanted, and the cells were washed three times and resuspended in 400 µl PBS/2mM EDTA. Samples were analyzed via flow cytometry as described above.
2.11. In vitro GFP protein knockdown
To assess protein knockdown following delivery of siRNA via polyplexes, H1299/GFP cells were utilized. One day prior to transfection, 50,000 H1299/GFP cells were seeded and incubated overnight at 37°C and 5% CO2 in 24-well plates (Corning Incorporated). Cells were transfected for 4 h in 37°C and 5% CO2 with 50 μL of CP, VIPER, or PEI polyplex solution containing 50 pmol GFP or scrambled sequence siRNA within a total volume of 350 μL of serum containing cell culture medium with or without 100 µM chloroquine (MP Biomedicals). After 4 h, 600 µL of fresh serum containing culture medium was added and cells were incubated for an additional 20 h. Subsequently, the cells were washed, trypsinized and analyzed via flow cytometry for the median fluorescence intensity (MFI) of GFP protein expression using 488 nm excitation and 530/30 nm bandpass emission filter set. Samples were run in triplicates, with each sample gated by morphology based on forward/sideward scattering for a minimum of 10,000 viable cells. Results are presented as mean values and standard deviation.
2.12. Live cell spinning disk microscopy
The kinetics or polyplex uptake versus GFP protein knockdown were assessed by live cell imaging using spinning disk confocal microscopy. For live cell imaging experiments, 30,000 H1299/GFP cells were seeded in µ-Slide 8 well coverslips and incubated for 24 h at 5% CO2 and 37°C. Cells were transfected in 300 µl fresh RPMI medium for 24 h in 37°C and 5% CO2 with 25 µl of VIPER or LF suspension prepared at N/P ratio 8 containing 50 pmol GFP or scrambled sequence siRNA. Spinning disk confocal images were acquired at 488 nm and 561 nm excitation on a Nikon TiE microscope equipped with a Perfect Focus System (PFS), a Yokogawa CSU-W1 spinning disk unit (50 μm pinhole size, 405/488/561/640 LD Quad dichroic mirror) and laser illumination by an Andor ALC600 laser-beam combiner (405 nm/488 nm/561 nm/640 nm), an Andor Borealis illumination unit and environmental chamber (Okolab BIO 1, Bold Line). During microscopic measurements cells were grown in a humidified atmosphere Containing 5% CO2 and at a temperature of 37 ° C
Images were captured with an Andor IXON 888 Ultra EMCCD camera using a Nikon CFI Plan Apo Lambda 40X NA 0.95 air objective. The microscope was controlled via NIS-Elements (version 4.51.01). Over 12 h, 5x5 tiled single-plane images of each well were acquired every 15 min. NIS-Elements was used to stitch the tiles into single large images.
2.13. In vitro GAPDH gene knockdown
For gene silencing experiments, 1,000,000 Jurkat cells were seeded in a 96 well plate and triplicate samples were transfected for 24 h in 37°C and 5% CO2 with 50 μL of polyplex solution containing 100 pmol hGAPDH or scrambled sequence siRNA within a total volume of 200 μL of serum containing cell culture media. Negative controls consisted of blank/untreated cells while positive control cells were transfected with Lipofectamine 2000 lipoplexes, which were prepared as discussed above. Cells were harvested and processed to isolate total RNA using the PureLinkTM RNA mini kit (Life Technologies) according the manufacturer’s protocol with DNase I digestion (Thermo Fisher Scientific). Synthesis of cDNA from total RNA and PCR amplification were performed with Brilliant III ultra-fast SYBR® green QRT-PCR master mix kit (Agilent Technologies, Santa Clara, CA, US) using a Stratagene Mx 3005P (Agilent Technologies). Cycle threshold (Ct) values were determined by MxPro software (Agilent Technologies). A standard curve including 5 points was made from a 1:5 serial dilution of an untreated sample and assigned concentrations of each point (1, 0.2, 0.004, 0.0008, 0.00016) were plotted vs. their corresponding Ct values. The gene expression of GAPDH was normalized by the expression of β-actin. Hs_GAPDH_2_SG primers for GAPDH and Hs_ACTB_2_SG primers for β-actin (Qiagen, Valencia, CA, US) were used in all experiments.
2.14. In vivo polyplex accumulation following pulmonary delivery
All animal experiments were approved by a Wayne State University Institutional Animal Care and Use Committee (IACUC). In order to measure polyplex accumulation following pulmonary delivery, a previously described mouse model was used with slight modifications.[16] Female balb/c mice were purchased from Charles River Laboratories (Boston, MA, US) and used at 5 weeks’ age. Briefly, mice were intratracheally instilled (under ketamine/xylazine anesthesia) with 50 µl of CP, VIPER or PEI polyplexes, prepared with 2 nmol TYE™-563 (IDT) or pHrodo™ (Thermo Fisher Scientific) labeled siRNA at N/P 8. Control mice were treated with the same volume of 5% glucose. After 24 h, all animals were sacrificed under i.p. ketamine/xylazine anesthesia and lungs were inflated with 4% paraformaldehyde, dissected and stored in 4% paraformaldehyde in the dark at 4°C. After 48 h, fixed lungs were rinsed with PBS and cryoprotected in a series of glycerol solutions at 4°C (15% - 30%) over 8 days, exchanging the glycerol solution every 48 h before they were embedded in OCT gel. Lungs were cut on a cryotome, and slices (15 µm) were counterstained with 5 µM of DRAQ5 (Thermo Fisher Scientific) for 5 min and rinsed with PBS. Slides were mounted with FluorSave™ mounting medium (EMD Millipore, Billerica, MA, US) and were imaged with a Leica TCS SPE-II laser scanning confocal microscope (Leica, Wetzlar, DE). Autofluorescence from lung tissue was used to visualize lung cell morphology and laser excitation/emission settings were adjusted for each fluorophore accordingly. The images were exported from the Leica Image Analysis Suite and processed with the Fiji distribution of ImageJ.[17]
2.15. In vivo GAPDH gene knockdown
In order to measure in vivo transfection efficiency following pulmonary delivery, balb/c mice (9 per group) were intubated with CP, VIPER or PEI polyplexes prepared with msGAPDH or scrambled siRNA at N/P 8 as described above. After 24 h, the mice were sacrificed and lungs were flash frozen using liquid nitrogen and stored at -80°C. To isolate complete RNA, the frozen tissue was placed into a mortar with a small amount of liquid nitrogen and grinded into a fine powder using the pestle. Following homogenization, the frozen lung powder was processed with the TRIzol™ Plus RNA Purification Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol to yield purified RNA. Samples were stored at -80°C and isolated mRNA was further quantified by qRT-PCR as described above. Mm_GAPDH_3_SG primers for GAPDH and Mm_ACTB_2_SG primers for β-actin (Qiagen) were used in all experiments.
2.16. Statistics
All results are given as mean value ± standard deviation (SD). One-way ANOVA with Bonferroni posthoc test, two-way ANOVA and calculation of area under the curve (AUC) were performed in GraphPad Prism software (Graph Pad Software, La Jolla, CA, US).
3. Results and Discussion
3.1. siRNA condensation efficiency
In order to effectively deliver siRNA into the cytoplasm of the cell, it is crucial to protect the sensitive backbone of the molecule from numerous sources of degradation such as nucleases. [18] A promising method of protection from this degradation involves the use of charge complexation with positively charged molecules (such as polyethylenimine) interacting with the negative charges found on the phosphate rich siRNA molecule. [19] Similar to the widely used polycation polyethylenimine (PEI), the virus-inspired polymer for endosomal release (VIPER) contains a polycation block that is able to electrostatically condense the negatively charged siRNA. [9] While crucial for the protection of the nucleic acid, numerous studies have demonstrated dose-dependent toxicity attributed to large polycationic segments. [20] As such, it is vital to limit the concentration of polycationic polymer to a level that allows for the efficient condensation of siRNA while minimizing free polymer that could cause unwanted toxicity.
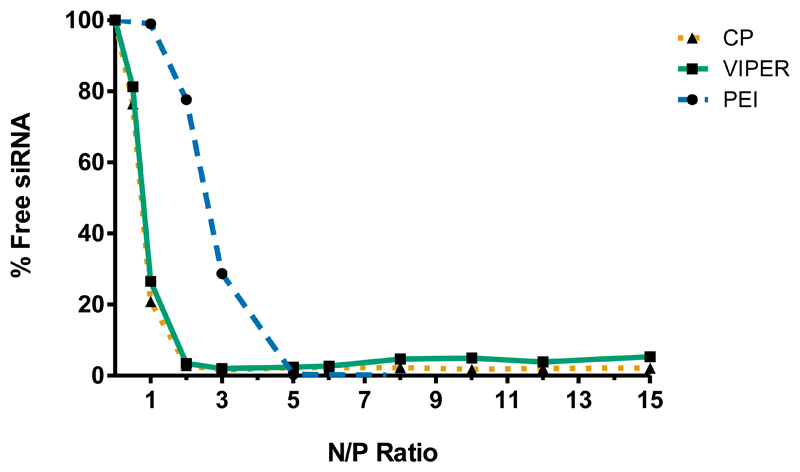
In order to determine the optimal amount of unmodified block copolymer (CP) or melittin-conjugated block copolymer (VIPER) needed to efficiently condense and protect the siRNA from nuclease degradation, SYBR® gold assays were performed at various N/P ratios (the molar ratio of nitrogen in amine to phosphate in siRNA) ranging from 0 to 15. In this assay, free or unbound siRNA is accessible to the intercalating nucleic acid dye SYBR® Gold and is subsequently quantified based on the fluorescence emitted. Results were compared to hyperbranched polyethylenimine (PEI 25 kDa) as a control. Figure 1 shows that both CP and VIPER polymers are able to completely condense siRNA at N/P 2 and higher. Conversely, PEI was able to achieve complete siRNA condensation at N/P 5 which is consistent with previous reports. [21] These data suggest that both CP and VIPER bind siRNA with higher affinity than PEI at low N/P ratios and more importantly, that the conjugation of melittin to the block copolymer does not interfere with its condensation efficiency. This observed difference between the block copolymers and PEI may be attributed to the polycation pDMAEMA present in CP and VIPER, which has been shown to have high affinity for nucleic acids while maintaining low toxicity profiles. [22]
Figure 1.
siRNA Condensation efficiency of CP and VIPER. SYBR® gold assays for both unmodified block copolymer (CP) and melittin conjugated block copolymer (VIPER) in comparison to the polyamine polyethylenimine (PEI). The polymers’ ability to electrostatically condense siRNA was analyzed from N/P ratios 1-15. Free siRNA represents 100% siRNA release.
3.2. Characterization of polyplex size, polydispersity and surface charge
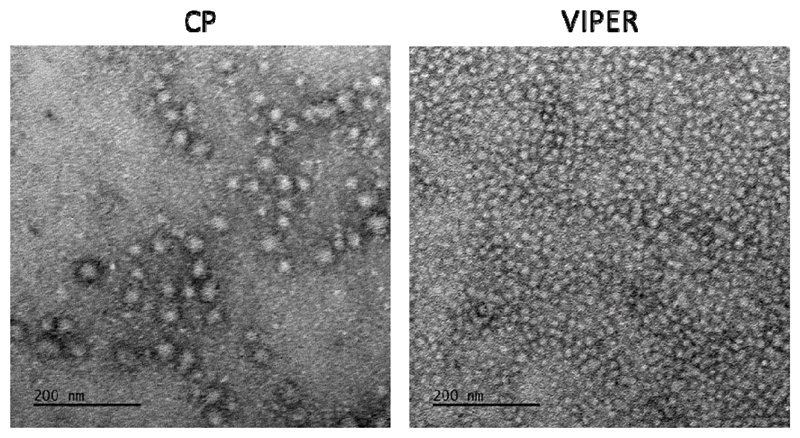
Polyplex surface charge and overall size are two of the major determinates for efficient nanoparticle drug delivery. [23] However, the optimal parameters for these characteristics vary greatly depending on the intended route of the particle and should be tailored to drive particle accumulation in vivo. [24] Frangioni et al. reported that positively charged particles larger than 34 nm readily accumulate in the lungs while zwitterionic particles smaller than 34 nm will reach lymph nodes quickly and are cleared. [25] In this study, CP and VIPER polyplexes were formed with 50 pmol of siRNA at various N/P ratios and were analyzed via dynamic light scattering and laser Doppler anemometry. As shown in Table 1, both CP and VIPER polyplexes decreased in size as the N/P ratio increased, resulting in diameters <100 nm at N/P 8. Interestingly, the CP polyplexes showed a substantial increase in dispersity (PDI) as the N/P ratio was increased, whereas the VIPER polyplexes had a more consistent PDI in all the formulations analyzed. TEM imaging of the polyplexes (N/P=8) revealed structures with spherical shape and confirmed the increased polydispersity observed by DLS (Figure 2). The difference in particle dispersity seen here may be an unintentional design feature caused by the presence of the melittin peptide in the VIPER block copolymer, which may provide additional polymer-polymer interactions that improve polyplex packaging or stability.
Table 1. CP and VIPER polyplex hydrodynamic diameter and zeta potential.
| Polymer | N/P Ratio | Particle size (nm) | Polydispersity index | Zeta potential (mV) |
|---|---|---|---|---|
| Unmodified block copolymer (CP) | N/P 1 | 236 ± 14.3 | 0.208 | 24.3 ± 4.9 |
| N/P 2 | 132 ± 16.1 | 0.608 | 36.2 ± 0.6 | |
| N/P 3 | 51 ± 2.8 | 0.713 | 42.6 ± 11.3 | |
| N/P 8 | 49 ± 4.4 | 0.666 | 52.0 ± 4.7 | |
| Melittin block copolymer (VIPER) | N/P 1 | 151 ± 0.4 | 0.221 | -5.3 ± 1.3 |
| N/P 2 | 115 ± 1.6 | 0.271 | 23.1 ± 0.6 | |
| N/P 3 | 142 ± 4.2 | 0.330 | 30.4 ± 2.5 | |
| N/P 8 | 62 ± 6.3 | 0.357 | 40.5 ± 9.6 |
Figure 2.
TEM images of CP and VIPER polyplexes. Transmission electron microscopy images of both unmodified block copolymer (CP) and melittin conjugated block copolymer (VIPER) polyplexes at N/P 8 were taken at 30000x magnification.
In addition to polyplex size and dispersity within suspension, another important characteristic to be evaluated is the surface charge of the particle. While neutral or negatively charged nanoparticles have been shown to increase circulation times, positively charged nanoparticles are well known to have a higher rate of nonspecific uptake by cells, leading to an overall higher accumulation.[26] Here, all measured polyplexes were positively charged except the VIPER polyplexes at N/P 1, which showed a slightly negative zeta potential of -5.3 mV, likely due to differences in packaging efficacy at low charge ratios. The positive surface charge of the CP and VIPER polyplexes at N/P ≥ 2 corroborates previous zeta potential measurements of these polymers complexed with DNA. [9] Due to their similar surface charge and size differences, further analysis of CP and VIPER polyplexes on a cell interaction level was carried out at N/P ratios ranging from 3 to 10.
3.3. In vitro cellular uptake of polyplexes
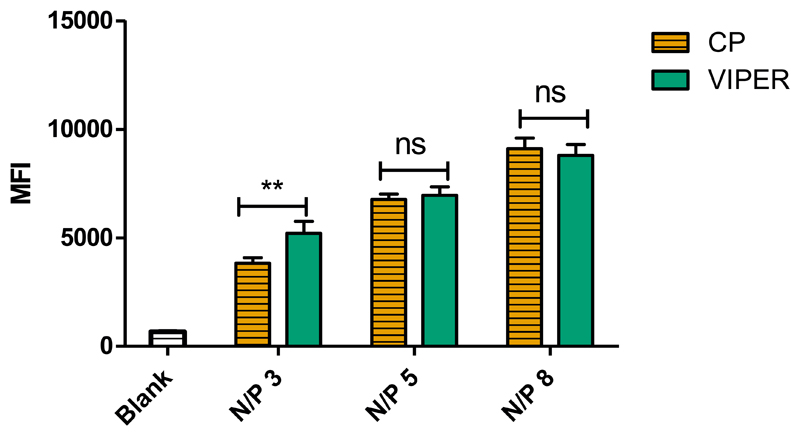
To determine the ability of VIPER polyplexes to deliver siRNA into cells, human non-small cell lung carcinoma cells (H1299) were transfected and cellular uptake was quantified by flow cytometry. VIPER polyplexes were formulated with Alexa Fluor 488 labeled siRNA at different N/P ratios (3,5,8) and H1299 cells were transfected for 24 h. The unmodified block copolymer (CP) was also tested at the same N/P ratios and served as a transfection control while cells treated with 5% glucose were included as a negative control. At N/P 3, significantly higher cellular uptake was observed for those samples transfected with VIPER polyplexes when compared to samples transfected with CP (Figure 3). In addition, the increase of N/P ratio corresponds with an increase in cellular uptake between both CP and VIPER polyplexes with no significant difference between the polymers used.
Figure 3.
Cellular uptake of CP and VIPER in H1299 cells. Median fluorescence intensity (MFI) was determined by flow cytometry to evaluate the cellular uptake in a human non-small cell lung carcinoma cell line (H1299) of unmodified block copolymer (CP) or melittin conjugated block copolymer (VIPER) polyplexes at N/P ratios of 3, 5 and 8 with 50 pmol Alexa Fluor-488 labelled siRNA. Blank samples consisted of H1299 cells treated with 5% glucose only. Data points indicate mean ± SD (n=3). Two-way ANOVA ** P < 0.01
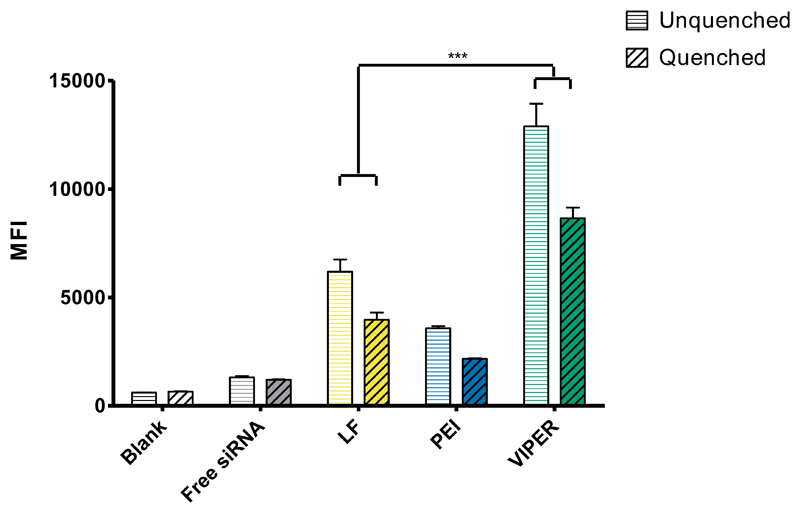
As mentioned above, cationic polyplexes are known to non-specifically interact with the cell surface due to their positive charge and induce adsorptive endocytosis. Therefore, differences between CP and VIPER were not expected, considering that their structural differences act on the endosomal level and not on the interaction with the cell membrane. In order to differentiate extracellular association from actual internalization of the VIPER polyplexes, however, we repeated the in vitro uptake experiment in human T lymphocyte cells with an additional 0.4% trypan blue quenching treatment. By treating the transfected cells with trypan blue before flow cytometry analysis, the fluorescence from membrane bound polyplexes is quenched. As shown in Figure 4, the MFI slightly decreased in all trypan blue treated groups; indicating that there are limited membrane bound VIPER polyplexes with a major portion of the polyplexes taken up intracellularly. In addition, there was significantly higher amount of cellular uptake of VIPER polyplexes compared to the commercial transfection reagent Lipofectamine 2000 (LF) and the polycation standard PEI which is an indication that VIPER polyplexes are promising candidates for transfecting lymphocytes.
Figure 4.
Cellular uptake of VIPER in Jurkat cells. Median fluorescence intensity (MFI) was determined by flow cytometry to evaluate the cellular uptake in human T lymphocytes of melittin conjugated block copolymer (VIPER) polyplexes and 25k polyethylenimine (PEI) polyplexes at N/P 10 with 50 pmol Alexa Fluor-488 labelled siRNA. Lipofectamine 2000 (LF) polyplexes/lipoplexes were prepared according to manufacturer’s protocol with 50 pmol Alexa Fluor-488 labelled siRNA. Data points indicate mean ± SD (n=3). One-way ANOVA ***P < 0.001
3.4. In vitro GFP knockdown
To evaluate the gene silencing efficiency of VIPER at the protein level, we utilized human non-small cell lung carcinoma cells (H1299) that stably express the reporter gene enhanced green fluorescent protein (GFP). Based on observations from our in vitro cellular uptake, H1299/GFP cells were transfected with CP and VIPER polyplexes formulated with siRNA against GFP or scrambled sequence at various N/P ratios (3, 5, 8 and 10) to determine the optimal parameters for the most efficient protein knockdown. In addition, the cells were treated with the drug chloroquine – a drug reported to aid in the endosomal release of siRNA.[27] After treatment, the median fluorescence intensity (MFI) of GFP in each cell sample was quantified via flow cytometry. In the initial experiment, CP and VIPER polyplexes were formulated with 100 pmol GFP siRNA at N/P 5 and 3, respectively, based on conditions optimized for plasmid delivery.[9] These low N/P ratio polyplexes resulted in no measurable knockdown of GFP in cells that were or were not treated with chloroquine, suggesting that endosomal entrapment of the polyplexes was not the cause of poor knockdown efficiency (Supplement 1). Conversely, when CP polyplexes were formulated at higher N/P ratios of 8 or 10, the samples that were treated with chloroquine show a significantly higher amount of GFP knockdown compared to those not treated, suggesting that CP polyplexes at N/P 8 or 10 were unable to be released from the endosome to elicit knockdown of GFP (Figure 5). However, low GFP expression was observed in both chloroquine treated and untreated cells that were transfected with VIPER polyplexes. Additionally, there was a significantly higher amount of GFP knockdown in the VIPER samples without chloroquine compared to the parallel samples transfected with LF, PEI and CP. This finding emphasizes that VIPER polyplexes efficiently escape the endosome. Finally, both CP and VIPER polyplexes containing scrambled siRNA (negative control, NC) did not reduce the MFI of GFP in their samples, indicating that the observed protein knockdown was not mediated by the polymer system or any non-specific effects, but was RNAi-mediated by the GFP siRNA delivered into the cytoplasm.
Figure 5.
GFP knockdown of CP and VIPER in H1299/GFP cells. Median fluorescence intensity (MFI) was determined by flow cytometry to evaluate the protein knockdown in human non-small cell lung carcinoma cells expressing GFP (H1299/GFP) after transfection with unmodified block copolymer (CP) or melittin conjugated block copolymer (VIPER) polyplexes at N/P ratios of 8 and 10 with 50 pmol GFP siRNA for 24h. Blank samples consisted of H1299/GFP cells treated with 5% glucose only. The positive control consisted of Lipofectamine 2000 (LF) lipoplexes with 50 pmol GFP siRNA, and 25k polyethylenimine (PEI) polyplexes at N/P 10 were used as an additional transfection standard. Data points indicate mean ± SD (n=3). Two-way ANOVA ***P < 0.001
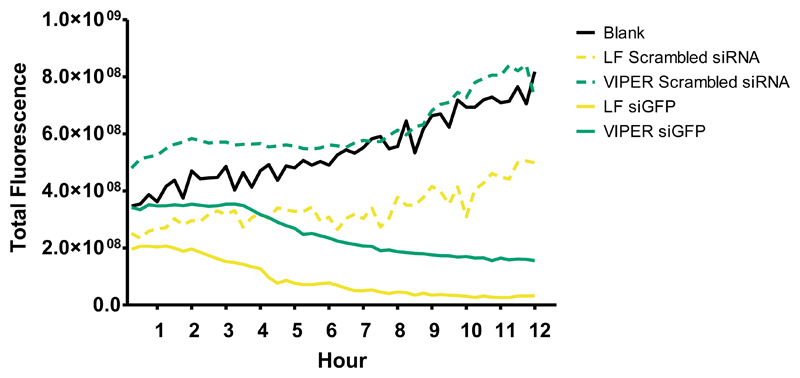
After optimization of the N/P ratio to achieve the most efficient total protein knockdown at a static time point, live cell microscopy was employed to investigate the kinetics of protein knockdown throughout the transfection process. Similar to the flow cytometry studies, H1299/GFP cells were transfected with VIPER polyplexes or Lipofectamine formulated with siRNA against GFP or scrambled sequence at a N/P ratio of 8. Immediately following the initial transfection, the treated cells were then visualized via spinning disk confocal microscopy and images were acquired every 15 minutes over a 12-hour period. The H1299/GFP cells that were untreated established a baseline level of total GFP fluorescence visualized (Figure 6). Importantly, those cells that were treated with VIPER polyplexes containing scrambled siRNA show analogous levels of total GFP fluorescence when compared to the untreated cells, which indicates that the VIPER transfection was well tolerated by the cells and does not cause any reduction of fluorescence due to toxicity. While Lipofectamine treated cells exhibit a strong reduction in total GFP fluorescence throughout the 12-hour transfection period, it is important to note that those cells treated with scrambled sequence siRNA loaded into Lipofectamine show a strikingly lower level of GFP expression compared the baseline established by the untreated cells suggesting particle toxicity as the cause of GFP protein downregulation. This observation supports the well-documented toxicity profiles associated with lipid based nanoparticle systems.[28] A striking decrease in GFP expression following VIPER/siGFP transfection is recorded beginning at the 4-hour time point with the reduction of total GFP fluorescence remaining robust throughout the next 8 hours. The majority of protein knockdown occurring after 4 hours is well supported in the literature, with many studies reporting the saturation of particle uptake taking place 4 – 6 hours after initial transfection.[29]
Figure 6.
Live cell microscopy of CP and VIPER GFP knockdown in H1299/GFP cells. Total fluorescence was visualized via live cell microscopy to evaluate the protein knockdown in human non-small cell lung carcinoma cells expressing GFP (H1299/GFP) after transfection with or melittin conjugated block copolymer (VIPER) polyplexes at N/P ratios of 8 with 50 pmol GFP siRNA for 12hr. Blank samples consisted of H1299/GFP cells treated with 5% glucose only. The positive control consisted of Lipofectamine 2000 (LF) lipoplexes with 50 pmol GFP siRNA.
3.5. In vitro GAPDH gene knockdown
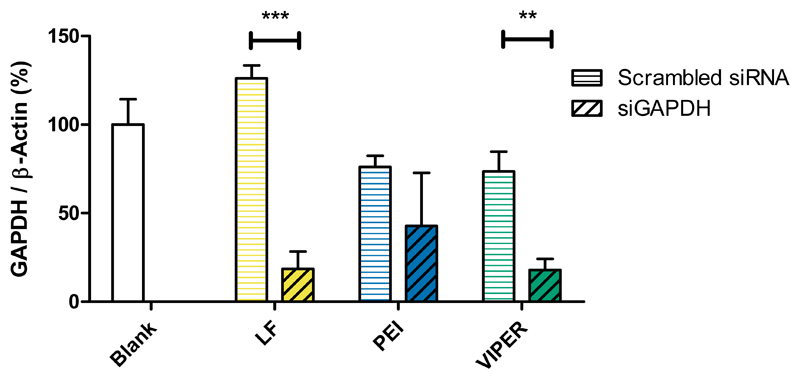
Following validation of reporter protein silencing mediated by VIPER polyplexes in vitro, we next evaluated their ability to silence the endogenously expressed housekeeping gene GAPDH in Jurkat cells. Per our GFP knockdown optimization studies, the cells were transfected with VIPER polyplexes containing hGAPDH siRNA (siGAPDH) or scrambled sequence siRNA (siNC) at N/P 10 for 24 h. Negative controls consisted of blank cells that were treated with 5% glucose only while positive control cells were transfected with Lipofectamine 2000. PEI (N/P 15) polyplexes were used as transfection standard. GAPDH gene expression was normalized to β-actin gene expression and quantified by real time PCR. Jurkat cells that were transfected with VIPER/siGAPDH polyplexes showed significantly higher levels of gene knockdown when compared to the blank cells or those that were treated with VIPER/siNC polyplexes (Figure 7). Similar levels of reduced GAPDH gene expression were observed in cells that were treated with either LF/siGAPDH or VIPER/siGAPDH polyplexes, whereas PEI polyplexes did not mediate sequence-specific GAPDH reduction. The comparable level of gene knockdown achieved by the VIPER polyplexes when compared to the commercial transfection reagent lipofectamine indicates that VIPER polyplexes at N/P 10 are able to efficiently deliver siRNA resulting in robust gene knockdown in vitro. These results were the basis for all in vivo experiments described below where Lipofectamine cannot be employed due to its toxicity.
Figure 7.
GAPDH gene knockdown of VIPER in Jurkat cells. GAPDH gene knockdown efficiency was evaluated in human T lymphocytes after 24 hr transfection with melittin conjugated block copolymer (VIPER) at N/P 10 with 100 pmol hGAPDH siRNA. Blank samples consisted of Jurkat cells treated with 5% glucose only. The positive control consisted of Lipofectamine 2000 (LF) lipoplexes with 100 pmol hGAPDH siRNA while 25k polyethylenimine (PEI) polyplexes at N/P 10 were used as an additional transfection standard. GAPDH expression was normalized with β-actin expression and quantified by real time PCR. Data points indicate mean ± SD (n=3). One-way ANOVA, **P < 0.01, ***P < 0.001.
3.6. In vivo polyplex accumulation following pulmonary delivery
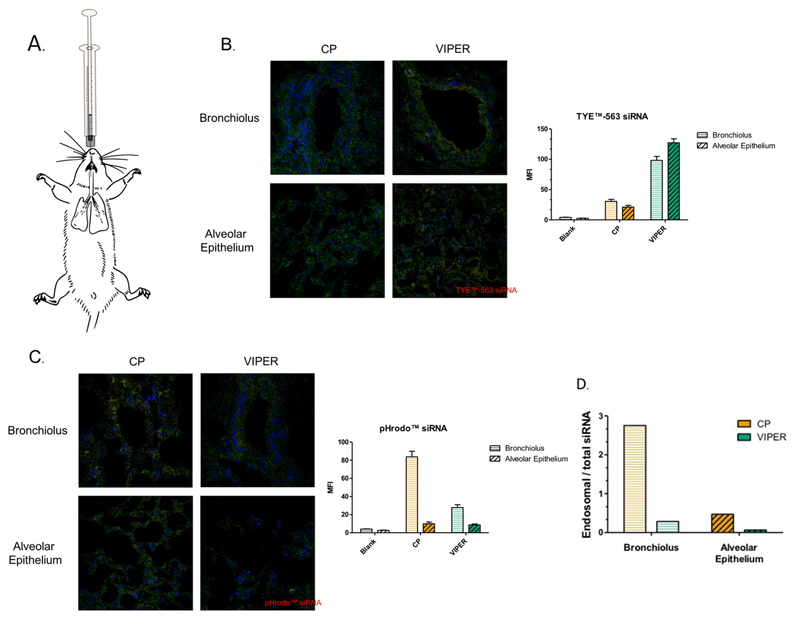
The delivery of therapeutic siRNA has long been proposed as an attractive treatment option for a variety of different pulmonary diseases with several nanoparticle-based siRNA treatments having entered different clinical phases.[30] To investigate the ability of VIPER to deliver siRNA via pulmonary delivery, a murine model for pulmonary delivery was used. [7] Mice were intratracheally administered CP or VIPER polyplexes that were made with either TYE™-563 or pHrodo™ labeled siRNA at N/P 8 (Figure 8A). The pHrodo™ dye is a commercially available fluorescent pH indicator that has been reported to dramatically increase in fluorescence as the acidity of the environment changes. [31] The following day, the animals were sacrificed and lungs were embedded in OCT gel following fixation with paraformaldehyde. Confocal scanning light microscopy of cryosections was used to qualitatively investigate the accumulation of siRNA within cells of the lung tissue of treated mice. Both the bronchioli and alveolar epithelia of mice treated with VIPER polyplexes show an increased amount of TYE™-563 siRNA in the bronchi and alveolar epithelium when compared to the lung tissue of mice treated with CP polyplexes (Figure 8B). It was therefore hypothesized that VIPER polyplexes efficiently reach the cytoplasm of bronchial and alveolar epithelial cells after endocytosis whereas CP polyplexes may be entrapped in endosomes, leading to self-quenching of the fluorescently labeled siRNA. Conversely, lung tissue of mice treated with VIPER polyplexes show a reduced amount of fluorescence from pHrodo™ labeled siRNA compared to those treated with CP polyplexes (Figure 8C). Therefore, in addition to the VIPER polyplexes mediating higher accumulation of TYE™-563 labeled siRNA in lung tissue, the reduced amount of pHrodo™ fluorescence in lungs treated with VIPER polyplexes indicates that these polyplexes are indeed able to escape the highly acidic environment of the endo/lysosomal compartment and deliver their payload into the pH neutral cytoplasm. As shown in Figure 8D, CP polyplexes accumulate to an almost three-fold higher extent in acidic environments of the bronchial tissue, such as the endo-lysosome, whereas VIPER polyplexes are found with an about three-fold higher abundance in the cytoplasm of the bronchioles than in acidic vesicles. The observation that entrapped CP polyplexes containing pHrodo™-labeled siRNA are detected especially in bronchial epithelium could be a result of a larger abundance of lysosomes in bronchial epithelial cells rather than in alveolar cells.
Figure 8.
In vivo accumulation of CP and VIPER in Balb/c mice lungs. A) Illustration of intratracheal administration of siRNA polyplexes into normal Balb/c mice. B & C) Confocal microscopy images of lung tissue treated with unmodified block copolymer (CP) or melittin conjugated block copolymer (VIPER) polyplexes at N/P 8 with 2 nmol TYE™-563 or pHrodo™ labeled siRNA for 24h. D) Ratio of endosomal siRNA to total accumulated siRNA in lung tissue treated with CP or VIPER polyplexes.
3.7. In vivo GAPDH knockdown
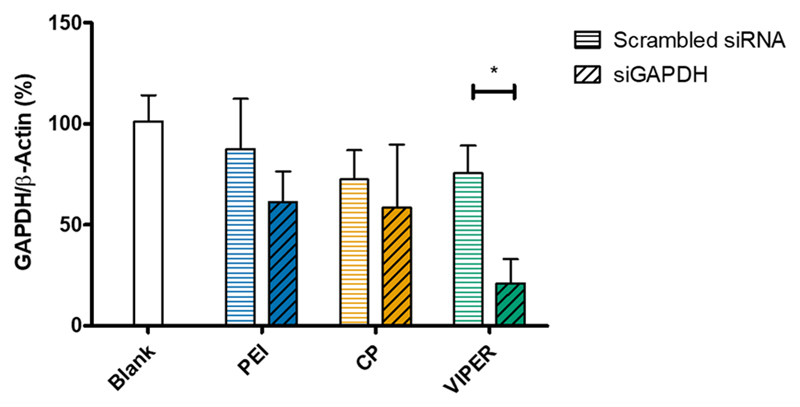
Following the investigation of polyplex accumulation and endosomal escape, mice were intratracheally administered CP, VIPER or PEI polyplexes that were made with either msGAPDH or scrambled sequence siRNA at N/P 8 to analyze gene knockdown efficiency. Untreated (blank) mice were instilled with the same volume of 5% glucose solution. Lipofectamine was not used in the in vivo experiment due to its known toxicity. The following day, the animals were sacrificed and the lungs were collected for total RNA isolation. Following qRT-PCR analysis, GAPDH mRNA was normalized by the expression of β-actin (Figure 9). The mice that were treated with VIPER/siGAPDH polyplexes showed a 76% reduction of GAPDH expression compared to blank mice which was significantly reduced compared to the GAPDH expression of mice treated with VIPER/siNC. In contrast, neither PEI nor CP polyplexes mediated sequence-specific gene knockdown but rather reduced GAPDH nonspecifically, which could be a result of polymer-related off target effects.[32] Most importantly, however, did the VIPER polyplexes mediate a level of in vivo gene knockdown that we have not observed before with any other siRNA delivery system. We lately reported 50% gene knockdown with micelleplexes prepared by microfluidics and had previously reported to observe 30% gene knockdown in vivo with similar and optimized micelleplexes.[33] However, to our knowledge, such a great in vivo gene knockdown after pulmonary administration of polyplexes to the lung is unprecedented.
Figure 9.
GAPDH gene knockdown of CP and VIPER in vivo. GAPDH gene knockdown efficiency was validated in whole lung RNA isolated from Balb/c mice that were instilled with melittin conjugated block copolymer (VIPER) or unmodified block copolymer (CP) at N/P 8 with 2 nmol msGAPDH siRNA. Blank samples consisted of Balb/c mice instilled with 5% glucose only. 25k polyethylenimine (PEI) polyplexes at N/P 8 with 2 nmol siRNA were instilled into Balb/c mice as a non-toxic transfection standard. GAPDH expression was normalized with β-actin expression and quantified by real time PCR. Data points indicate mean ± SD (n=6). Two-way ANOVA, *P < 0.05.
4. Conclusion
Herein we present the use of the VIPER polymer system for the encapsulation and delivery of siRNA in vitro and in vivo. Our present work demonstrates the impressive ability of the VIPER system to condense and protect siRNA at low N/P ratios while physical characterization revealed uniform particles with >200 nm hydrodynamic diameters. Utilizing flow cytometry, real time PCR and live cell microscopy, transfection conditions were optimized and the VIPER polyplexes were demonstrated to mediate efficient transcript and protein knockdown. Upon the successful optimization of transfection parameters in vitro, we investigated pulmonary delivery of the VIPER polyplexes in a murine model and demonstrate high accumulation of the particles within both the bronchial and alveolar epithelium along with the ability for the particles to escape endosomal entrapment. Lastly, we show that the VIPER polyplexes mediated a significant decrease of GAPDH expression in vivo after pulmonary delivery when compared to its unconjugated form. Collectively, these findings based on cell culture and animal models demonstrate the ability of the VIPER system to mediate significant gene and protein knockdown in vitro and in vivo.
Supplementary Material
Acknowledgements
This work was supported by the Wayne State University Start-Up Grant and European Research Council [grant number ERC-2014-StG – 63783]; the National Institute of Health [grant numbers 2R01NS064404, P30CA22453]; the National Institute on Drug Abuse [grant number DA034783]; the Nanosystems Initiative Munich (NIM); and Wayne State School of Medicine for GRA support of Daniel Feldmann.
References
- [1].Kwok WW, Schuening F, Stead RB, Miller AD. Retroviral transfer of genes into canine hemopoietic progenitor cells in culture: a model for human gene therapy. Proc Natl Acad Sci U S A. 1986;83:4552–4555. doi: 10.1073/pnas.83.12.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kumar SR, Markusic DM, Biswas M, High KA, Herzog RW. Clinical development of gene therapy: results and lessons from recent successes. Mol Ther Methods Clin Dev. 2016;3 doi: 10.1038/mtm.2016.34. 16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Narayanan G, Cossu G, Galli MC, Flory E, Ovelgonne H, Salmikangas P, Schneider CK, Trouvin JH. Clinical development of gene therapy needs a tailored approach: a regulatory perspective from the European Union. Hum Gene Ther Clin Dev. 2014;25:1–6. doi: 10.1089/humc.2013.230. [DOI] [PubMed] [Google Scholar]
- [4].Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- [5].Scollay R. Gene therapy: a brief overview of the past, present, and future. Ann N Y Acad Sci. 2001;953:26–30. doi: 10.1111/j.1749-6632.2001.tb11357.x. [DOI] [PubMed] [Google Scholar]
- [6].Lachelt U, Wagner E. Nucleic Acid Therapeutics Using Polyplexes: A Journey of 50 Years (and Beyond) Chem Rev. 2015;115:11043–11078. doi: 10.1021/cr5006793. [DOI] [PubMed] [Google Scholar]
- [7].Xie Y, Kim NH, Nadithe V, Schalk D, Thakur A, Kilic A, Lum LG, Bassett DJ, Merkel OM. Targeted delivery of siRNA to activated T cells via transferrin-polyethylenimine (Tf-PEI) as a potential therapy of asthma. J Control Release. 2016;229:120–129. doi: 10.1016/j.jconrel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- [9].Cheng Y, Yumul RC, Pun SH. Virus-Inspired Polymer for Efficient In Vitro and In Vivo Gene Delivery. Angew Chem Int Ed Engl. 2016;55:12013–12017. doi: 10.1002/anie.201605958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang Y, Zhou K, Huang G, Hensley C, Huang X, Ma X, Zhao T, Sumer BD, DeBerardinis RJ, Gao J. A nanoparticle-based strategy for the imaging of a broad range of tumours by nonlinear amplification of microenvironment signals. Nat Mater. 2014;13:204–212. doi: 10.1038/nmat3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu L, Zheng M, Librizzi D, Renette T, Merkel OM, Kissel T. Efficient and Tumor Targeted siRNA Delivery by Polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol)-folate (PEI-PCL-PEG-Fol) Mol Pharm. 2016;13:134–143. doi: 10.1021/acs.molpharmaceut.5b00575. [DOI] [PubMed] [Google Scholar]
- [12].Devi GR. siRNA-based approaches in cancer therapy. Cancer Gene Ther. 2006;13:819–829. doi: 10.1038/sj.cgt.7700931. [DOI] [PubMed] [Google Scholar]
- [13].Scholz C, Wagner E. Therapeutic plasmid DNA versus siRNA delivery: common and different tasks for synthetic carriers. J Control Release. 2012;161:554–565. doi: 10.1016/j.jconrel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- [14].Schellinger JG, Pahang JA, Johnson RN, Chu DS, Sellers DL, Maris DO, Convertine AJ, Stayton PS, Horner PJ, Pun SH. Melittin-grafted HPMA-oligolysine based copolymers for gene delivery. Biomaterials. 2013;34:2318–2326. doi: 10.1016/j.biomaterials.2012.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Behe M, Kissel T. In vivo SPECT and real-time gamma camera imaging of biodistribution and pharmacokinetics of siRNA delivery using an optimized radiolabeling and purification procedure. Bioconjug Chem. 2009;20:174–182. doi: 10.1021/bc800408g. [DOI] [PubMed] [Google Scholar]
- [16].Jones SK, Sarkar A, Feldmann DP, Hoffmann P, Merkel OM. Revisiting the value of competition assays in folate receptor-mediated drug delivery. Biomaterials. 2017;138:35–45. doi: 10.1016/j.biomaterials.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Scherman D, Rousseau A, Bigey P, Escriou V. Genetic pharmacology: progresses in siRNA delivery and therapeutic applications. Gene Ther. 2017;24:151–156. doi: 10.1038/gt.2017.6. [DOI] [PubMed] [Google Scholar]
- [19].de Martimprey H, Vauthier C, Malvy C, Couvreur P. Polymer nanocarriers for the delivery of small fragments of nucleic acids: oligonucleotides and siRNA. Eur J Pharm Biopharm. 2009;71:490–504. doi: 10.1016/j.ejpb.2008.09.024. [DOI] [PubMed] [Google Scholar]
- [20].Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- [21].Elsayed M, Corrand V, Kolhatkar V, Xie Y, Kim NH, Kolhatkar R, Merkel OM. Influence of oligospermines architecture on their suitability for siRNA delivery. Biomacromolecules. 2014;15:1299–1310. doi: 10.1021/bm401849d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Neu M, Fischer D, Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J Gene Med. 2005;7:992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- [23].He C, Hu Y, Yin L, Tang C, Yin C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials. 2010;31:3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- [24].Jiang W, Kim BY, Rutka JT, Chan WC. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- [25].Choi HS, Ashitate Y, Lee JH, Kim SH, Matsui A, Insin N, Bawendi MG, Semmler-Behnke M, Frangioni JV, Tsuda A. Rapid translocation of nanoparticles from the lung airspaces to the body. Nat Biotechnol. 2010;28:1300–1303. doi: 10.1038/nbt.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Thurston G, McLean JW, Rizen M, Baluk P, Haskell A, Murphy TJ, Hanahan D, McDonald DM. Cationic liposomes target angiogenic endothelial cells in tumors and chronic inflammation in mice. J Clin Invest. 1998;101:1401–1413. doi: 10.1172/JCI965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Erbacher P, Roche AC, Monsigny M, Midoux P. Putative role of chloroquine in gene transfer into a human hepatoma cell line by DNA/lactosylated polylysine complexes. Exp Cell Res. 1996;225:186–194. doi: 10.1006/excr.1996.0169. [DOI] [PubMed] [Google Scholar]
- [28].Kedmi R, Ben-Arie N, Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials. 2010;31:6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- [29].Cartiera MS, Johnson KM, Rajendran V, Caplan MJ, Saltzman WM. The uptake and intracellular fate of PLGA nanoparticles in epithelial cells. Biomaterials. 2009;30:2790–2798. doi: 10.1016/j.biomaterials.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Merkel OM, Rubinstein I, Kissel T. siRNA delivery to the lung: what's new? Adv Drug Deliv Rev. 2014;75:112–128. doi: 10.1016/j.addr.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Han J, Burgess K. Fluorescent indicators for intracellular pH. Chem Rev. 2010;110:2709–2728. doi: 10.1021/cr900249z. [DOI] [PubMed] [Google Scholar]
- [32].Merkel OM, Urbanics R, Bedocs P, Rozsnyay Z, Rosivall L, Toth M, Kissel T, Szebeni J. In vitro and in vivo complement activation and related anaphylactic effects associated with polyethylenimine and polyethylenimine-graft-poly(ethylene glycol) block copolymers. Biomaterials. 2011;32:4936–4942. doi: 10.1016/j.biomaterials.2011.03.035. [DOI] [PubMed] [Google Scholar]
- [33].Endres T, Zheng M, Kilic A, Turowska A, Beck-Broichsitter M, Renz H, Merkel OM, Kissel T. Amphiphilic biodegradable PEG-PCL-PEI triblock copolymers for FRET-capable in vitro and in vivo delivery of siRNA and quantum dots. Mol Pharm. 2014;11:1273–1281. doi: 10.1021/mp400744a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.