Abstract
Metastases, the spread of cancer cells to distant organs, cause the majority of cancer-related deaths. Few metastasis-specific driver mutations have been identified, suggesting aberrant gene regulation as a source of metastatic traits. However, how metastatic gene expression programs arise is poorly understood. Here, using human-derived metastasis models of renal cancer, we identify transcriptional enhancers that promote metastatic carcinoma progression. Specific enhancers and enhancer clusters are activated in metastatic cancer cell populations and the associated gene expression patterns are predictive of poor patient outcome in clinical samples. We find that the renal cancer metastasis-associated enhancer complement consists of multiple co-activated tissue-specific enhancer modules. Specifically, we identify and functionally characterize a co-regulatory enhancer cluster, activated by the renal cancer driver HIF2A and an NF-kappaB-driven lymphoid element, as a mediator of metastasis in vivo. We conclude that oncogenic pathways can acquire metastatic phenotypes through cross-lineage co-option of physiological epigenetic enhancer states.
Keywords: Renal cancer, metastasis, enhancer, functional genomics
Introduction
Cancers develop the ability to spread, or metastasize, from their site of formation to distant organs through a multi-step selection process (1). A variety of mechanisms are known to increase the probability of successful completion of one or more metastasis steps (2,3). However, the mechanisms by which cancer clones acquire metastatic traits have remained poorly understood. For example, genetic analyses of both model systems and patient samples have thus far failed to identify mutational signatures of metastasis (4–7). On the other hand, metastatic progression in various experimental and clinical contexts is associated with specific gene expression programs, indicating the existence of underlying gene regulatory mechanisms that provide cancer cells with metastatic capabilities (5). How these aberrant gene expression programs arise is unknown.
Worldwide renal cell carcinomas (RCC) resulted in an estimated 338,000 new cancer diagnoses and 144,000 cancer deaths in 2012 (8). The most common form of RCC, accounting for 70-75% of the cases, is clear cell renal cell carcinoma (ccRCC) (9). Genetically, ccRCCs are characterized by inactivation of the von Hippel-Lindau tumor suppressor (VHL), which is lost in up to 90% of ccRCCs (10,11). VHL loss leads to normoxic stabilization of the hypoxia-inducible factors HIF1A and HIF2A (also known as EPAS1), which in turn drive the expression of multiple target genes involved in ccRCC development (12). Of the two HIF isoforms, HIF2A is more important for ccRCC progression (12), whereas HIF1A may have tumor suppressive functions (13). In addition to supporting tumor initiation and early progression, HIF2A is also critical for ccRCC metastasis. For example, HIF2A drives the expression of the chemokine receptor CXCR4 which promotes metastatic progression in ccRCC and other tumor types (14–19). Furthermore, VHL loss can also lead to the activation of other canonical oncogenic pathways such as NF-kappaB (20,21), which is associated with ccRCC progression and resistance to drug-induced apoptosis (22–25). The VHL-HIF2A pathway has thus been unequivocally linked to ccRCC initiation and progression at multiple levels. Yet, the molecular mechanisms that shape HIF2A transcriptomic output during ccRCC evolution remain incompletely understood.
Transcriptional enhancers are distal DNA elements that play an important role in gene regulation (26). Enhancer activation landscapes are highly tissue specific, implying that enhancers and enhancer clusters are critical determinants of cell fate and identity (27,28). Abnormal enhancer activation has also been linked to various disease states, including cancer (29). Specifically, altered enhancer activity can modulate cancer risk and activate oncogenes (30–38), suggesting that aberrant enhancer usage may also be critical for cancer progression and metastasis. Recently, genome-wide chromatin profiling has also linked VHL loss and the consequent HIF2A stabilization to the maintenance of enhancer activation landscapes in ccRCC (39). However, whether and how specific enhancers contribute to metastatic cancer progression remains largely unknown.
To further understand the mechanisms of metastasis gene activation, we set out to test whether perturbed enhancer function underlies metastatic carcinoma progression. Combining high-throughput enhancer profiling by chromatin immunoprecipitation and sequencing (ChIP-seq) and chromatin conformation capture by Hi-C in metastasis models of ccRCC, a malignancy with significant metastatic potential (40), we identified enhancers associated with metastatic progression. Using CRISPR interference (CRISPRi) and CRISPR/Cas9 genome editing we demonstrate the functional importance of distal enhancer elements for metastatic colonization in vivo. We further show how a specific metastasis-promoting enhancer cluster is activated by the co-regulatory action of two signaling pathways, the ccRCC core driver HIF2A and the NF-kappaB pathway that acts via a tissue restricted lymphoid enhancer element. Finally, cross-species analysis revealed that the metastatic ccRCC enhancer complement consists of multiple developmental enhancer programs co-opted from different normal cell lineages. We propose a model whereby metastatic transcriptional programs in carcinoma cells arise through cross-lineage co-option of developmentally distinct physiological enhancer activation states.
Results
Enhancer activation patterns in metastatic ccRCC
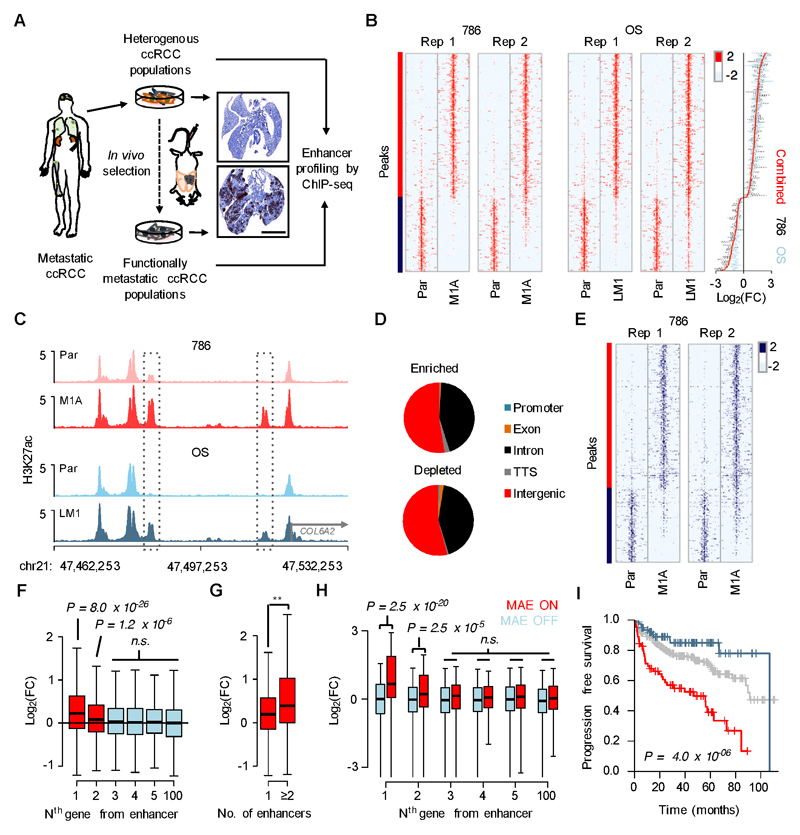
To identify metastasis-associated enhancers (MAEs), we performed ChIP-seq using an antibody for acetylated histone H3 lysine 27 (H3K27ac), a chromatin modification associated with active gene regulatory elements (41), in four different VHL mutant models of ccRCC: the non-metastatic human cell lines 786-O and OS-RC-2, and the corresponding metastatic subpopulations 786-M1A (M1A) and OS-LM1 (LM1), respectively (Fig. 1A). In immunocompromised mice, the M1A and LM1 cells form tumors that histologically resemble human ccRCCs and they display a substantially increased metastatic phenotype when compared to their parental populations (19). At the molecular level, these systems harbor clinically relevant mutations in well-characterized ccRCC genes (42) and their gene expression profiles resemble those seen in ccRCC patients with poor clinical outcome (19). Recent evidence suggests that their response to targeted therapy is also similar to that seen in patients (43,44). Approximately 25,000 enhancers were detected in each sample, of which a shared set of 314 H3K27ac-enriched and 161 H3K27ac-depleted regions showed conserved changes across the replicates and the two model systems (Fig. 1B-C, Supplementary Fig. S1A-C and Supplementary Tables S1-2). The majority of elements were located in introns and intergenic regions >10kb from known transcription start sites (Fig. 1D and Supplementary Fig. S1D), supporting their putative role as enhancers. Enrichment of a second enhancer-associated chromatin mark, monomethylated histone H3 lysine 4 (H3K4me1) was also detected in the same enhancer regions (Fig. 1E and Supplementary Fig. S1E-F), suggesting that MAEs were not poised enhancers but rather represented de novo activated regions (41).
Figure 1. Enhancer activation landscape in metastatic renal cancer.
A, A schematic of the experimental approach. Human vimentin immunohistochemistry of mouse lungs 40 days after tail vein inoculation of 786-O (top) and M1A (bottom) cells. Scale bar 5mm. B, Left, heatmaps showing normalized H3K27ac ChIP-seq signal at 314 H3K27ac-enriched (red side bar) and 161 H3K27ac-depleted (blue side bar) genomic regions in metastatic ccRCC subpopulations (M1A and LM1) compared to their corresponding parental lines. A 10kb region centered to the H3K27ac peak is shown. Right, median fold change in ChIP-seq signal for all 475 enhancer elements. C, Examples of genomic regions with increased H3K27ac signal in metastatic cells. D, Genomic distribution of enriched and depleted H3K27ac regions relative to known transcripts. E, Heatmap showing normalized H3K4me1 ChIP-seq signal, genomic regions as in panel B. F, Relative mRNA expression of transcripts at different positions relative to altered H3K27ac regions. Data from two RNA-seq experiments for both 786 and OS systems combined, whiskers 1.5 x interquartile range. Wilcoxon rank-sum test with Bonferroni correction. G, Relative mRNA expression of transcripts with one or ≥2 associated altered H3K27ac regions. Data from two RNA-seq experiments for both 786 and OS systems combined, whiskers 1.5 x interquartile range. ** P = 0.002, Wilcoxon rank-sum test. H, mRNA expression fold change (tumor/normal) of transcripts at different positions relative to active (MAE ON) and inactive (MAE OFF) H3K27ac-enricehd MAEs. RNA-seq data from ten normal and matched human ccRCC samples, whiskers 1.5 x interquartile range. Wilcoxon rank-sum test with Bonferroni correction. I, Progression free survival in the TCGA ccRCC cohort, patients categorized based on the sum of Z-scores of mRNAs correlating with altered H3K27ac regions in the model systems, top 20% in red, bottom 20% in blue, middle 60% in grey. P-value derived from a Cox proportional hazards model in which sum of Z-scores is used as a continuous variable. N=400
MAEs often appeared relatively close to each other (e.g. Fig. 1C). Indeed, the distribution of distances between both enriched and depleted MAEs followed a bimodal pattern with an overrepresentation of MAEs being <100kb apart from each other, when compared to an expected genome wide distribution of similarly sized groups of H3K27ac enriched elements (Supplementary Fig. S2A-B). The clustering of MAEs suggested that metastatic programming could be linked to super enhancers, i.e. genomic regions with particularly high enrichment of H3K27ac that have been linked to the control of cell identity (27). We found no evidence for the de novo formation of super enhancers in the metastatic cells, however, MAEs tended to be constituent enhancers within super enhancer loci (Supplementary Fig. S2C).
To test the association between enhancer activity and gene expression, we performed RNA-seq mRNA profiling. In general, the expression of the most proximal gene correlated best with MAE activity (Fig. 1F). A subtler correlation was seen on the second most proximal gene, and no general effect was detected for genes further away (Fig. 1F). MAE clusters, defined as two or more MAEs proximal to the same gene, were more strongly associated with gene expression than single MAEs (Fig. 1G and Supplementary Fig. S2D), but super enhancer-associated MAEs were not more significantly correlated with gene expression when compared to other MAEs (Supplementary Fig. S2E). Pathway analysis performed on the MAE-associated gene set highlighted functional classes associated with cancer and tumor progression (Supplementary Fig. S3 and Supplementary Tables S3-4). Importantly, the enhancer activation landscapes in our model systems strongly correlated with those present in a set of human ccRCC samples, confirming that MAE activation patterns were relevant in vivo (Supplementary Fig. S4A-B). To further test the clinical relevance of the MAEs identified in our experimental systems, we analyzed H3K27ac patterns and corresponding RNA-seq transcriptomic profiles in a patient cohort of ten matched cases of normal kidney and ccRCC, nine of which harbored confirmed VHL mutations (39). In general, H3K27ac-enriched MAEs showed stronger H3K27ac signal in ccRCCs when compared to normal kidney (Supplementary Fig. S4C). All ten ccRCCs showed activation of a subset of H3K27ac-enriched MAEs when compared to matched normal kidney but the precise MAE activation landscape varied between the cases (Supplementary Fig. S4D). Importantly, comparison of the genes near active and inactive MAEs, respectively, revealed a strong correlation between gene expression and proximal MAE activity (Fig. 1H), closely mirroring the data from model systems (Fig. 1F). Finally, the expression pattern of genes correlating with proximal MAE activation in our model systems was associated with poor patient outcome in the large TCGA clinical ccRCC data set (Fig. 1I) (11). Thus, metastatic ccRCC populations show clustered enhancer activation patterns that correlate with clinically relevant gene expression traits.
Enhancers mediate metastatic colonization
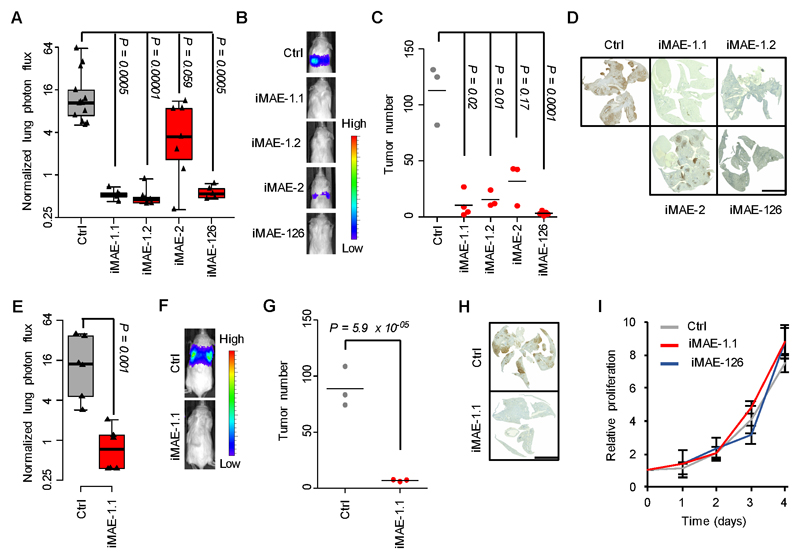
To identify the functionally relevant regions within MAEs, we performed ChIP-seq using an antibody for the enhancer-associated transcriptional cofactor p300 (Fig. 2A) (45). Of the 314 H3K27ac-enriched MAEs, 205 contained at least one p300 peak. We chose the two most statistically significant p300-containing MAEs, MAE-1 and MAE-2 for functional analysis by lentivirally delivered CRISPRi (46) in M1A cells (Fig. 2B-C). Additionally, we focused on MAE-126 as it belonged to the same MAE cluster as MAE-1 (Fig. 2D). ChIP-qPCR and ChIP-seq confirmed specific CRISPRi-mediated reduction in H3K27ac and p300 (Fig. 2E-F and Supplementary Fig. S5A-I). We then inoculated the CRISPRi cells into the tail vein of immunocompromised mice. Targeting MAE-1 or MAE-126 strongly reduced the lung colonization potential of M1A cells; MAE-2 inhibition showed a non-significant trend (Fig. 3A-B). Histological analysis revealed that MAE inhibition had a significant negative effect on the number of lung metastatic colonies, indicating a role for MAE-1 and MAE-126 in the early steps of metastatic colonization at the distant site (Fig. 3C-D and Supplementary Fig. S6A). The functional relevance of MAE-1 was confirmed in an additional metastatic clone (Fig. 3E-H and Supplementary Fig. S6B-C). MAE-1 and MAE-126 inhibition did not affect cell proliferation in vitro (Fig. 3I) and targeting MAE-1 had only a small and statistically non-significant effect on subcutaneous tumor growth (Supplementary Fig. S6D), indicating phenotypic specificity for MAE-1/MAE-126 as a mediator of metastatic colonization. ChIP-qPCR confirmed efficient enhancer targeting in vivo (Supplementary Fig. S6E). Orthotopic implantation of cancer cells gave concordant results: MAE-1 inhibition did not affect growth in the kidney but metastatic colonization in the lungs was reduced (Supplementary Fig. S6F-I). Clinical ccRCC specimens also demonstrated evidence of MAE-1/MAE-126 activity (Supplementary Fig. S6J). Collectively, these experiments demonstrate that specific MAEs are functional mediators of metastatic colonization.
Figure 2. Specific inhibition of MAE activity by CRISPRi.
A, Average ChIP-seq signal for the 314 H3K27ac-enriched MAEs in the M1A cells. B, The 10 most significantly enriched p300-containing MAEs, ranked by H3K27 fold change (FC) shown as grey bars, P-value shown in red. C, Strategy for CRISPRi-mediated repression of enhancer activity. A lentiviral vector expressing two sgRNAs from independent U6 promoters was used to direct dCas9-KRAB to p300 peaks within MAEs. D, H3K27ac ChIP-seq signal in the MAE-1/MAE-126 locus on chromosome 2. E, H3K27ac ChIP-seq signal showing effects of CRISPRi-mediated targeting of MAE-1 and MAE-126. F, Genome wide effects of CRISPRi-mediated enhancer targeting on H3K27ac enrichment.
Figure 3. Enhancers promote metastatic colonization.
A, Effects of CRISPRi-mediated targeting of MAE-1, MAE-2 and MAE-126 on lung metastatic fitness of M1A cells. Normalized lung photon flux in mice after 7 weeks of tail vein inoculation of 300,000 cells. P-value calculated by one-tailed Wilcoxon rank-sum test with Bonferroni correction. N=13 for Ctrl, N=5 for iMAE-1.1, N=7 for iMAE-1.2, N=7 for iMAE-2, N=5 for iMAE-126. Whiskers extend to data extremes. B, Representative bioluminescence images from the experiment shown in A. C, Histological quantification of lung metastatic foci from representative lungs of the experiment shown in A. N=3 for Ctrl, N=4 for iMAE-1.1, N=3 for iMAE-1.2, N=3 for iMAE-2, N=5 for iMAE-126. Horizontal bars represent sample mean. P-values for log-transformed data calculated by one-sided Student’s t-test with Bonferroni correction. D, Representative human vimentin immunohistochemistry of mouse lungs quantified in C. Scale bar 5mm. E, Normalized lung photon flux in mice after 9 weeks of tail vein inoculation of 300,000 M2B cells. P-value calculated by one-tailed Wilcoxon rank-sum test. N=6 for both groups. Whiskers extend to data extremes. F, Representative bioluminescence images from the experiment shown in E. G, Histological quantification of lung metastatic foci from representative lungs of the experiment shown in E. N=3 for both groups. Horizontal bars represent sample mean. P-value for log-transformed data calculated by one-sided Student’s t-test. H, Representative human vimentin immunohistochemistry of mouse lungs quantified in G. Scale bar 5mm. I, In vitro proliferation of M1A cells transduced with the indicated constructs. N=3 for each time point. Whiskers represent S.D. Two-sided Student’s t-test. P=0.82 and 0.69 for iMAE1.1 and iMAE-126, respectively.
A co-regulatory enhancer cluster supports metastasis via CXCR4 activation
The human genome is organized into topologically associating domains (TADs) that are thought to limit the range over which enhancers can act (26). To identify putative target genes for MAE-1 and MAE-126, we determined the TAD structure at the MAE-1/MAE-126 locus using Hi-C (47). We did not observe differences in the TAD structure between 786-O and M1A cells in the immediate MAE-1/MAE-126 region, indicating that perturbation of the higher order chromatin structure was unlikely to cause MAE-1 or MAE-126 activation in the metastatic cells (Fig. 4A). However, other genomic loci, such as that immediately distal to the MAE-1/MAE-126 did show evidence for altered TAD structure in the metastatic cells (Fig. 4A and Supplementary Fig. 7A). While in this particular case the TAD change was not associated with altered gene expression within this locus, it suggests that TAD alterations could be relevant for metastasis gene activation in other loci. Both MAE-1 and MAE-126 resided in the same 600kb TAD that also included two protein coding genes, DARS and CXCR4, of which CXCR4 was located closer to the MAE-1/MAE-126 cluster (Fig. 4B). Only CXCR4 expression was reduced in the CRISPRi cells targeting MAE-1 and MAE-126 (Fig. 4C-D), supporting the observation that the most proximal genes were the most likely MAE targets (Fig. 1F and 1H). Loss of CXCR4 expression was also evident at the protein level (Fig. 4E). The DARS locus, on the other hand, harbored strong H3K27ac signal already in the parental cells, possibly explaining the insensitivity of DARS expression to MAE-1/MAE-126 perturbation (Supplementary Fig. S7B). In a genome-wide analysis, CXCR4 was the most significantly downregulated gene in the MAE-1/MAE-126 CRISPRi cells (Supplementary Fig. S7C and Supplementary Table S5). We also detected increased interaction between MAE-1, MAE-126 and the CXCR4 promoter in the M1A cells when compared the non-metastatic parental cells using 3C (Supplementary Fig. S7D).
Figure 4. The co-regulatory MAE-1/MAE-126 enhancer cluster supports CXCR4 expression.
A, Heatmap showing Hi-C signal and predicted TADs (green triangles) in a 3.6Mb region flanking the MAE-1/MAE-126 locus in 786-O and M1A cells. TAD containing the MAE-1/MAE-126 locus highlighted with an asterisk. B, A close-up of the TAD highlighted in panel A showing known genes and their position in relation to MAE-126 and MAE-1. C, RNA-seq data showing the effects of CRISPRi-mediated targeting of MAE-1 and MAE-126 on the expression of genes within 10Mb from MAE-1. N=6 for Ctrl, N=4 for iMAE1.1, N=4 for iMAE-1.2, N=2 for iMAE-126, technical replicates. D, mRNA expression as measured by qRT-PCR in M1A cells transduced with the indicated CRISPRi constructs. Mean of three experiments. Error bars S.E.M. E, Western blot in M1A and M2B cells transduced with the indicated CRISPRi constructs. A representative experiment of three is shown. F-G, SUZ12 (F) and H3K27me3 (G) ChIP followed by qPCR analysis at the CXCR4 locus in response to CRISPRa-mediated enhancer activation. Mean of two experiments. Error bars S.E.M. H, CXCR4 mRNA expression as measured by qRT-PCR in 786-O cells expressing the indicated constructs. Mean of three experiments. Error bars S.E.M. I, CXCR4 protein expression in 786-O cells as measured by immunoblotting. A representative experiment of three is shown.
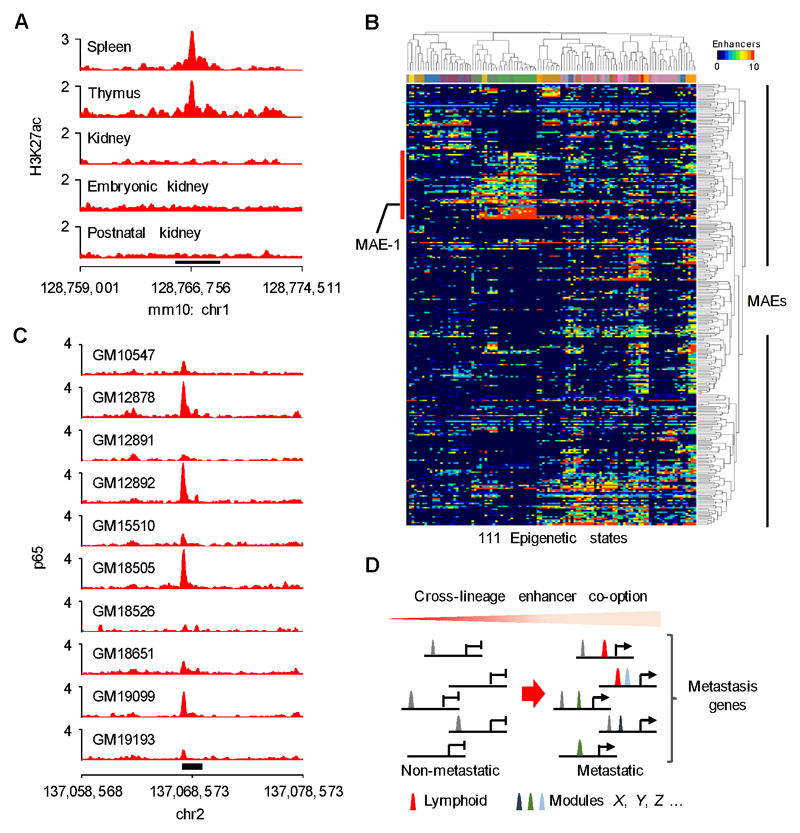
Figure 7. Cross-lineage enhancer co-option in metastatic ccRCC.
A, H3K27ac ChIP-seq signal in the orthologous mouse Mae-1 region. Black bar indicates Mae-1. B, Heatmap showing the activity of 294/314 MAEs in 111 physiological tissues from the Roadmap Epigenomics project. Column color code indicates normal tissue type, for a description of associated epigenetic states see Supplementary Figure 15B. Red bar highlights an enhancer module active in various lymphoid tissues. C, ENCODE p65 ChIP-seq data showing binding at the MAE-1 locus (black rectangle) in human lymphoblastoid cell lines. D, Model: Metastasis genes are activated via cross-lineage co-option of enhancer modules that collaborate with previously activated enhancers (grey peaks) to increase metastatic fitness.
To evaluate the specificity of the CXCR4 regulatory landscape in ccRCC, we used H3K27ac ChIP-seq to identify putative enhancers in the CXCR4 locus in an experimental system of triple-negative breast cancer metastasis derived from MDA-MB-231 cells (17,48). As previously reported, the bone metastatic 1833-BoM cells in this series expressed high levels of CXCR4 mRNA (Supplementary Fig. S8A). We found no evidence of MAE-1 activity in these cells, however, there was a prominent H3K27ac peak, MAE-Br, upstream of CXCR4 in a region that was not active in ccRCC cells (Supplementary Fig. S8B). Comparative CRISPRi-mediated targeting of MAE-Br, MAE-1 and MAE-126 in 1833-BoM and M1A cells, respectively, demonstrated that MAE-Br was only active in the breast cancer cells whereas MAE-1 and MAE-126 supported CXCR4 expression only in M1A cells (Supplementary Fig. S8C-F). These experiments confirmed the specificity of CRISPRi-mediated enhancer targeting and suggested that metastatic clones arising from different cancer types may use alternative enhancer configurations to activate shared metastasis genes.
Previously, the Polycomb Repressive Complex 2 (PRC2) has been shown to repress CXCR4 expression in non-metastatic ccRCC clones (19). Subsequently, however, it has been reported that PRC2 does not play an instructive role in gene silencing, but rather it accumulates at inactive promoters to strengthen a silenced state (49,50). We therefore tested whether MAE-1 activation in non-metastatic ccRCC could evict PRC2 from the CXCR4 promoter and induce gene expression. In line with the hypothesis, recruitment of the dCas9-VPR activator (51) to the MAE-1 locus was sufficient to decrease SUZ12 binding and histone H3 lysine 27 trimethylation (H3K27me3) at the CXCR4 promoter, with associated induction of CXCR4 expression (Fig. 4F-I). These results, together with previous observations (19), support a model whereby the PRC2 complex contributes to CXCR4 silencing and consequent suppression of metastatic potential when sufficient signals for gene activation are not present.
We then tested whether CXCR4 was the functional mediator of the pro-metastatic activity of the MAE-1/MAE-126 cluster in ccRCC. Re-expression of CXCR4 completely rescued the lung colonization phenotype in MAE-1-targeted M1A cells (Fig. 5A-D and Supplementary Fig. S9A-B). The MAE-1/MAE-126 cluster thus supports metastatic colonization by activating the expression of the chemokine receptor CXCR4, a gene linked to metastatic progression through clinical association analysis, as well as experimental approaches in multiple cancer types, including ccRCC (16–19).
Figure 5. CXCR4 is the functional mediator of MAE-1/MAE-126-driven metastatic colonization.
A, Normalized lung photon flux in mice 5 weeks after tail vein inoculation of 300,000 M1A cells expressing the indicated constructs. N=9 for Ctrl-EV, N=8 for iMAE-1.1-EV, N=9 for iMAE-1.1-CXCR4. P-value calculated by one-tailed Wilcoxon rank-sum test with Bonferroni correction. Whiskers extend to data extremes. B, Representative bioluminescence images from the experiment shown in A. C, Histological quantification of lung metastatic foci from representative lungs of the experiment shown in A. N=3 for all groups. Horizontal bars represent sample mean. P-values for log-transformed data calculated by one-sided Student’s t-test with Bonferroni correction. D, Representative human vimentin immunohistochemistry of mouse lungs quantified in C. Scale bar 5mm.
HIF2A and NF-kappaB co-regulate the MAE1/MAE-126 enhancer cluster
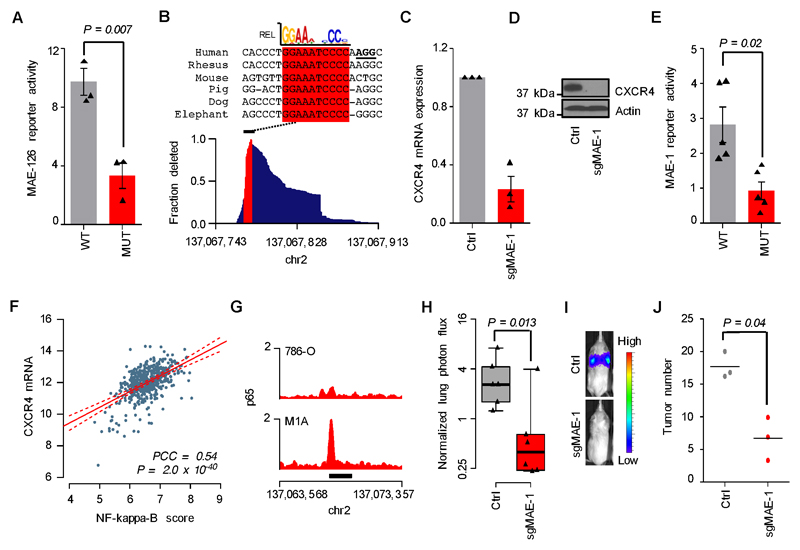
The transcription factor HIF2A, a critical oncogenic driver in ccRCC (12), is necessary but not sufficient for CXCR4 expression in ccRCC (Supplementary Fig. S10A-E) (19). We therefore tested whether HIF2A contributed to the activation of MAE-1/MAE-126. ChIP-seq revealed that HIF2A and its dimerization partner HIF1B strongly bound MAE-126, at a region that contained a HIF2A binding motif, in both non-metastatic and metastatic cells, whereas no significant binding was observed elsewhere in the CXCR4 locus, including MAE-1 (Supplementary Fig. S11A-D). Moreover, mutating the HIF2A binding site decreased MAE-126 reporter activity (Fig. 6A), and CRISPR/Cas9-based mutational targeting of this site in the endogenous MAE-126 locus abolished CXCR4 mRNA and protein expression in M1A cells (Supplementary Fig. S11E-F). The contribution of HIF2A to CXCR4 expression was thus mediated via MAE-126, but HIF2A signaling alone was insufficient for MAE-1 activation.
Figure 6. Mechanisms of MAE-1/MAE-126 activation in metastatic ccRCC.
A, Transient reporter assay showing the effects of mutated HIF motif on MAE-126 enhancer activity in M1A cells. Mean of three experiments. Error bars S.E.M. Two-sided Student’s t-test. B, Bottom, genomic MAE-1 deletion profile in M1A cells targeted by sgMAE-1. Top, mammalian sequence alignment of the most commonly deleted region, a conserved RELA/p65 motif highlighted in red. sgMAE-1 PAM sequence underlined in black. C, CXCR4 mRNA expression in M1A cells as measured by qRT-PCR. Mean of three experiments. Error bars S.E.M. D, CXCR4 protein expression in M1A cells as measured by immunoblotting. A representative experiment of three is shown. E, Transient reporter assay showing the effects of mutated RELA/p65 motif on MAE-1 enhancer activity in M1A cells. Mean of five experiments. Error bars S.E.M. Two-sided Student’s t-test. F, Correlation of CXCR4 mRNA expression with NF-kappaB activity in the TCGA ccRCC data set. N=506; PCC, Pearson’s correlation coefficient. G, p65 ChIP-seq data in the MAE-1 locus. Black bar indicates MAE-1. H, Effects of CRISPR/Cas9-mediated targeting of MAE-1 on lung metastatic fitness of M1A cells. Normalized lung photon flux in mice after 9 weeks of tail vein inoculation of 300,000 cells. N=6 for both groups. One-tailed Wilcoxon rank-sum test. Whiskers extend to data extremes. I, Representative bioluminescence images from the experiment shown in H. J, Histological quantification of lung metastatic foci from representative lungs of the experiment shown in H. N=3 for both groups. Horizontal bars represent sample mean. P-values for log-transformed data calculated by one-sided Student’s t-test.
We then explored the mechanisms by which MAE-1 activity is controlled. Transient reporter assays showed increased MAE-1 activation in metastatic cells when compared to the parental lines (Supplementary Fig. S12A-B), suggesting that the MAE-1 DNA sequence determined its activity at least partially independently of the chromatin context. Sequence analysis identified an evolutionarily conserved binding motif for the NF-kappaB transcriptional effector p65/RELA centered at the peak of the p300 bound MAE-1 region (Supplementary Fig. S12C), suggesting that the NF-kappaB pathway was responsible for MAE-1 activity. In line with this possibility, CRISPR/Cas9-based mutational targeting of this site abolished CXCR4 mRNA and protein expression (Fig. 6B-D). Mutating the putative p65/REL binding site decreased MAE-1 reporter activity (Fig. 6E). We observed a close correlation between NF-kappaB activity and CXCR4 mRNA levels as well as poor patient outcome in the large TCGA ccRCC data set (Fig. 6F and Supplementary Fig. S12D). The metastatic cells showed higher level of NF-kappaB activity at the transcriptome level when compared to the non-metastatic cells (Supplementary Fig. S12E), and ChIP-seq analyses demonstrated p65 binding at the MAE-1 locus in M1A cells (Fig. 6G and Supplementary Fig. S12F). Importantly, mutating the MAE-1 locus decreased the metastatic colonization potential of M1A cells (Fig. 6H-J and Supplementary Fig. S12G-H), providing direct genetic evidence for the role of MAE-1 in metastatic colonization. In agreement, metastatic ccRCC cells displayed significantly higher activation of NF-kappaB (Supplementary Fig. S13A-D) and inhibition of NF-kappaB pathway activity through expression of a super repressor mutant (Supplementary Fig. S13E-G) resulted in downregulation of CXCR4 in the M1A cells (Supplementary Fig. S13H). Conversely, expression of a constitutively active IKK-2 mutant, a kinase that can activate the NF-kappaB pathway but also other pathways, resulted in increased NF-kappaB activity, MAE-1 activation and an increase in CXCR4 expression in 786-O cells (Supplementary Fig. S14A-F). Taken together, the expression of the ccRCC metastasis gene CXCR4 is controlled by the combined action of two independent enhancers, MAE-1 and MAE-126. MAE-126 is activated by the canonical VHL-HIF2A pathway already at the pre-metastatic stage, and MAE-1 is subsequently induced and regulated by the NF-kappaB pathway, with possible contribution from other MAE-1/MAE-126-binding factors.
Cross-lineage enhancer co-option in metastatic cells
The highly conserved p65/RELA motif in MAE-1, and its sensitivity to mutation, suggest that MAE-1, and possibly other MAEs, have a role in normal gene regulation and are not simply de novo cancer enhancers. Consistent with this idea, H3K27ac ChIP-seq data from multiple tissue types in the mouse ENCODE data set (52) revealed that the majority of MAEs belong to tissue-restricted enhancer modules representing multiple developmentally distinct cell linages (Supplementary Fig. S15A). The orthologous mouse Mae-1 locus is active in the spleen and thymus, whereas no general activation is seen in embryonic, postnatal or adult mouse kidney (Fig. 7A). Furthermore, interrogation of the relationship between ccRCC MAEs and 2,328,936 normal human enhancers identified by the Roadmap Epigenomics project (53) revealed that 94% of MAEs co-localized with normal enhancers belonging to several different physiological regulatory modules, such as the prominent lymphoid regulatory module that harbors MAE-1 (Fig. 7B and Supplementary Fig. S15B). Similar analysis of the enhancers activated in the metastatic breast cancer clones 1833-BoM and 4175-LM2 gave concordant results with the CXCR4-associated MAE-Br being part of the lymphoid enhancer cluster (Supplementary Fig. S16A-D). Pathway analysis of the MAE-associated genes also identified several functional categories involved in immune cell biology (Supplementary Fig. 3). Finally, p65 was found to bind MAE-1 in several human lymphoblastoid cell lines which are derived from the B-cell lineage, further supporting the role of the NF-kappaB pathway in the activation of MAE-1 (Fig. 7C) (54). Thus, MAE-1 exemplifies a functionally conserved and tissue-restricted lymphoid enhancer co-opted by aggressive ccRCC cells during metastatic progression.
Discussion
Here we show that metastatic cancer clones activate a distinct set of enhancers and that these drive the expression of metastasis-associated genes. We provide functional evidence for the importance of specific distal enhancers as mediators of metastatic colonization and demonstrate how the activity of these enhancers is supported in metastatic cells. Furthermore, we find that the metastasis-associated enhancer landscape in ccRCC consists of multiple superimposed enhancer modules aberrantly co-opted from diverse physiological cell lineages, providing clues to the mechanisms of metastasis gene activation more generally (Figure 7D).
Despite significant efforts, metastasis driver mutations have thus far not been identified (4,6,7). Yet, as demonstrated by extensive transcriptional profiling of human cancer data sets and experimental systems, metastatic progression is associated with recurrent gene expression patterns (55). How these patterns emerge has remained largely unknown. Our results support a model whereby metastatic transcriptional programs arise from de novo combinatorial activation of multiple distinct and developmentally distant transcriptional modules. These results are in line with recent reports that have demonstrated extensive chromatin remodeling in association with metastatic progression in other tumor types as well (56–58). The complex patterns of metastasis-associated enhancers could explain why individual mutational events do not seem to be sufficient for the establishment a metastatic state. Also, our results and those by Morrow et al. (57) suggest that metastatic gene expression programs in human systems are driven by multiple transcriptional effectors, not individual factors as seen in mice (56,58). This discrepancy may reflect the differences in the kinetics of metastatic progression in mice and humans, respectively.
A specific example of cross-lineage enhancer co-option as a promoter of metastasis is provided by our discovery of the pro-metastatic enhancer cluster that is supported by the ccRCC core driver HIF2A and an NF-kappaB-dependent lineage-restricted lymphoid enhancer. Both of the two main constituent enhancers of this cluster, MAE-1 and MAE-126, are needed for its metastasis-promoting function. Given the tissue specific nature of VHL loss-driven tumorigenesis, it is likely that other factors in addition to HIF2A and NF-kappaB also contribute to MAE-1/MAE-126 function. The identification of NF-kappaB as a regulator of a pro-metastatic enhancer is in line with previous observations that have linked high NF-kappaB activity to cancer progression and poor patient survival in ccRCC (23,25). However, the level of NF-kappaB activity that directly follows VHL inactivation (20,21) is insufficient for the activation of the pro-metastatic program. Rather, our model suggests that further enhancement of NF-kappaB is needed for the activation of MAE-1 and possibly other pro-metastatic target loci. The detailed mechanisms of such NF-kappaB hyperactivation remain to be investigated.
Renal cancers can invade the venous vasculature as tumor thrombi. Yet, even in cases in which vascular involvement has been macroscopically confirmed, a significant fraction never forms distant metastasis (59,60). Thus, in line with evidence from other experimental and clinical cancer contexts (3), early invasion and intravasation do not seem to be the major rate limiting steps of renal cancer progression. We show that the MAE-1/MAE-126 cluster mediates metastatic colonization by regulating the expression of CXCR4, a gene encoding a G-protein coupled chemokine receptor that supports metastatic progression in several different cancer types by increased survival and chemotactic migration (16–19). Whether the MAE-1/MAE-126 cluster also contributes to earlier steps of metastasis remains unclear, however. In line with previous data (19), we find that PRC2, a critical multi-protein complex involved in gene silencing (61), is linked to the CXCR4 repressed non-metastatic cell state. That MAE-1 activation is sufficient for PRC2 eviction from CXCR4 promoter suggests that PRC2 contributes to the maintenance of CXCR4 repression when sufficient activating signals are not present, but it does not determine CXCR4 silencing. This model is in line with data from recent mechanistic analyses of PRC2 function (49,50), and it suggests that reduced PRC2 function could increase transcriptional variability which in turn could favor tumor progression (19,62). Additionally, our comparative functional dissection of the mechanisms underlying CXCR4 expression suggest that alternative regulatory modules may drive its expression in different cancer types. Thus, depending on the tissue of origin, different enhancer configurations may converge on shared metastatic mediators. Analysis of the regulatory basis of metastasis, and the mechanisms that maintain transcriptional stability in non-metastatic clones, is therefore needed for a comprehensive understanding of the origins of metastatic programming.
In genetically engineered mouse models, the transition from normal homeostasis to neoplastic growth is characterized by aberrantly activated tissue regeneration programs that in conjunction with perturbed cell identity networks support the establishment of oncogenic enhancers (63). Our findings of cross-lineage enhancer co-option at the metastatic stage complement this model and suggest the possibility that tumor development from benign to metastatic is driven by progressive lineage infidelity that facilitates the activation of increasingly aggressive cancer phenotypes. The biological implications of such broad perturbations in cellular identity and possible resultant conflicts in co-existing cell fate programs (64) remain to be explored.
The recent findings that modulation of enhancer activity can alter cancer risk (31–33,38), that cancer-specific enhancer elements activate oncogenes (30), and that cancer-associated enhancer clusters are susceptible to pharmacological inhibition (65,66) have raised interest in understanding the mechanisms by which gene regulatory elements contribute to cancer progression and metastasis (56–58). Our work demonstrates that in renal cancer metastatic transcriptional programs are supported by a combination of multiple super-imposed physiological enhancer modules co-opted by metastatic cancer clones. Such cross-lineage enhancer co-option as a source of metastatic traits may be applicable to other cancer types as well. The complex regulatory origins of metastatic traits suggested by this model could have implications for therapeutic strategies that aim at reducing metastasis in patients.
Methods
Cell lines
The renal cancer cell lines 786-O TGL (786-O), OS-RC-2 TGL (OS-RC-2) and their metastatic derivatives 786-M1A (M1A), 786-M2B (M2B) and OS-LM1 (LM1), respectively, have been previously described (19) and they were obtained from J. Massagué (MSKCC, New York) in 2014. The breast cancer cell line MDA-MB-231 and its bone and lung metastatic derivatives 1833-BoM and 4175-LM2, respectively, have been described (17,48) and they were obtained from J. Massagué (MSKCC, New York) in 2014. The identity of the renal cancer cell lines has been confirmed by Sanger sequencing-based detection of known unique homozygous VHL mutations in April 2017. The breast cancer cell lines have been exome sequenced as previously reported and they carry the expected mutational complement (42). All cancer cell lines have been confirmed negative for mycoplasma by biannual tests using the MycoAlert™ Mycoplasma Detection Kit (Lonza, LT07-318). At the time of the study, none of the cell lines were listed in the ICLAC database of commonly misidentified cell lines. For lentivirus production, we used HEK293T cells. Renal cancer cells were cultured either in RPMI-1640 supplemented with 10% FBS, penicillin (100U mL -1) and streptomycin (µg ml−1) or DMEM/F12 supplemented with B27 (Invitrogen), streptomycin (µg ml−1) and EGF/FGF (Peprotech 10ng/mL). Other cells were grown in DMEM, supplemented with 10% FBS, penicillin (100U mL -1) and streptomycin (µg ml−1). Cell lines were used in experiments within 10 passages from thawing.
Chromatin IP
For ChIP-seq sub-confluent cells were crosslinked for 10min at room temperature in 1% formaldehyde in growth media, followed by 5 min quenching with 0.125M glycine. Cells were washed twice with PBS, the supernatant was aspirated and the cell pellet was frozen in liquid nitrogen and stored at -80°C. For tumor ChIP, samples were homogenised in PBS using a Precellys instrument (Bertin) before cross-linking. 100µl of Protein A/G magnetic beads (Thermo, 26162) were blocked with 0.5% BSA in PBS, followed by incubation with antibody at 4°C for a minimum of 4h. The following antibodies were used: H3K27ac (Abcam, ab4729), H3K4me1 (Abcam, ab8895), p300 (Santa Cruz, sc-585), SUZ12 (Abcam ab12073), H3K27me3 (Millipore 17-622), HIF1B (Novus, NB100-110), HIF2A (Abcam ab199) and p65 (Abcam ab16502). Control experiments were done using rabbit polyclonal IgG (Abcam, ab27478). Cross-linked cells were resuspended and sonicated in lysis buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA pH 8.0, 0.1% SDS, and 1% Triton X-100). Sonication was performed in a Bioruptor (Diagenode) for 14 cycles (30” on/30” off) at max output to obtain fragments of 100-500bp. Sonicated lysates were cleared and incubated overnight at 4°C with antibody-bound magnetic beads. Beads were sequentially washed three times with low salt buffer (50mM HEPES pH7.5, 140mM NaCl, 1% Triton) and once with high salt buffer (50mM HEPES pH7.5, 500mM NaCl, 1% Triton). DNA was eluted in elution buffer (50mM NaHCO3, 1%SDS) and cross-links were reversed for 3h (65°C, 1000rpm shaking). DNA was purified using the QuickClean II PCR Extraction Kit (Genescript L00419-100) according to the manufacturer’s recommendations and eluted with 100µl H2O. ChIP-qPCR primers are listed in Supplementary Table S6.
ChIP-seq library preparation
Purified ChIP DNA was used to prepare Illumina multiplexed sequencing libraries with the KAPA Hyper Prep Kit (KR0961) Illumina platforms sample preparation protocol (v1.14). After adapter ligation, libraries were size selected to 150-350bp using Agencourt AMPure XP beads (Beckman Coulter, A63880) according to the protocol recommendations. Size selected libraries were amplified for 15 cycles using the KAPA HiFi HotStart ReadyMix. PCR libraries were then pooled in equimolar concentrations and sequenced.
RNA-seq
RNA was extracted in triplicate from sub-confluent cells using the RNeasy Mini Kit (Qiagen 74104) following the manufacturer’s recommendations. RNA quality was assessed with the RNA Nano 6000 kit (Agilent 5067-1511) and Agilent Bionalyzer 2100 instrument. Purified RNA (1µg) was used for the preparation of Illumina multiplexed sequencing libraries following the KAPA Stranded mRNA-seq Kit (KR0960) Illumina platforms sample preparation protocol (v3.15). After adapter ligation, libraries were cleaned using Agencourt AMPure XP beads (Beckman Coulter, A63880) according to the manufacturer’s recommendations. Purified adapter-ligated DNA was amplified for 15 cycles. PCR libraries were then pooled in equimolar concentrations and sequenced.
ChIP-seq and RNA-seq data analysis
All ChIP-seq and RNA-seq sequencing was performed on Illumina HiSeq 2500 or HiSeq 4000 systems using 50bp single-end runs. For ENCODE data, fastq files were downloaded and processed locally. Raw ChIP-seq sequencing reads were aligned to hg19 or mm10 using bowtie2 (67) and the resulting sam files were converted into sorted bam files using samtools (68). Genome wide correlation between replicates was calculated using 1000bp windows; regions of no signal were excluded. For cell lines, H3K27ac peaks were called using MACS2 (69) with options --broad and -p 1e-8 and the corresponding input samples as controls. Overlap between peaks were identified using the intersect function in bedops (70). For the identification H3K27ac regions specifically enriched/depleted in the metastatic ccRCC populations, replicate ChIP-seq experiments for each cell line were combined and peaks were called using MACS2 with the corresponding parental/metastatic sample as a control. Overlaying the 786 and OS systems resulted in a shared set of 416 enriched and 223 depleted H3K27ac regions in the metastatic clones, respectively. Promoter-associated H3K27ac peaks within this set were filtered out by removing regions closer than 1000bp to H3K4me3 peaks as determined previously (19), resulting in a final set of 314 enriched and 161 depleted metastasis-associated enhancers (MAEs). The genomic distribution of these regions relative to known transcription start sites was determined using Homer (71). MEME suite modules Tomtom (72) and FIMO (73) were used for motif identification within sequences of interest. For peak visualization we used the R packages BSgenome.Hsapiens.UCSC.hg19, GenomicAlignments and GenomicRanges (74). Briefly, reads were extended to the modal length of 250bp and duplicates were removed after which genome coverage was calculated for the region of interest. For final ranking of MAEs we subtracted the signal from the corresponding input sample and used DESeq2 (75) to calculate a fold change and p-value for H3K27ac enrichment. Overlap with Roadmap Epigenomics enhancers was determined using bedops. A minimum of 20% overlap with 314 enriched MAEs was needed for an element to be assigned to a MAE. The activity of a MAE in the 111 Roadmap epigenetic states was determined as the sum of active Roadmap enhancers overlapping with each of the MAEs. Correlation between cell lines and clinical samples was assessed by averaging cell line and clinical sample H3K27ac data across 38,625 H3K27ac-enriced regions identified in the ccRCC cell lines.
RNA-seq sequencing reads were aligned to hg19 using RSEM (76) and bowtie2 with default settings. For determining the association between the expression of transcripts that correlated with MAE activity and clinical outcome we used ccRCC RNA-seq data from the TCGA project (11). Briefly, we calculated sums of z-scores (MAE activation score) using all genes that were first or second most proximal to a MAE and showed a corresponding average gene expression fold change of +/-1.5 in the metastatic M1A and LM1 cells when compared to the corresponding parental populations. Pathway analysis was performed using Ingenuity Pathway Analysis, B-H multiple testing corrected P-value of <0.001 was considered significant. The P-value for clinical association was determined using a Cox proportional hazards model where the MAE activation score was treated as a continuous variable. The NF-kappaB score was calculated as a sum of z-scores of a previously compiled set of ccRCC-associated NF-kappaB target genes (25).
The sequencing data that support the findings of this study have been deposited to the Gene Expression Omnibus under the accession code GSE98015. The ENCODE data was downloaded from https://www.encodeproject.org. The Roadmap Epigenomics data was downloaded from http://www.roadmapepigenomics.org.
CRISPRi, CRISPRa and CRISPR/Cas9 mutagenesis
Lentivirus were produced by transfecting HEK293T with standard packaging vectors using Lipofectamine 2000 (Thermo, 11668027). Viral supernatants were harvested 48h following transfection and filtered through a 0.45 mm PVDF syringe filter. Virus supernatant was then used for infection in the presence of polybrene (Millipore, 8 µg/ml). For CRISPRi enhancer inactivation cells were first stably transduced with a lentiviral construct expressing KRAB-dCas9-mCherry (Addgene 60954, (77)). For CRISPRa-mediated enhancer activation, cells were transduced with a lentiviral construct expressing dCas9-VPR-mCherry (A. Obenauf, Research Institute of Molecular Pathology, Vienna). sgRNA pairs with separate U6 promoters targeting the p300 enhancer peak were cloned in tandem into the pKLV-U6gRNA plasmid (Addgene 50946, (78)) and lentivirally transduced into cells expressing either KRAB-dCas9-mCherry or dCas9-VPR-mCherry, for CRISPRi or CRISPRa, respectively. Double positive cells for the mCherry and BFP markers were selected by fluorescence-activated cell sorting (FACS). Control cells expressed a non-targeting sgRNA pair. CRISPRi-mediated reduction in H3K27ac and p300 was tested by ChIP-qPCR. Genome-wide assessment of CRISPRi was performed by ChIP-seq. Briefly, enrichment in a combined set of H3K27ac regions was determined and difference between the CRISPRi and control sample was assessed using DESeq single sample mode (79). For CRISPR/Cas9 mutagenesis, cells were stably transduced with a modified version of the Cas9 expressing plasmid Lenti-Cas9 Blast (Addgene 52962, (80)) in which the blasticidin resistance gene was replaced with the fluorescent marker mCherry. The sgRNAs targeting the HIF binding site of MAE-126, the evolutionary conserved region in MAE-1 or HIF2A locus were cloned into pKLV-U6gRNA and stably transduced into the cells. Double positive cells for mCherry and BFP were then FACS selected. sgRNAs were designed as previously described (81). A full list of the sgRNAs used in this study is available in Supplementary Table S6.
Human samples
Anonymized ccRCC RNA-seq data and associated clinical annotations were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/). Four tumor samples for ChIP-seq analyses were collected from nephrectomy specimens. The study was conducted in accordance with the Declaration of Helsinki. All patients were recruited for tissue donation by providing written consent and after study approval from an Institution Review Board (NRES Committee East of England - Cambridge Central [REC 03/018]). Raw ccRCC and corresponding normal kidney RNA-seq and ChIP-seq (H3K27ac and inputs) data were downloaded from the NCBI Short Read Archive (Bioproject ID PRJNA396588 and PRJNA340239) (39). Illumina adapters and poor quality sequence was removed and the data was processes as described above for cell lines. H3K27ac signal in H3K27ac-enriched MAEs was normalized to the corresponding input control for all samples, after which each tumor was compared to the corresponding normal sample. A fold change of >3 was used as a cut-off for calling a MAE active. Gene expression fold-change over the corresponding normal tissue was calculated for genes in the proximity of all MAEs in all samples after which the ‘MAE on’ and ‘MAE off’ groups were compared.
Animal studies
All animal experiments were performed in accordance with protocols approved by the Home Office (UK) and the University of Cambridge ethics committee (PPL 70/7990). For experimental lung metastasis assays, we inoculated 3 x 105 tumor cells, resuspended in 100ul of PBS, into the lateral tail vein of 5-7 weeks old male NOD/SCID or NSG mice, obtained from Charles River. Tumor growth was monitored by IVIS bioluminescence imaging (PerkinElmer). At the experimental endpoint lungs were harvested for immunohistochemistry. For the orthotropic tumor assay, a 1cm incision was made above the left kidney of 6-week-old male NOD/SCID mice (Charles River). The kidney was exposed and 1 x 106 tumor cells, resuspended in 25ul of Matrigel (BD), were injected under the renal capsule. Tumor growth was monitored by IVIS bioluminescence imaging (PerkinElmer). At the experimental endpoint lungs were harvested for IVIS imaging. For subcutaneous tumor growth assays, 5 x 105 cells, resuspended in Matrigel (BD), were injected in the flanks of male NSG mice (Charles River). Tumor growth was monitored by caliper measurement. Tumor volume was calculated using the equation V=(L x W2) x 0.5, where L is the length and W is the width of the tumor. After 6 weeks, animals were euthanized and tumors were harvested.
Histology and immunohistochemistry
Lungs and subcutaneous tumors were collected and fixed overnight in 4% paraformaldehyde, washed, embedded in paraffin and sectioned. H&E staining was performed by standard methods. Human Vimentin (Cell Signaling, Cat. 5741, 1:100) staining was performed in a Bond-Max instrument (Leica) using Bond Polymer Refine Detection reagents (Leica) according to the manufacturer’s protocol (IHC Protocol F).
Hi-C
In-nucleus Hi-C was performed on 20 million cells per cell line as previously described (82), with only minor modifications: DNA extraction and all purifications were performed using Agencourt Ampure XP beads (Beckman Coulter) and ligations were performed in a volume of 1ml. Final libraries were amplified using 6 cycles of PCR and 2 samples pooled per sequencing lane. 150 bp paired end sequencing was performed on the Illumina HiSeq 4000 instrument.
Hi-C data analysis
For Hi-C data analysis, reads were aligned, filtered and summarized into frequency matrices with HiC-Pro (83), using the default settings and by setting the minimum and maximum fragment sizes to 100 and 600, respectively. The interaction matrices for each experiment were normalized with ICE (Iterative Correction and Eigenvector Decomposition) using the implementation in the iced python package included in HiC-Pro, with 100 iterations. Topologically associating domains (TADs) were calculated with TADbit (84) from interaction matrices based on 40kb genomic bins. HiCPlotter (85) was used for creating figures incorporating the normalized matrices, genes and enhancer information tracks, as well as the TAD distribution for each experiment. For plotting purposes, the normalized matrices corresponding to replicates of the same experiment were summed together.
3C
Samples for 3C were crosslinked, lysed and HindIII digested similarly to Hi-C samples (82). After digestion, ligation was performed using T4 DNA Ligase (NEB) at 16°C overnight. DNA was purified using Agencourt Ampure XP beads (Beckman Coulter) and quantified (Quant-iT Pico Green, Life Technologies). qPCRs were performed, in triplicate, using JumpStart™ Taq ReadyMix™ for High Throughput qPCR (Sigma-Aldrich) with 400ng 3C library and 0.3μM each primer. 25μl reactions were performed according to the manufacturers protocol but using a Tm of 62°C and 50 cycles. Primer efficiencies were determined for each primer pair by standard curve generation on serial dilutions of a digested and ligated sample using the above conditions. The ratio of interactions for each sample was calculated using: Ratio metastatic/parental = ([Efficiency of test primers - Ct(test metastatic)-Ct(test parental)]/Efficiency of control primers - Ct(control metastatic)-Ct(control parental)]).
mRNA and protein detection
Cell line and tumor total RNA was extracted using RNAzol (Sigma) according to the manufacturer’s protocol. RNA (1ug) was used to generate cDNA with the High-Capacity cDNA Reverse Transcription Kit (Thermo). For qPCR, we used the StepOnePlus instrument (Thermo) with pre-designed TaqMan gene expression assays (Thermo): CXCR4 (Hs00607978_s1), DARS (Hs00154683_m1), IL6 (Hs00174131_m1), IL8 (Hs00174103_m1) and TBP (Hs00427620_m1). TBP served as housekeeping control and data was analyzed using the double delta Ct method. For immunoblotting sub-confluent cells were pelleted, washed with cold PBS and lysed with RIPA buffer. Nuclear and cytoplasmic extracts were obtained using the NE-PER Nuclear and Cytoplasmic extraction reagents (Thermo) according to the recommendations of the manufacturer. Proteins were separated by SDS-PAGE, transferred to PVDF membranes and blotted with antibodies against CXCR4 (Abcam, ab124824, 1:500), HIF2α (Novus, NB100-122, 1:500), VHL (Cell Signaling, 2738, 1:500), phospho-p65 (Cell Signaling, 3033, 1:1000), p65 (Santa Cruz, sc8008, 1:1000), TBP (Abcam, ab63766, 1:5000), B-tubulin (Abcam, ab6046, 1:10000) and ß-actin (Sigma, A5441, 1:5000), ß-tubulin (Sigma, T5201, 1:5000). Secondary antibodies were HRP-conjugated (Dako, 1:5000).
cDNA expression
We cloned the CXCR4 cDNA into a modified pLVX-Puro plasmid (Clonetech) where the puromycin resistance gene was replaced with a hygromycin resistance gene. The empty vector was used as a control. Lentivirus were generated in HEK293T cells as described above. Hygromycin selection started the following day (Invivogen, 900 µg/ml). CXCR4 expression was determined by qPCR. For NF-kB inactivation in metastatic ccRCC cells, we cloned the IKBalpha-mut super repressor cDNA (Addgene 15291, (86)) into the pLVX-Puro vector (Clonetech). Similarly, for NF-kB activation in parental ccRCC cells, the IKK-2 S177E S181E mutant (Addgene 11105, (87)) was cloned into the pLVX-Puro vector. For VHL re-introduction in metastatic cells, HA-VHL (Addgene 19234, (88)) was cloned into the pLVX-Puro vector. Empty vector was used as a control. Lentivirus were generated in HEK293T cells as described above. Puromycin selection started the following day (Invivogen, 4 µg/ml). NF-kB activity was determined by transient reporter assay experiments and expression of downstream target genes (IL-6 and IL-8).
Reporter assays
Enhancer activity was determined by luciferase reporter assays. The region comprising the p300 peak of MAE-126 and MAE-1 (approximately 900bp) was cloned into the pNL1.1[Nluc] vector (Promega, N1001) containing a NanoLuc Luciferase downstream of a minimal promoter using the following primers: 5’-ACGTGCTAGCGTTGGAGGGGAGATCTAGGC-3’ and 5’- ACGTCTCGAGGCCTGTCCATCCACTACCTA-3’ for MAE-126, and 5’-ACGTGCTAGCGTGATGACCTTTTCCACTTGCT-3’ and 5’-ACGTCTCGAGGTCCAGATTCCGACCTTCC-3’ for MAE-1. Genomic DNA from 786-O cells was used as template. Parental and metastatic ccRCC cells were co-transfected with 2µg of enhancer-NanoLuc reporter plasmid and 200ng of pRL-TK Renilla luciferase control vector (Promega E2241) as a normalization control using an Amaxa Nucleofector (program T-20, reagent V, Lonza). After 48h of incubation NanoLuc luciferase activity was measured using the NANO-Glo Luciferase assay (Promega, N1110) and Renilla luciferase activity was measured with the Renilla luciferase assay system (E2810). Mutation of the HIF2A binding site in pNL1.1[Nluc]-MAE-126 was generated using the QuickChange II site-directed mutagenesis kit (Agilent) with the following oligos: 5’- CTCAGCCCTGGCATTTCATCTTTTAGGCGCCGTTCTTAAAGGG-3’ and 5’- CCCTTTAAGAACGGCGCCTAAAAGATGAAATGCCAGGGCTGAG -3’. Mutation of the p65 binding site in pNL1.1[Nluc]-MAE-1 was generated using the QuickChange II site-directed mutagenesis kit (Agilent) with the following oligos: 5’-GCAGAGCCTTGGGGATTTAAAGGGTGGGGTGATGAATC-3’ and 5’-GATTCATCACCCCACCCTTTAAATCCCCAAGGCTCTGC-3’. To determine NF-kB activity, five tandem NF-kB response elements were cloned into pNL1.1[Nluc] vector using the following oligos: 5’-CTAGCTTGGGGACTTTCCGCTGGGGACTTTCCGCTGGGGACTTTCCGCTGGGGACTTTCCGCTGGGGACTTTCCGCAC-3’ and 5’-TCGAGTGCGGAAAGTCCCCAGCGGAAAGTCCCCAGCGGAAAGTCCCCAGCGGAAAGTCCCCAGCGGAAAGTCCCCAAG-3’. Cells were co-transfected with 50ng of pNL1.1[Nluc]-NF-kB(5x) or respective empty vector control and 10ng of the Renilla luciferase plasmid (pRL-TK). NanoLuc and Renilla luciferase activities were measured as described above.
Proliferation assays
For in vitro proliferation assays 1x104 cells were seeded in triplicate in 12-well plates (one plate per day up to 5 days). Each day cells were resuspended and counted using a Vi-Cell XR instrument (Beckman Culture).
Statistical analysis
Statistical analyses were conducted in R and GraphPad Prism. Sample sizes are denoted in figure legends. P-values lower than 0.05 were considered statistically significant. No statistical method was used to predetermine sample size. For animal bioluminescence data, one-tailed Wilcoxon rank-sum test was used and for experiments with ≥2 tests a Bonferroni correction was applied. No animals were excluded from the analyses. For histological tumor count analyses the experimental groups were blinded, for other experiments the experimental groups were not randomised or blinded. For parametric tests data normality was assessed using the Shapiro-Wilk test, variance between the groups was not assumed to be equal. Log transformation was applied as indicated in the figure legends. For tumor count data, a one-tailed Student’s t-test with the Welch approximation of degrees of freedom was used. Bonferroni correction was applied for experiments with ≥2 tests. For reporter assays, a two-tailed Student’s t-test with the Welch approximation of degrees of freedom was used. For testing the association between MAE activation and mRNA expression of the nearby genes, a one sample two-sided Wilcoxon rank-sum test was used. For survival analysis, P-value was derived using a Cox proportional hazards model in which sums of Z-scores we used as a continuous variable. For visualization of survival data, Kaplan-Meier curves were used. For qRT-PCR three independent experiments are shown, each of which is the average of three technical replicates. Boxplots represent median and 25th and 75th percentiles, the whiskers extend to data extremes or 1.5 x interquartile range as indicated in the figure legends.
Supplementary Material
Significance.
Renal cancer is associated with significant mortality due to metastasis. We show that in metastatic renal cancer, functionally important metastasis genes are activated via co-option of gene regulatory enhancer modules from distant developmental lineages, thus providing clues to the origins of metastatic cancer.
Acknowledgements
We thank A. Speed for technical assistance, P. Jones for discussion, A. Obenauf for discussion and the CRISPR activator construct, and V. Gnanapragasam for administration of the DIAMOND study which allowed access to clinical RCC samples. Infrastructure for the Cambridge Urological Bio-repository was funded by the Cambridge Biomedical Research Campus and CRUK Cambridge Centre. The Human Research Tissue Bank is supported by the NIHR Cambridge Biomedical Research Centre. This work was supported by the Medical Research Council (MC_UU_12022/7).
Grant support: This work was supported by the Medical Research Council (MC_UU_12022/7). Infrastructure for the Cambridge Urological Bio-repository was funded by the Cambridge Biomedical Research Campus and CRUK Cambridge Centre. The Human Research Tissue Bank is supported by the NIHR Cambridge Biomedical Research Centre.
Footnotes
Conflicts of interest: The authors declare no potential conflicts of interest.
References
- 1.Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nature Reviews Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 2.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell. 2017;168:670–91. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turajlic S, Swanton C. Metastasis as an evolutionary process. Science. 2016;352:169–75. doi: 10.1126/science.aaf2784. [DOI] [PubMed] [Google Scholar]
- 5.Vanharanta S, Massagué J. Origins of metastatic traits. Cancer Cell. 2013;24:410–21. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet. 2017;49:358–66. doi: 10.1038/ng.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 9.Shuch B, Amin A, Armstrong AJ, Eble JN, Ficarra V, Lopez-Beltran A, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol. 2015;67:85–97. doi: 10.1016/j.eururo.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 10.Young AC, Craven RA, Cohen D, Taylor C, Booth C, Harnden P, et al. Analysis of VHL Gene Alterations and their Relationship to Clinical Parameters in Sporadic Conventional Renal Cell Carcinoma. Clin Cancer Res. 2009;15:7582–92. doi: 10.1158/1078-0432.CCR-09-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaelin WG. Von Hippel-Lindau disease. Annu Rev Pathol. 2007;2:145–73. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 13.Shen C, Beroukhim R, Schumacher SE, Zhou J, Chang M, Signoretti S, et al. Genetic and functional studies implicate HIF1alpha as a 14q kidney cancer suppressor gene. Cancer Discov. 2011;1:222–35. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–11. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 15.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–6. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 16.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 18.Zhang XH, Jin X, Malladi S, Zou Y, Wen YH, Brogi E, et al. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell. 2013;154:1060–73. doi: 10.1016/j.cell.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanharanta S, Shu W, Brenet F, Hakimi AA, Heguy A, Viale A, et al. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat Med. 2013;19:50–6. doi: 10.1038/nm.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An J, Rettig MB. Mechanism of von Hippel-Lindau protein-mediated suppression of nuclear factor kappa B activity. Mol Cell Biol. 2005;25:7546–56. doi: 10.1128/MCB.25.17.7546-7556.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Minamishima YA, Yan Q, Schlisio S, Ebert BL, Zhang X, et al. pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-kappaB agonist Card9 by CK2. Mol Cell. 2007;28:15–27. doi: 10.1016/j.molcel.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi H, Ohh M. The von Hippel-Lindau tumor suppressor protein sensitizes renal cell carcinoma cells to tumor necrosis factor-induced cytotoxicity by suppressing the nuclear factor-kappaB-dependent antiapoptotic pathway. Cancer Research. 2003;63:7076–80. [PubMed] [Google Scholar]
- 23.Sourbier C, Danilin S, Lindner V, Steger J, Rothhut S, Meyer N, et al. Targeting the nuclear factor-kappaB rescue pathway has promising future in human renal cell carcinoma therapy. Cancer Research. 2007;67:11668–76. doi: 10.1158/0008-5472.CAN-07-0632. [DOI] [PubMed] [Google Scholar]
- 24.Oya M, Ohtsubo M, Takayanagi A, Tachibana M, Shimizu N, Murai M. Constitutive activation of nuclear factor-kappaB prevents TRAIL-induced apoptosis in renal cancer cells. Oncogene. 2001;20:3888–96. doi: 10.1038/sj.onc.1204525. [DOI] [PubMed] [Google Scholar]
- 25.Peri S, Devarajan K, Yang DH, Knudson AG, Balachandran S. Meta-analysis identifies NF-kappaB as a therapeutic target in renal cancer. PLoS One. 2013;8:e76746. doi: 10.1371/journal.pone.0076746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long HK, Prescott SL, Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell. 2016;167:1170–87. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin CY, Erkek S, Tong Y, Yin L, Federation AJ, Zapatka M, et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016;530:57–62. doi: 10.1038/nature16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sur I, Taipale J. The role of enhancers in cancer. Nature Reviews Cancer. 2016;16:483–93. doi: 10.1038/nrc.2016.62. [DOI] [PubMed] [Google Scholar]
- 30.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–7. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 32.Sur IK, Hallikas O, Vaharautio A, Yan J, Turunen M, Enge M, et al. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–3. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- 33.Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528:418–21. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med. 2014;20:1130–7. doi: 10.1038/nm.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webster DE, Barajas B, Bussat RT, Yan KJ, Neela PH, Flockhart RJ, et al. Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition. Genome Res. 2014;24:751–60. doi: 10.1101/gr.166231.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016;48:176–82. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tak YG, Hung Y, Yao L, Grimmer MR, Do A, Bhakta MS, et al. Effects on the transcriptome upon deletion of a distal element cannot be predicted by the size of the H3K27Ac peak in human cells. Nucleic Acids Res. 2016;44:4123–33. doi: 10.1093/nar/gkv1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schodel J, Bardella C, Sciesielski LK, Brown JM, Pugh CW, Buckle V, et al. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet. 2012;44:420–5. doi: 10.1038/ng.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao X, Tan J, Lim KJ, Koh J, Ooi WF, Li Z, et al. VHL Deficiency Drives Enhancer Activation of Oncogenes in Clear Cell Renal Cell Carcinoma. Cancer Discov. 2017;7:1284–305. doi: 10.1158/2159-8290.CD-17-0375. [DOI] [PubMed] [Google Scholar]
- 40.Schlesinger-Raab A, Treiber U, Zaak D, Holzel D, Engel J. Metastatic renal cell carcinoma: results of a population-based study with 25 years follow-up. Eur J Cancer. 2008;44:2485–95. doi: 10.1016/j.ejca.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 41.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacob LS, Vanharanta S, Obenauf AC, Pirun M, Viale A, Socci ND, et al. Metastatic Competence Can Emerge with Selection of Preexisting Oncogenic Alleles without a Need of New Mutations. Cancer Research. 2015;75:3713–9. doi: 10.1158/0008-5472.CAN-15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho H, Du X, Rizzi JP, Liberzon E, Chakraborty AA, Gao W, et al. On-target efficacy of a HIF-2alpha antagonist in preclinical kidney cancer models. Nature. 2016;539:107–11. doi: 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–7. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jermann P, Hoerner L, Burger L, Schubeler D. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc Natl Acad Sci U S A. 2014;111:E3415–21. doi: 10.1073/pnas.1400672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell. 2014;55:347–60. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, E PRI, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–8. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–64. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao B, Barrera LA, Ersing I, Willox B, Schmidt SC, Greenfeld H, et al. The NF-kappaB genomic landscape in lymphoblastoid B cells. Cell Rep. 2014;8:1595–606. doi: 10.1016/j.celrep.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–52. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 56.Roe JS, Hwang CI, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, et al. Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell. 2017;170:875–88. doi: 10.1016/j.cell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morrow JJ, Bayles I, Funnell APW, Miller TE, Saiakhova A, Lizardo MM, et al. Positively selected enhancer elements endow osteosarcoma cells with metastatic competence. Nat Med. 2018;24:176–85. doi: 10.1038/nm.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, Gruner BM, et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell. 2016;166:328–42. doi: 10.1016/j.cell.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ciancio G, Manoharan M, Katkoori D, De Los Santos R, Soloway MS. Long-term survival in patients undergoing radical nephrectomy and inferior vena cava thrombectomy: single-center experience. Eur Urol. 2010;57:667–72. doi: 10.1016/j.eururo.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 60.Swierzewski DJ, Swierzewski MJ, Libertino JA. Radical nephrectomy in patients with renal cell carcinoma with venous, vena caval, and atrial extension. Am J Surg. 1994;168:205–9. doi: 10.1016/s0002-9610(94)80069-3. [DOI] [PubMed] [Google Scholar]
- 61.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wassef M, Rodilla V, Teissandier A, Zeitouni B, Gruel N, Sadacca B, et al. Impaired PRC2 activity promotes transcriptional instability and favors breast tumorigenesis. Genes Dev. 2015;29:2547–62. doi: 10.1101/gad.269522.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ge Y, Gomez NC, Adam RC, Nikolova M, Yang H, Verma A, et al. Stem Cell Lineage Infidelity Drives Wound Repair and Cancer. Cell. 2017;169:636–50. doi: 10.1016/j.cell.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.David CJ, Huang YH, Chen M, Su J, Zou Y, Bardeesy N, et al. TGF-beta Tumor Suppression through a Lethal EMT. Cell. 2016;164:1015–30. doi: 10.1016/j.cell.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–20. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012;28:1919–20. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–8. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–61. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 79.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–7. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagano T, Varnai C, Schoenfelder S, Javierre BM, Wingett SW, Fraser P. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biol. 2015;16:175. doi: 10.1186/s13059-015-0753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Serra F, Baù D, Filion G, Marti-Renom MA. Structural features of the fly chromatin colors revealed by automatic three-dimensional modeling. bioRxiv. 2016 doi: 10.1101/036764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akdemir KC, Chin L. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 2015;16:198. doi: 10.1186/s13059-015-0767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–79. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 87.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, et al. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–6. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 88.Li L, Zhang L, Zhang X, Yan Q, Minamishima YA, Olumi AF, et al. Hypoxia-inducible factor linked to differential kidney cancer risk seen with type 2A and type 2B VHL mutations. Mol Cell Biol. 2007;27:5381–92. doi: 10.1128/MCB.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.