Abstract
Transcriptional enhancers function as docking platforms for combinations of transcription factors to control gene expression. How enhancer sequences determine nucleosome occupancy, transcription factor recruitment, and transcriptional activation in vivo remains unclear. Using ATAC-seq across a panel of Drosophila inbred strains we found that SNPs affecting Grainyhead binding sites causally determine the accessibility of epithelial enhancers. We show that deletion or ectopic expression of Grh causes loss or gain of DNA accessibility, respectively. However, while Grh binding is necessary for enhancer accessibility, it is insufficient to activate enhancers. Finally, we show that human Grh homologs, GRHL1/2/3, function similarly. We conclude that Grh binding is necessary and sufficient for the opening of epithelial enhancers, but not for their activation. Our data support the model that complex spatiotemporal expression patterns are controlled by regulatory hierarchies in which pioneer factors, such as Grh, establish tissue-specific accessible chromatin landscapes upon which other factors can act.
Each cell type in our body expresses a unique set of genes. Deciphering the regulatory programs that govern these transcriptional states requires predictive models that can link the genome sequence with the recruitment of transcription factors and other DNA binding proteins; then link this protein binding to chromatin state, the chromatin state with enhancer function, and the enhancer function with target gene expression. A better understanding of the DNA regulatory code will advance our interpretation of non-coding sequence variation, and may ultimately provide new ways to detect and manipulate cell states, for example in cancer cells or for regenerative medicine.
About a decade ago, the main bottleneck for deciphering gene regulation was to locate all the enhancers involved in the control of a certain cell state. Today that problem is largely solved thanks to advances in epigenomics1–3. Indeed, high-throughput techniques such as ChIP-seq4 against transcription factors, co-factors, and histone modifications; DNaseI-seq5 and ATAC-seq6 for open chromatin profiling; and DNA methylation profiling7, have delivered genome wide enhancer predictions for a multitude of healthy and diseased tissues and cell types2,3.
Now that we can systematically profile genomes, epigenomes, and transcriptomes, the next challenge is to discover the rules that link DNA sequence with chromatin state and enhancer function. The function of an enhancer is mainly determined by the specific combination of transcription factor binding sites it contains8,9. Deciphering how motif combinations and their architectures encode a particular output function has been facilitated by Massively Parallel enhancer-Reporter Assays (MPRA)10–12, which are used to test the function of a library of candidate sequences. Such assays can directly test which sequences yield what output, but are less well suited to include the chromatin state into the equation. Furthermore, only a few studies have addressed, on a genome-wide scale, how enhancers generate spatiotemporal expression patterns in vivo, because this requires the creation of a transgenic animal for each enhancer to be tested13,14. Most enhancer models, including the billboard and enhanceosome models15 assume that the cooperative binding of transcription factors causes an enhancer to become accessible and concurrently active15,16. Active enhancers always have bound TFs and are nucleosome-free17, however, little is known about the potential different roles of individual transcription factors with respect to nucleosome occupancy versus enhancer output. Some transcription factors can displace nucleosomes more efficiently than others, and these are often referred to as “pioneer factors” or chromatin openers18–20. Examples of pioneer factors in mammalian genomes include SOX2, PU.1, FOXA1, and TP5318,21–23. For TP53 it was recently shown that upon binding, it not only outcompetes nucleosomes, but also activates gene expression22,24. For the other pioneer factors, it is less clear whether they can activate enhancers or whether additional factors (e.g., OCT4 in the case of SOX2)25 are required to activate an enhancer. In Drosophila, the only pioneer factor known thus far is Zelda, which establishes the chromatin landscape in early Drosophila embryos during the maternal-to-zygotic transition26,27.
Here we set out to study how the three layers, namely sequence information, chromatin accessibility and enhancer function are linked in epithelial cells. To this end we used a combination of chromatin accessibility (bulk, cell-sorted, and single-cell ATAC-seq), natural variation, machine-learning, evolutionary variation and in vivo enhancer-reporter assays in epithelial tissues in Drosophila. Our results provide strong evidence for a hierarchical model of enhancer control that is conserved across Metazoa.
Results
Natural variation in chromatin accessibility predicts potential chromatin regulators
To identify DNA features that are essential for establishing chromatin accessibility in a specific tissue, we profiled open chromatin across a cohort of inbred Drosophila strains. Particularly, we performed 30 ATAC-seq6 experiments on epithelial tissues (eye-antennal imaginal discs), covering 23 distinct strains from the Drosophila Genetic Reference Panel (DGRP)28,29 (see Methods). The open chromatin profiles are highly similar (ρ = 0.76-0.96) within this cohort of eye-antennal discs and differ substantially (ρ 0.25-0.50) from the open chromatin profiles of non-epithelial tissues like the adult brain30 (Fig. 1a, Supplementary Fig. 1).
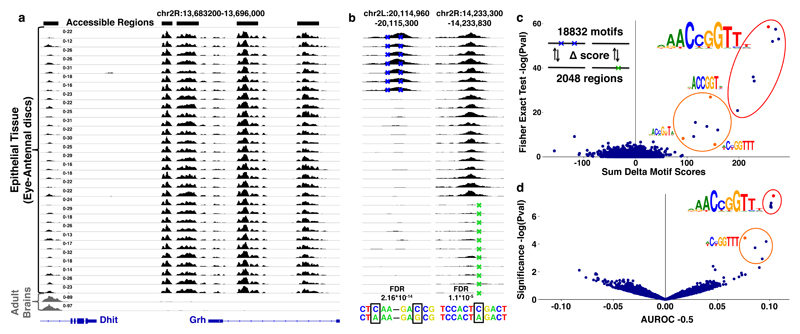
Figure 1. Variation in chromatin accessibility across Drosophila inbred lines.
(a) Accessible chromatin profiles obtained by ATAC-seq on 32 independent tissue samples. The first 30 profiles are from epithelial tissues (eye-antennal discs), the last two from adult brains. (b) Example of two regions with caQTLs: the first has two caQTLs; chr2L:g.20115168G>C and chr2L:g.20115092A>C that correlate with a gain in accessibility (FDR = 2.16*10-14), in the second region the caQTL chr2R:g.14233638C>A significantly correlates with a loss in accessibility (FDR = 1.1*10-5) (see Methods). (c) Delta Motif plot, showing the cumulative effect of the 4289 caQTLs on the CRM score of 18832 motifs, plotted on the x-axis (see Methods). The y-axis shows the enrichment of motifs affected by caQTLs compared to control SNPs (one-sided Fisher’s exact test (log transformed)). Significantly enriched motifs are encircled and examples (red or orange) are visualized. (d) AUROC plot, showing the AUROC statistics for motif matching (18832 motifs) between the 2048 accessible and inaccessible sequences. On the x-axis are the AUROC -0.5 values plotted for each motif. The y-axis shows the significance (log transformed pval). Significantly enriched motifs are encircled and examples (red or orange) are visualized.
We identified 30774 accessible regions across the 30 eye-antennal disc samples (see Methods). To link variation in chromatin accessibility with underlying sequence variation (cis-variation), we applied a Generalized Linear Model (GLM) on all 297000 SNPs that were present in accessible regions. This analysis identified 4289 (~1.5%) SNPs that correlated significantly (FDR<0.05) with the accessibility change of their encompassing region (Fig. 1b), termed chromatin accessibility QTLs (caQTLs31, see Methods). The 4289 caQTLs were located in 2048 regions with variable accessibility between the different inbred lines (examples Fig. 1b).
One possibility to explain how one or a few SNPs can have such dramatic effects on chromatin accessibility would be that a SNP may break (or create) a recognition sequence of a key transcription factor. To identify such potential factors, we scored every region that contained one or more caQTLs with a curated collection of more than 18 thousand transcription factor binding motifs (see Supplementary Table 1 and Methods). Two independent methods then identified motifs for one transcription factor, namely Grainyhead (Grh), to concordantly change with chromatin accessibility (Fig. 1c and d). Overall, changes in a Grh motif were significantly more associated with caQTLs than with SNPs that have no effect on accessibility (Fisher’s exact test Padj=6.75*10-22) and could directly explain the variable accessibility of 70 regions (Fig. 1c). These findings thus predict that a Grh binding site can causally determine the in vivo accessibility of an enhancer-size region.
Grainyhead plays a key role in the chromatin landscape of epithelial cells
Interestingly, Grainyhead is a highly conserved transcription factor with essential roles in epithelial cell fate specification and wound healing across Metazoa32–36. From the 30774 accessible regions of the eye-antennal discs, 10.5% (3246 regions) have at least one Grh binding site. These 3246 potential Grh target regions are located near a large set of 1786 genes that are strongly enriched for GO37 terms like epithelium development (pval=10-56) (Supplementary Table 2). The Grh target regions contain 22.2% of the mapped reads and are in general the most accessible regions of the epithelial chromatin landscape (Welch’s t-test pval=2.251*10-166, Fig. 2a). We hypothesised that Grh target regions are the most accessible because Grh stably binds its target sites in a large fraction of cells in the eye-antennal discs. Supporting this hypothesis is the fact that Grh proteins are ubiquitously expressed in basically all cells of the eye-antennal and wing imaginal discs (Fig. 2b-c).
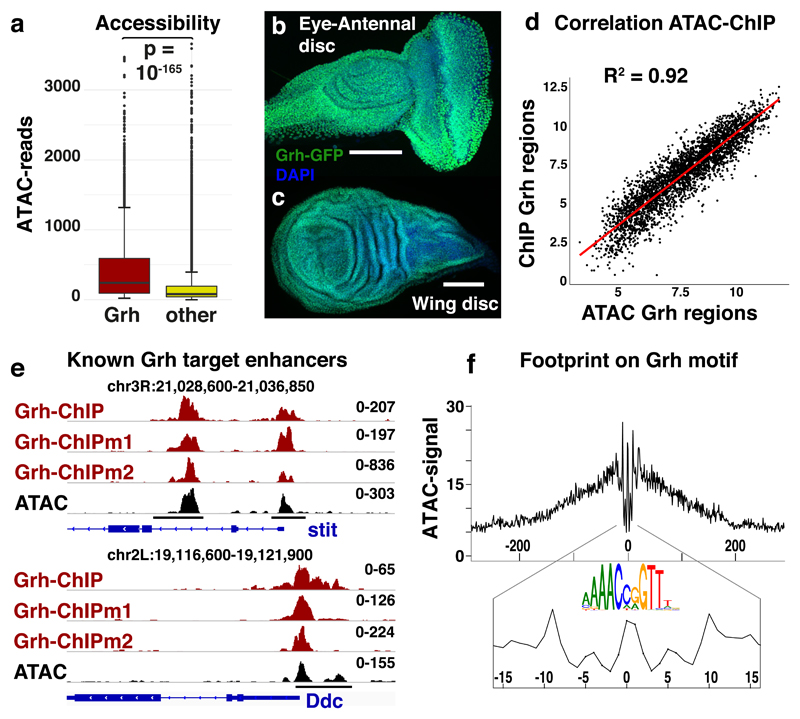
Figure 2. Grainyhead plays a key role in the epithelial chromatin landscape.
(a) Box-and-whisker plots visualizing the normalized reads for accessible regions containing a Grh motif (n=3246) and all others accessible regions (n=27528). The median (centre lines), 25th and 75th percentiles (box edges), data points within 1.5 times the interquartile range from the edge (whiskers) and outliers (data points) are shown. The Welch two sample t-test (two-sided) was used to evaluate the difference (pval = 2.251*10-166). (b-c) Grh-GFP protein expression in eye-antennal and wing imaginal discs, using a chimeric Grh-GFP fusion line39 (reproducible GFP pattern in at least 5 discs, scale bar = 100μm) (d) Correlation plot between the accessibility (ATAC) and Grainyhead occupancy (ChIP(mentation)) of the 3246 Grh target regions (Spearman’s ρ = 0.919). (e) Tracks of Grh-ChIP-seq, Grh-ChIPmentation-seq and ATAC-seq in eye-antennal discs. Grh dependent enhancers (black bars), that control the expression of stit44 (chr3R:21028800-21037000) and Ddc43 (chr2L:19116600-19121750) upon wounding, are shown. (f) DNA footprint on the Grh motif; the averaged ATAC-seq signal over all 3246 accessible Grh regions, centred on their Grh motif, is shown.
To test whether Grh actually binds to the predicted Grh binding sites inside the accessible regions of the eye-antennal discs, we performed ChIPmentation38 against Grh-GFP39 and also re-analyzed published anti-Grh ChIP-seq data40. The same Grh motifs that concordantly changed with chromatin accessibility (Fig 1c,d), were the strongest enriched in the Grh-ChIP peaks (Normalized enrichment score (NES)=12.30)41,42. Interestingly, the ChIP-seq signal across the 3246 Grh target regions correlated quantitatively (ρ=0.92) with the ATAC-seq signal (Fig. 2d) and covered previously characterised Grh dependent enhancers43,44 (Fig 2e).
We then evaluated the occupancy of the Grh binding sites by an independent assay by performing an in vivo DNA footprinting analysis6,45 (see Methods) and found that Grh binding sites had a protection profile that was as strong as nucleosomal or silent DNA (Fig. 2e). These findings indicate that whenever a region with a Grh motif is accessible, Grh is stably bound there. Overall, these data suggest that Grh plays a key role, both in breadth (many target regions) and in depth (the highest peaks) in the accessible chromatin landscape of epithelial cells.
Grainyhead binding sites are essential for enhancer activity
So far, we showed that changes in a Grh binding site could generate or destroy the accessibility of an enhancer-sized region in developing epithelia. To test whether these regions are indeed functional enhancers we cloned four enhancer pairs, each representing different SNP sequences, one with and one without the caQTL that affected a Grh binding site. These regions were individually cloned into GFP reporter vectors46 and stably integrated into the same position in the fly genome (see Methods) (Fig. 3a). Notably, the accessibility profile, assayed by ATAC–seq, of these integrated fragments was entirely determined by their sequence, independent of the local 3D chromatin context (Fig. 3a-d).
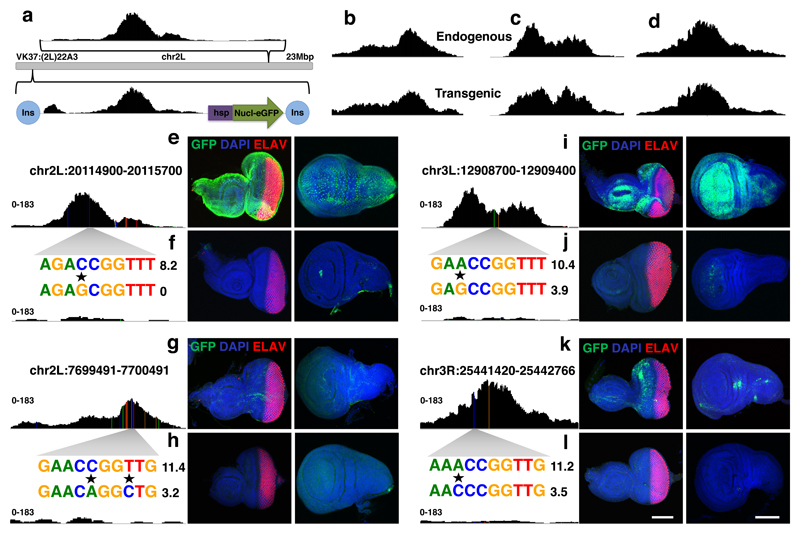
Figure 3. Activity of Grainyhead enhancers in imaginal discs.
(a) Stable insertion of pairs of cloned region into the genome using the VK37 integration site on chromosome 2L. (a-d) Peak height and shape of each region in their endogenous locus and after genomic insertion into the VK37 integration site, the ATAC-seq experiments were done once for each line. (e-l). GFP-expression (green) driven by the four tested enhancer pairs in eye and wing discs, differentiated photoreceptors are stained by ELAV (red) and nuclei are marked by DAPI (blue) (reproducible GFP pattern in at least 3 discs, scale bar = 100μm). Variable accessibility of the endogenous regions is shown, the accessible ones are each time presented at the top (e,g,i,k), while the inaccessible counterparts are presented underneath (f,h,j,l), the ATAC-seq experiments were done once for each line. A zoom in of the affected sequence is shown (caQTLs are marked by a star) together with the Grh PWM affinity score.
Next, by examining the expression pattern of the GFP reporter gene in eye and wing imaginal discs, we tested the direct effect of the caQTLs on the activity of these potential enhancers. Strikingly, the accessible sequences with an intact Grh binding site drove GFP expression in specific and reproducible patterns for all four fragments (Fig. 3e-g-i-k). Their counterparts on the other hand, lacking Grh sites, were predominantly inactive (Fig. 3f-h-j-l). Thus, the Grh binding sites are necessary for both enhancer accessibility and activity.
Grainyhead regulates enhancer accessibility but not activity
Our transgenic reporters showed a clear correlation between the presence of a Grh binding site and GFP expression. The four expression patterns were however distinct from each other and not ubiquitous (Fig 3e-g-i-k). Taking into account that Grh binding nevertheless determined chromatin accessibility and that Grh is ubiquitously expressed in the discs, we hypothesized that Grh binding opens or “primes” its target enhancers, without necessarily activating them. Regions that did not yield any noticeable transcriptional activity despite of being accessible or bound by TFs had also been reported in other studies10,47.
To test whether Grh binds to enhancers without activating them, we measured the reporter activity of 21 additional Grh target enhancers (Supplementary Table 3), of which 15 showed activity in the eye-antennal disc (Fig. 4a). Interestingly, there was no correlation (ρ = 0.05) between the accessibility of these Grh bound enhancers and the number of cells in which the GFP reporter was active (Fig. 4b). Furthermore, using single-cell ATAC-seq, we found that the actual number of single cells, where the Grh bound enhancers were accessible, did correlate with their bulk ATAC-seq signals (ρ = 0.82, Fig. 4c, supplementary Fig. 2). These results already suggest that Grh binding opens its target enhancers, without necessarily activating them.
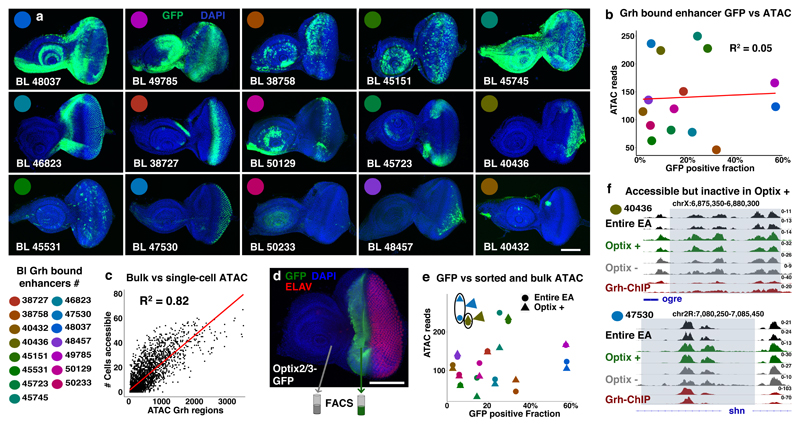
Figure 4. Accessibility of Grainyhead enhancers does not imply activity.
(a) Confocal images showing the activity (GFP, green) of 15 Grh bound Janelia-Gal4 enhancers14 in the eye-antennal discs (reproducible GFP pattern in at least 4 discs per shown image, scale bar = 100μm). (b) Plot visualizing the relative activity (GFP% of encompassing eye-antennal disc) and accessibility (ATAC-seq averaged over 30 independent eye-antennal disc samples) of the 15 Grh bound enhancers. (c) Correlation plot showing the accessibility of the 3246 Grainyhead regions (normalized ATAC-reads) on the x-axis versus the actual number of single cells where the region is accessible (Spearman’s ρ = 0.82). (d) Confocal image showing the reproducible (5 discs, scale bar = 100μm) expression pattern of the Optix2/3 enhancer48 (GFP, green; cell nuclei DAPI, blue; and differentiated photoreceptors (ELAV, red)) in the eye-antennal disc. (e) Plot visualizing the relative activity (GFP%) and accessibility (ATAC-seq, circle: entire eye-antennal discs, triangle: Optix positive subpopulation) of the 15 Grh bound enhancers. (f) Accessible chromatin and Grh-ChIP profiles of two Grh bound enhancers, 40436 and 47530, both are accessible in the Optix positive subpopulation, but show no activity there (2 biological replicates for each sample).
Next, to demonstrate that these enhancers can indeed be accessible in cells where they are not active, we FAC-sorted and performed ATAC-seq on a specific subpopulation of cells from the eye-antennal disc48 (Fig. 4d, supplementary note 1). This revealed that Grh-bound enhancers were similarly accessible in the sorted subpopulation compared to the rest of the tissue (Fig 4e) and that multiple enhancers, like 40436 and 47530, were accessible in the subpopulation without having GFP reporter activity in that domain (Fig. 4f). These findings indicate that Grh binding opens or “primes” its target enhancers, without activating them.
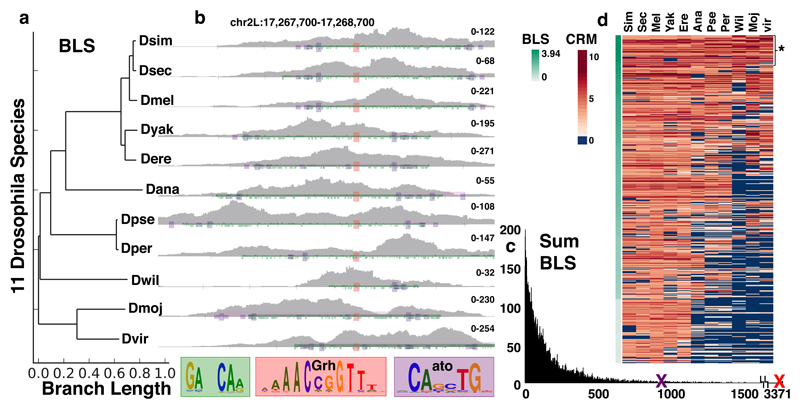
Evolutionary comparison of Grainyhead enhancers identifies candidate activators
The tested Grh-bound enhancers showed a large variety of expression patterns, suggesting that multiple other TFs can cooperate with Grh. To identify candidate co-TFs, we performed a cross-species motif analysis, since this is a powerful approach to identify functional TF binding sites49. We thus performed ATAC-seq in the eye-antennal disc of 10 additional Drosophila species, and selected conserved enhancers that have a Grh motif and are accessible in the other species (example Fig. 5b). These two criteria were nearly 100% predictive of enhancers that are actually bound by Grh (Fig. 2d,e). Next, we scanned these enhancers for conserved motifs from our library of 18832 TF PWMs, using a Branch Length Score (BLS)49 (Fig. 5a) and identified several co-conserved TF binding motifs (Fig. 5c, Supplementary table 5). Interestingly, one of the top co-conserved motifs is (CANNTG), an E-box motif for which the best candidate activator is Atonal, a bHLH factor that is active in a subset of cells in imaginal discs46. These E-boxes were conserved in 92 Grh enhancers (Fig. 5d*, example Fig. 5b) which were located near Ato-induced genes46 (Supplementary Fig. 3), suggesting that the [Grh+Ato] enhancers are functional. Furthermore, from twenty experimentally validated Atonal target enhancers46, we found six to be co-bound by Grh and Ato (Supplementary Fig. 3), and identified four new Grh target enhancers that were active in Atonal expressing cells (Fig. 4a, BL48037, BL46823, BL38727 and BL50129). We conclude from these results that the activity of enhancers primed through Grh binding requires the recruitment of additional factors like Atonal.
Figure 5. Identification of co-regulator motifs through evolutionary conservation.
(a) Branch Length Scores for the 11 Drosophila species. (b) ATAC signal from eye-antennal discs of 11 Drosophila species on a conserved Grh-Atonal enhancer (two eye-antennal disc ATAC-seq replicates per specie). The Grh motifs are shown in red, Atonal in purple and repeats in green. (c) Histogram of the cumulative Branch Length Scores for the 18k motifs. The top Grh motif (red X) has a score of 3371, the top Atonal motif (purple X) has a score of 878. (d) Heatmap showing the CRM scores for the Atonal motif across the conserved Grh enhancers ordered by BLS score. The top 480 enhancers are shown, with the 92 most conserved Grh-Ato enhancers (BLS > 3) marked by *.
Loss-of-function and gain-of-function experiments demonstrate that Grainyhead is a pioneer factor
The data presented so far suggests that Grh is a pioneer TF for epithelial cells. The main function attributed to pioneer TFs is that they can bind their recognition sites within closed chromatin, making their target regions accessible and available for other proteins to bind18–20. This means that removing a pioneer factor should directly reduce the accessibility of its target regions and vice versa that these regions should become accessible upon ectopic expression of the factor.
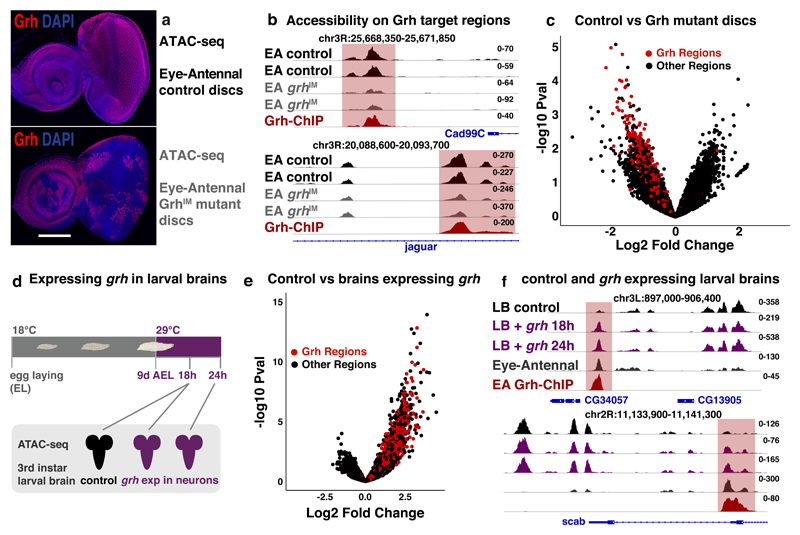
We tested both predictions and first investigated how lack of Grh proteins impacts the open chromatin landscape of epithelial cells. We thus performed bulk ATAC-seq on eye-antennal discs that were largely mutant for grh (grhIM mutant clones)50 (Fig. 6a, Supplementary Fig. 5). Comparing the accessible chromatin landscape of grhIM mutant discs with matching controls identified 1076 regions with reduced accessibility (logFC<-0.5, pval<0.1) in the grhIM mutants. Interestingly, only Grh motifs (from our 18K motifs) were strongly enriched in these regions (i-cisTarget42 NES=21). Known Grh target enhancers near epithelial genes, like Cad99C or jar, lost accessibility in the grh mutant discs (Fig. 6b). Overall, the 3246 Grh target regions were significantly less accessible in the Grh mutant tissue (Fig. 6c, Welch’s t-test pval=3.28*10-8). In summary, these results demonstrate that loss of Grh directly reduces the accessibility of its target regions.
Figure 6. The pioneering role of Grh.
(a) Confocal images of control (top) and discs largely mutant for Grh (bottom), Grh C-terminal antibody68 staining (red), nuclei DAPI (blue) (reproducible results for 5 discs, scale bar = 100μm). (b) Tracks visualizing ATAC of control (black) and Grh mutant (grey) eye-antennal disc and Grh-ChIP (dark-red) in eye-antennal disc (2 biological replicates each). Grh target regions near epithelial genes Cad99C and jar lose accessibility. (c) Volcano plot showing the change in accessibility (DESeq2 log2FC and –log10 pval) of the 30774 regions in control versus Grh mutant eye-antennal discs, red dots mark the 3246 Grh regions. (d) Ectopic grh expression in the neurons of developing larvae. (e) Volcano plot showing the change in accessibility (DESeq2 log2FC and –log10 pval) of 37668 accessible regions in control larval brains versus larval brain with ectopic grh expression, red dots mark the 3246 Grh regions. (f) Tracks visualizing ATAC on control larval brains (black), larval brains with 18 and 24h grh expression (purple) (ATAC-seq experiments were done once for each time point) and eye-antennal discs (dark grey, 30 biological replicates), and Grh-ChIP on eye-antennal discs (dark-red, 3 biological replicates). Genomic loci near CG34057 and scab are shown with Grh target region (red).
Next, we tested whether Grh was able to bind to nucleosomal DNA, making its target regions accessible, as this is a crucial feature of pioneer factors18,19. To evaluate this, we analyzed larval brains, which mainly comprises of neurons and glial cells that do not express Grh, and where we found that the majority (74.3%) of Grh target regions were inaccessible. If Grh is a true pioneer factor of the epithelial epigenome, it should be able to bind to its target regions within this closed chromatin, making them accessible. To test this, we ectopically expressed the epithelial isoform of grh (grhN)34 for 18 and 24 hours in all neurons of 3rd instar Drosophila larval brains (Fig. 6d), using a pan-neuronal ELAV driver51, performed ATAC-seq, and compared their accessible chromatin landscape with that of wild-type brains. Remarkably, basically all 3246 Grh target regions, identified in the eye-antennal discs, specifically gained accessibility in response to ectopic Grh expression (Fig. 6e, Welch’s t-test pval=1.9*10-10), while the neuronal cistrome did not change (supplementary note 2). Furthermore, only Grh motifs were significantly enriched (NES=26.6) in the 1774 regions with the strongest accessibility gain (logFC > 0.5, pval < 0.05) (Fig. 6f shows two examples). Potential target genes near these regions were enriched for GO terms that are normally not present in brain tissues, like embryonic dorsal epidermis (pval=9.6*10-50) and epithelium development (pval=9.3*10-29). Thus, consistent with our caQTL analysis and in vivo enhancer reporters, these results demonstrate that Grh is a pioneer factor, sufficient to directly and specifically open its target regions.
Functional Grainyhead motifs are embedded in a specific DNA context
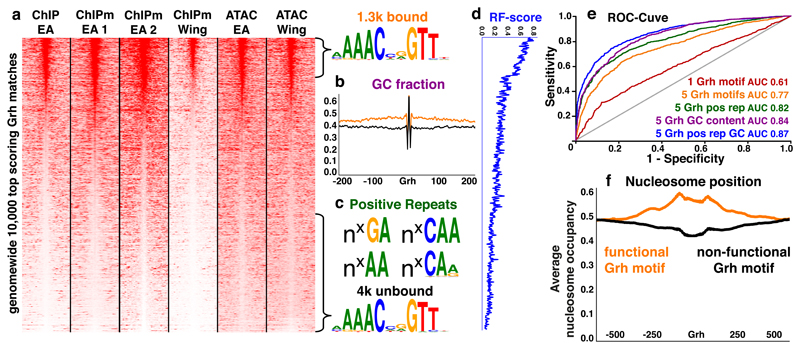
Having established that Grh is a pioneer factor, capable of binding and opening its target regions in multiple tissues, we sought to investigate which sequence elements determine in vivo Grh binding. We first assessed whether the genomic Grh binding pattern was simply determined by the affinity for its binding sites. We collected the 10,000 highest scoring Grh motif matches in the genome, and ordered them according to their Grh occupancy (ChIP-seq meta-analysis, see Methods). The Grh-ChIPmentation and ATAC signal in wing imaginal discs clearly follows this ranking, indicating that despite their different cell fates, Grh binds to the same target regions in both epithelial tissues (Fig. 7a). From these 10,000 highest scoring Grh motif matches, we identified a set of 1,300 regions with recurrent Grh binding, and a set of 4,000 regions with non-functional (not bound nor accessible) Grh motifs (Fig. 7a). By combining five different Grh PWMs into a Random Forest model52 (Supplementary Fig. 5), we could distinguish functional from non-functional sites relatively well (AUROC=0.76, Padj<0.001), suggesting that the affinity for a Grh motif is already predictive for Grh binding.
Figure 7. Discovery of additional sequence features instructive for the in vivo binding of Grainyhead.
(a) Seqminer69 plots visualising the Grh-ChIP(mentation) and ATAC-seq signals on the 10k highest scoring Grh binding sites genome wide (ranked using order statistics see Methods). Similar de novo Grh motifs were found on the top 1300 bound regions and bottom 4000 unbound regions. (b) Average GC fraction of the 1300 bound regions and 4000 unbound regions, showing 200 bp up and downstream of the Grh motif. (c) Repeat sequences of variable length (n) enriched around (±300bp) the 1300 functional Grh motifs. (d) Random Forest score for the encompassing Grh regions (smoothed over 15 regions). (e) Receiver Operating Characteristic curves assessing the performance of a Random Forest classifier to discriminate between 1300 Grh bound and 4000 unbound sequences. (f) Predicted nucleosome affinity55 for the 1300 functional (orange) and 4000 non-functional (black) Grh regions.
Next, we examined the local sequence context around the Grh motifs (±300bp) to identify additional features that may affect Grh binding. We found that functional regions had a higher GC content (Fig. 7b), which may increase the flexibility of the DNA53. Additionally, di- and tri-nucleotide repeat sequences (GA, AA, CAA and CAa/g) were enriched around functional Grh motifs (Fig. 7c). Combining all these features in a Random Forest classifier52 allowed us to quantitatively predict functional Grh binding sites (Fig. 7d) (AUROC=0.870, Padj<0.001, Fig. 7e, Precision-Recall Supplementary Fig. 5). Hence, a suitable Grh motif in a local “favourable” sequence context, is predictive for its in vivo binding.
Since nucleosome positioning is also strongly affected by the local sequence context54, we investigated whether there is a difference in nucleosome affinity between the functional and non-functional motifs. We found that the predicted nucleosome occupancy profiles55 differed significantly, with a pronounced dip in nucleosome occupancy at non-functional motifs, and a wider increase around functional Grh motifs (Fig. 7f). This indicates that Grh preferentially binds to DNA sites in regions that have a high intrinsic affinity for nucleosomes (i.e., likely to be bound by nucleosomes in the absence of Grh), similar to other pioneer factors like PU.1, FOXA, SOX2 and TP5318,20–22,24,56. Thus, in vivo Grh binding is highly predictive and associated with nucleosome displacement.
Grainyhead homologs play similar roles in human
We have identified Grh as the principal pioneer factor of the epithelial accessible chromatin landscape in Drosophila. The mammalian homologs are three Grainyhead-like transcription factors (GRHL1, GRHL2, GRHL3)57 with known and conserved cell adhesion target genes58–60. Since the DNA-binding domain of Grainyhead proteins is highly conserved across Metazoa32,33, we investigated whether Grh could have retained its pioneering functions in mammals.
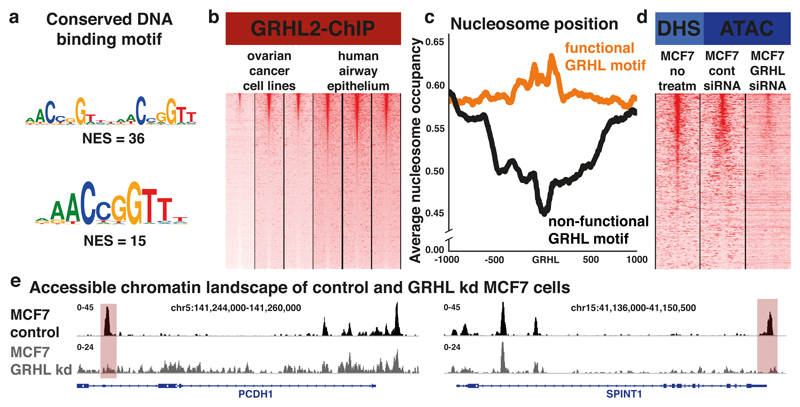
First, using publically available ChIP-seq datasets for GRHL258,59, we confirmed that the DNA binding motif for Grh is conserved between Drosophila and mammals (Fig. 8a). We then identified a core set of regions that are recurrently bound by GRHL2 (Fig. 8b, ChIP-meta-analysis, see Methods) and compared the predicted nucleosome occupancy profiles55 of functional versus non-functional GRHL2 motifs. We found that GRHL2 binding occurs more frequently in regions with a high preference for nucleosomes (Fig. 8c), very similar to the Grh binding profiles observed in Drosophila (Fig. 7f). Additionally, the crucial amino acids for GRHL1 to specifically engage the DNA61 were all conserved in the DNA-binding domain of Drosophila Grh (Supplementary Fig. 6). These findings suggest that part of the interaction mechanisms between Grh proteins and the DNA is conserved from Drosophila to human.
Figure 8. Human GRHLs have similar properties as their Drosophila homolog.
(a) Top enriched motifs found in the GRHL2-ChIP peaks. (b) Seqminer plots visualizing the GRHL2-ChIP signal of six ChIP experiments on OVCAR3, OVCA429, PEO159 and airway epithelium d29, d36 and d4458 respectively, regions are ordered using a ChIP-meta-analysis (see Methods). (c) Predicted nucleosome preference of regions with functional GRHL motifs (red) and regions with non-functional GRHL motifs (black), centred on the GRHL motif. (d) Seqminer plots visualizing the accessible chromatin of MCF7 on the GRHL bound regions, public DHS data on non-treated cells, omni-ATAC on MCF7 cells 48h after non-targeting siRNAs, omni-ATAC on MCF7 cells 48h after mix of GRHL targeting siRNAs. (e) Omni-ATAC-seq tracks of the MCF7 cell line, control (black) and 48h after treatment with GRHL targeting shRNAs (Grey) (Omni-ATAC-seq experiments were done once for each sample). Predicted GRHL target regions (red) near two epithelial genes, PCDH1 and SPINT1 are shown.
To determine whether the GRHL factors also play a key role in setting up the mammalian epithelial chromatin landscape, we analysed the accessible chromatin landscape of the epithelial-like breast cancer cell line MCF7, using DHS data from ENCODE62. The entire accessible chromatin landscape of MCF7 cells was significantly enriched for GRHL motifs (NES=3.7), which were present in 22.8% of the accessible regions. These accessible GRHL target regions are located near 1337 expressed genes that were strongly enriched for GO terms like epithelium development (FDR=1.43*10-65) and cell junction (FDR=4.21*10-49). Combined with previous studies59,58,36, these results indicate that the GRHL transcription factors play a key role in the mammalian epithelial accessible chromatin landscape.
Finally, we investigated whether the GRHL transcription factors also have a direct impact on chromatin accessibility. The GRHL1 and GRHL2 genes are highly expressed in MCF7 cells63 and we had identified a set of putative GRHL target regions. To test whether the accessibility of these regions is due to GRHL binding, we performed omni-ATAC-seq on control MCF7 cells and MCF7 cells in which all three GRHL genes were knocked down for 48 hours using siRNAs (Fig. 8d, see Methods). We found that the regions with reduced accessibility upon GRHL knockdown were strongly enriched for GRHL target regions (Fisher’s Exact Test pval=2.11*10-47) and GRHL binding sites (NES=12.8). These regions included GRHL regulated enhancers near target genes coding for proteins involved in cell adhesion, such as PCDH1 and SPINT1 (Fig. 8e). These findings suggest that the function of Grh as pioneer factor of the epithelial chromatin landscape is conserved between Drosophila and mammals.
Discussion
We performed to our knowledge the first in vivo caQTL study that links sequence variation with changes in chromatin accessibility and enhancer activation. Previous caQTL studies have been performed in human lympoblastoid or iPS cells31,64. Performing a caQTL study in a complex, developing tissue, with a diversity of spatiotemporal expression patterns, enabled us to link regulatory variation with developmental enhancers. We discovered that SNPs that change a Grainyhead binding site can causally determine the in vivo accessibility of an enhancer-size region. Even though these enhancers also contain binding sites for other TFs, their accessibility can be attributed to Grh binding. Indeed, accessibility to epithelial cell enhancers is instigated when Grh is ectopically introduced in non-epithelial cells and abolished when Grh is removed from the epithelial tissue. We found that functional Grh binding sites are generally located in regions with a high affinity for nucleosomes, suggesting that in the absence of Grh its target enhancers are repressed due to strongly bound nucleosomes. Such an elegant scenario, where pioneer target enhancers are kept “off” in other cell types by nucleosome binding, was recently proposed as the “default off” model20.
Grh is ubiquitously expressed in the imaginal disc, and its stable binding across multiple cell types is in agreement with a recent study in the Drosophila embryo35, where Grh was found stably bound to its target genes throughout embryonic development. Even though Grh is bound to its targets in all cells of a tissue, the tested Grh bound enhancers were active in different patterns or at different stages. This observation is reminiscent of Zelda in the embryo19,26,27 where the spatiotemporal enhancer activity of Zelda targets is induced by binding of combinations of other factors, such as the maternal activator Bicoid and the gap genes Hunchback, Giant, and Kruppel65. Also for Grh we discovered many potential co-regulatory transcription factors, like for example Atonal, which is expressed and active in well-defined subparts of the eye-antennal disc46. On the other hand, there are some Grh target genes, like coracle34, Fasciclin334 and grh itself, that are ubiquitously expressed in epithelia. Identifying the co-factors or additional mechanisms behind this ubiquitous expression could be an interesting future challenge.
Importantly, only a subset of the enhancers that are active in imaginal discs are dependent on Grh. Many eye-specific enhancers such as Optix or p53 target enhancers (see Supplementary note 1 and 3), likely become accessible due to binding of the respective factor. Thus, multiple regulatory programs run in parallel within the same tissue, each with their array of target enhancers that are nucleosome-free either due to the binding of pioneer/lineage factors, or through the cooperative binding of multiple TFs16. The possibility that multiple regulatory layers are simultaneously active in the same cell also became apparent in neurons upon ectopic expression of Grh, which triggered ectopic accessibility of the entire epithelial cistrome, without affecting the neuronal cistrome (supplementary note 2). This further suggests that pioneer factors work in parallel, where each pioneer finds and opens its specific target regions, jointly establishing the accessible chromatin landscape upon which other factors can act.
A consequence of the hierarchical enhancer model is that the co-regulatory factors can also be repressors, which could explain why key transcription factors, like Grh, have been given both repressive and activating roles35,58. For many such factors, it was unclear how one TF could activate some target genes while repressing others at the same time. We show in this paper that Grh merely primes its target regions, making them accessible for other factors to bind. The open enhancers can then be subject to binding by either transcriptional activators or repressors that exert their effects on gene expression, explaining why Grh removal simultaneously leads to the reduced expression of some target genes, while increasing the expression of others.
Mammalian GRHL factors have recently been implicated in a number of human diseases, all involving issues with epithelial cell fate. In ovarian cancer for example, GRHL2 expression counteracts epithelial to mesenchymal transition and increases the overall survival of patients59,60. Mutations in the GRHL factors are also linked to pulmonary fibrosis66 and several craniofacial disorders67. We found that mammalian GRHL homologs are required for setting up the accessible epithelial chromatin landscape, and that the biochemistry of GRHL binding and nucleosome displacement are highly analogous to Drosophila. Our findings could help to better understand the role Grh (GRHLs) in human disease.
In conclusion, we set out to determine how sequence information is linked to chromatin accessibility and enhancer activity in vivo. We identified Grh as a pioneer factor of the epithelial chromatin landscape in Drosophila, with likely conserved roles across metazoa. Furthermore, our results support a model in which pioneer factors, such as Grh, sit on top of regulatory hierarchies, establishing tissue-specific accessible chromatin landscapes upon which other factors can act.
Online Methods
Fly husbandry and genotypes
From the Drosophila Genetics Reference Panel, we have used these lines (Bloomington numbers): 25174, 25185, 25186, 25187, 25193, 25194, 25199, 25208, 28123, 28153, 28202, 28206, 28222, 28224, 28243, 28260, 28262, 28265, 28275, 29651, 29652, 55026, 55031. For the Grainyhead ChIP we used: w[1118]; PBac{y[+mDint2] w[+mC]=grh-GFP.FPTB}VK0003339 (Bloomington 42272). For cell sorting we used the Optix2/3-GFP48 transgenic line. For generating the grh mutant eye-antennal discs we recombined the grhIM mutation50 on the FRT42 chromosome to generate the following stock: ey3.5-flp/ ey3.5-flp; FRT42 grhIM/CyO--, mhc-RFP and crossed them to w; FRT42, ey-flp; FRT42 cell-lethal/CyO, mhc-RFP and ey3.5-flp / ey3.5-flp; FRT42 Ubi-GFP/CyO. For the ectopic grh expression in larval brain we crossed these lines: ELAV-gal4;tub-gal80ts X UAS-grhN34. All Drosophila lines were raised on a yeast based medium and kept in an incubator at 25°C. For the ectopic expression of Grh, crosses were raised for ~10 days at 18°C, until the larvae were early 3rd instar, and then shifted to 29°C for 18 or 24 hours to inactivate Gal80, permitting Gal4 to activate the expression of UAS- grhN in ELAV expressing neurons.
Irradiation
3rd instar larvae received a dose of 40 grey of ionizing radiation (160 KeV, 25 mA for 10 minutes) using the small animal research irradiator (RS-2000). Two hours later ATAC-seq was performed on these 15 irradiated DGRP lines (25174, 25185, 25186, 25193, 25194, 25208, 28123, 28202, 28222, 28260, 28262, 28265, 28275, 29652, 55031).
Whole tissue ATAC-seq
We applied exactly the same ATAC protocol for eye-antennal discs as previously described6,70. For each sample, we dissected either 10 eye-antennal discs, 4 wing discs or 2 brains from wandering third instar larvae.
Cell sorted and single cell ATAC-seq
Eye-antennal discs were dissected in PBS and placed in SF900 medium, once 200 eye-antennal discs were dissected, the SF900 was removed and replaced with 400µl Trypsin, 0.05% EDTA for dissociation. The eye-antennal discs were then incubated at 37°C for 1 hour with agitation, samples were mixed every 20 minutes with a pipette. After dissociation, cells were pelleted by centrifugation at 800 xg for 5 minutes at 4°C, washed with PBS and re-pelleted using the same parameters. Finally, the cells were resuspended in 400 µl PBS and filtered using a 40µm cell strainer and stained with propium iodide (final conc 1µg/ml) to exclude dead cells. The cells were sorted on a BD Aria I, selecting against the presence of PI and for the presence of GFP.
For the sorted ATAC-seq samples, 50,000 cells were sorted into Eppendorfs containing 50 µl PBS. Cells were pelleted by centrifugation at 800 xg for 5 minutes at 4°C and ATAC-seq was performed as previously described6,70. For single cell ATAC-seq, as many cells as possible were sorted into an Eppendorf, these were then pelleted by centrifugation at 800 xg for 5 minutes at 4°C and resuspended at a concentration of 1000 cells/µl. Single cell ATAC-seq was performed as previously described71, using the 5-10µm OpenApp IFCs on the Fluidigm C1 and with no cell washing step.
ChIPmentation
Grh-GFP (Bloomington stock 42272)39 3rd instar larvae were dissected in ice-cold PBS and the carcasses with attached discs were fixed for 25 minutes at room temperature by mixing in 1 ml crosslinking solution. The carcasses were washed and the eye-antennal imaginal discs were dissected and sonicated until chromatin fragments reach an average size of 500 bp. The sonicated chromatin was spun at top speed for 10 minutes at 4°C. As a preclearing step, 20 µl of protein A/G magnetic beads (Merck, Millipore) was added, incubated for 1 hour at 4°C and removed by centrifugation at 3000 rpm for 2 minutes. The anti-GFP antibody (ab290, Abcam) was added to a fixed chromatin aliquot and incubated overnight at 4°C. Immunocomplexes were recovered by adding protein A/G magnetic beads to the sample followed by incubation for 3 hours at 4°C. The magnetic beads with precipitated chromatin were washed. Beads were resuspended in elution buffer, RNase was added to the immunoprecipitated chromatin and incubated for 30 minutes at 37°C. The immunoprecipitated DNA was purified. To incorporate sequencing adapters, we combined the purified cDNA with 4 µl of Nextera TD buffer (Illumina) and 1 µl of Nextera Tn5 enzyme (Illumina) on ice and incubated at 55°C for 5 min. The tagmented cDNA was purified again on a MinElute column and eluted in 20 µl EB buffer. To PCR amplify the fragments, we added 25 µl of NEBnext PCR master mix (Bioke), 5 µl of Nextera primer mix and incubated at 72°C for 5 min, then at 98°C for 30 seconds, followed by 15 cycles of 98°C for 10 seconds, 63°C for 30 secs and 72°C for 3 min. We purified the PCR amplicons with 55 µl AMPure beads (Analis).
ATAC and ChIPmentation-seq analysis
Adapted sequences were trimmed from the raw reads using fastq-mcf (default parameters using a list containing the common Illumina adapters). Cleaned reads went to a quality control step using FastQC from Babraham Bioinformatics.
We avoided mapping artefacts, as we mapped the ATAC-seq reads for each line to its personalized genome. All experiments were mapped to the Drosophila melanogaster Flybase r5.13 genome72, since this was the version used in the DGRP project to call the variance. In a first step the reads were mapped using bowtie273 onto the personalized genomes of each DGRP lines or on the standard Flybase r5.13 genome for the ChIPmentation and non-DGRP ATAC samples. For each of the 23 DGRP lines we adapted the consensus genome (r5.13) using seqtk mutfa74, each time including their SNPs that were previously called form whole genome sequencing28,29. After the first mapping round, additional SNPs were called on the ATAC reads using SAMtools75 mpileup -B –f r5.13.fasta DGRP_lineX.bam | varscan.sh mpileup2snp --output-vcf 1. Newly called homozygous SNPs (several thousand per line) were added to the existing vcf files using VCFtools76. The genomes were again updated to obtain a final personalized genome for every DGRP line, strongly reducing mapping errors and increasing the sensitivity of subsequent analyses. Cleaned reads were mapped onto the final genomes using bowtie273 again and Samtools75 was used for sorting and indexing.
Peaks were called on the mapped reads using macs277 callpeak (-g dm –nomodel (for ATAC only) –keep-dup all –call-summits). The narrow peak files (bed format) for all the DGRP lines were merged into a single file that contained a total of 33771 regions, accessible in at least one DGRP line. After filtering out chrU, Uextra, Het, mitochondria and the chromosome borders, we end up with 30774 accessible regions for the eye-antennal discs. For every ATAC-seq sample we count the number of reads falling into each accessible region using featureCounts78.
The raw counts matrices (see GEO processed files) were further processed in R version 3.2.2. DESeq279 was used to normalize the raw counts matrices using the sizeFactor for each sample. All differential accessible regions sets were obtained using DESeq2, following the standard procedure for differential mRNA. The contrasts were; 2 Optix positive vs 2 Optix negative sorted populations, 2 grhIM / cell lethal mutants vs 2 wild type / cell lethal eye-antennal discs, 2 ectopic grh expressing brains vs control and 8 irradiated DGRP lines vs their non-radiated controls.
Identifying enriched motifs
We used i-cisTarget41,42, a sequence based motif enrichment tool that takes conservation into account, to identify motifs that were enriched in our sets of accessible regions (online version, input bed files, genome dm3, 18k motif database). For every input set of region, the enrichment of each motif was calculated as an area under the recovery curve, observed over 136K candidate regulatory regions. We selected the motifs with the highest Normalized Enrichment Scores (NES)42 for our given peak sets.
For all 30774 accessible regions in the eye-antennal discs Trl/GAGA-factor motifs were the top enriched (NES=7.6) followed by Grh motifs (NES=5.7).
For all three GFP-Grh-ChIP(mentation) peak-sets the top enriched motifs were for Grh (or GRHL) transcription factors (NES=11.1-12.3). Grh motifs were also discovered de novo in the Grh-ChIP-seq peaks, using Homer80 de novo motif (pval 10-370) or RSAT81 peak-motifs (sig=12.58, eval=2.6*10-13).
For the 434 Optix specific regions the top motifs were for sine occulis / Optix (NES=19.9).
For the larval brains, we analysed all accessible regions and the 1774 regions that specifically went up in the brains with ectopic grh expression, results see in Fig. 6f.
Single cell ATAC-seq analysis
Data alignment was performed using bowtie273 on the Drosophila's melanogaster genome version dm3 (r5.13). ScATAC data was deduplicated using picard tools markDuplicates82. Out of the 96 single cells, 28 were removed from the data set based on the reads' distributions around TSSs. ChromVAR83 was used to quantify the number of fragments falling within each of the potentially accessible regions. The resulting matrix, (with cells as columns, regions as rows and values as counted fragments in a region within a cell) was binarized, taking a value of 1 if at least one fragment was quantified within the region in the cell and 0 otherwise.
Correlation and Variability between accessible regions
The counts matrix of the 30 DGRP ATAC samples and 2 Adult brain samples (public data obtained from GSE8397530 and processed like our own ATAC data) containing a total of 34768 accessible regions (DGRP and Brain merged) was used. We used the corrplot84 package in R to calculate and visualise the spearman correlations between every sample pair.
GLM and Chromatin Accessibility QTLs
The 30774 accessible regions were filtered before performing the GLM to reduce the effects of noise and mapping bias. 1453 regions with a high repeat content (> 25% RepeatMasker85 (UCSC)) were removed using bedtools.2.26.086 intersectBed –v –f 0.25. Next, 1513 regions with a low coverage for every DGRP line were removed (coverage of the region below 0.2pb for every DGRP lines), ending up with 27808 accessible regions. For each region, we extracted the normalized ATAC-seq reads for these 30 DGRP lines (25174_IR, 25185_IR, 25186_IR, 25187, 25193_IR, 25194_IR, 25199, 25208_IR, 28123, 28123_IR, 28153, 28202_IR, 28206, 28222_IR, 28224, 28243, 28260, 28260_IR, 28262, 28262_IR, 28265, 28265_IR, 28275, 28275_IR, 29651, 29652, 29652_IR, 55026, 55031, 55031_IR) and linked each region to the annotated and additionally called SNPs for these lines (see ATAC-seq analysis). 297000 SNPs were assigned to their encompassing region using bedtools.2.26.0 intersectBed on the extended vcf file.
For each region, we now have the normalized reads for each of the 30 lines as one vector and all SNPs called inside this region as a binary matrix for the 30 lines (present =1, absent=0, unknown=NA). We searched for correlating region-SNP vectors using the generalized linear model function in R (version 3.2.2)87. The p-values were adjusted using the Benjamini-Hochberg procedure in R. With a FDR of 0.05 we identified 4289 highly correlating SNP-region pairs referred to as caQTLs (Chromatin Accessibility Quantitative Trait Loci).
Delta Motif scores
To single out motifs that correlate significantly with the open chromatin changes, a Delta motif score was calculated for every of the 18832 unique motifs in our collection. The sequence for each of the 2048 variable regions, that contained at least one caQTL, was extracted using bedtools getfasta. Next, we mutated these sequences with their encompassing caQTLs according to their effect on the open chromatin using seqtk mutfa. For each of the 2048 regions we now have 2 sequences, one for the accessible chromatin and one for the less accessible/closed chromatin. We scored every time both sequences with the 18832 motifs using cluster buster –m 0 –c 0 and retained for every motif the highest CRM score for each sequence. By subtracting the CRM score of the less accessible/closed region from the encompassing accessible region we obtained a delta motif score for that region. For every motif, we summed all delta scores from the 2048 regions to obtain a cumulative delta score for each motif (plotted on the x-axis of fig. 1c).
We calculated a Delta motif score, following the same procedure, on 66360 SNPs that were present in an accessible region but had no effect on chromatin accessibility (GLM FDR > 0.95). We then calculated for each motif whether it was significantly more affected (|Delta score| > 3) by caQTLs compared to the non-correlating SNPs, using the Fishers exact test88 in R (log10 of the pval is plotted on the y-axis of fig. 1c).
AUROC statistics
In this method, used by the DREAM5 consortium89, we calculated the AUROC statistics for motif matching between the 2048 accessible and inaccessible sequences (see above). For every one of the 18832 motifs, we first selected all the regions where the motif was present (CRM score > 4 in the accessible and/or inaccessible sequence in at least 50 regions) and then calculated AUROC statistics using the pROC90 package in R. We then calculated for each motif its ability to discriminate accessible versus inaccessible sequences by comparing the AUROC to a random classifier (0.5). We calculated a p-value using stratified bootstrapping (n=2000) again with the pROC90 package in R. For representing the data in Figure 1d we subtracted from every AUROC value 0.5 (random classifier) and log transformed the p-value.
DNA footprint
We merged all 30 ATAC-seq samples, resulting in a data set of more than 350 Mio mapped reads. We adapted the mapped reads to represent the actual binding of the transposons, according to the original ATAC-seq paper6. To obtain the footprint on the grh motif, we selected all Grh target regions (3246), centred each region on the best Grh motif and plotted the ATAC-seq signal, normalized by the number of regions.
Ranking Grainyhead regions using Order Statistics
Drosophila eye-antennal discs: We combined the publicly available Grh-ChIP-seq40 data together with two new inhouse Grh-ChIPmentation datasets. Human: We combined 6 publicly available GRHL2-ChIP-seq58,59 datasets from epithelial cell lines.
First, we score the entire Drosophila Melanogaster genome (dm3 r5.53) / Human reference genome (hg19) with the top enriched Grainyhead motif using Cluster-Buster91. We selected the top 10000 highest scoring motifs (none overlapping) and generated a bed file, extending the motif location by 300 base pairs both up- and downstream. Next, for each region in the bedfile we counted the number of reads coming from the three ChIP experiments, using bedtools multiBamCov86. We obtained a final ranking base on the ChIP signal from all three experiments using order statistics. This ranking of regions with a Grainyhead motif was used to plot all other data in Fig. 3a (Grh-ChIPmentation in wing discs, ATAC in both eye-antennal and wing discs) using seqminer69.
Random forest
We trained and evaluated random forest classifier52 models (ensemble of 151 decision trees), using the scikit-learn Python package92. The max_features parameter was set to the square root of the total number of features used to train the model. We evaluated the performance of our models by generating Precision Recall cures and calculating the AUC (Fig. 3f). This was each time done on left out data, to be precise we trained the model on half of the positive (650) and half of the negative (2000) sequences and scored the unseen other half (and vice versa).
Feature discovery was done by comparing the sequence of the top 1300 bound versus the bottom 4000 unbound regions (each time 612 basepair sequences, centered on the Grainyhead motif). We ran RSAT peak motif81 to discover oligo’s, motifs and dyads that were significantly enriched in the bound versus the unbound Grainyhead regions. We identified several repeats (Fig. 3d) that were significantly enriched in the bound regions around the Grh motif.
We took these repeat elements (GA, CAr, CAA, AAA, A, CAr_GA, CAr_GA_AAA) and used Cister94, a tool for detecting cis-elements clusters with optimized parameters for the repeats (-a 0 -w 1000), to score the entire sequence (612 bp). We then integrated the Cister output over the sequence to obtain one score (feature) per repeat element. This way we can take the high variability of repeat lengths into account, longer repeats result in a higher feature score.
The score for the five Grainyhead motifs (Supplementary Fig. 5) was obtained using Cluster-Buster91, we used each of the five Position Weight Matrices to score the Grainyhead sequences and retained the top CRM score.
The GC fraction was calculated by selecting the DNA sequence one nucleosome up- and downstream of the Grainyhead motif (+-147 base pairs) and simply counting the occurrence of G’s and C’s divided by the total number of base pairs.
The final features that were used for training and evaluating the random forest were the following: (AUROC on 1300 positives and 1300 negatives (balances set) and AUPRC on 1300 positives and 4000 negatives (unbalanced set)):
-
a)
No training, random ordering (grey PR curve, AUROC = 0.5, AUPRC = 0.245, Padj = 1)
-
b)
One Grh motif (brown PR curve, AUROC = 0.613, AUPRC = 0.345, increase versus (a) Padj < 0.001)
-
c)
5 Grh motifs top CRM scores (red PR curve, AUROC = 0.766, AUPRC = 0.55, increase versus (b) Padj < 0.001)
-
d)
Integrated Cister score for the repeats (GA, CAr, CAA, AAA, A, CAr_GA, CAr_GA_AAA) over the entire region (612 bp) and closer to the Grainyhead motif (one nucleosome around the motif (306 bp) (green PR curve, AUROC = 0.82, AUPRC = 0.634, increase versus (c) Padj < 0.001)
-
e)
GC fraction for each region (one nucleosome (+-147 base pairs) around the Grh motif) (purple PR curve, AUROC = 0.842, AUPRC = 0.674, increase versus (c) Padj < 0.001)
-
f)
All previous features (Grh motifs, repeats and GC) combined (blue PR curve, AUROC = 0.87, AUPRC = 0.736, increase versus (d) Padj < 0.001, increase versus (e) Padj = 0.03)
Significance (Pval) of the improvement in AUROC or AUPRC was calculated using bootstrapping versus random or the specified previous RF result.
DNA shape and nucleosome predictions
Every time we compared the 1300 bound Grainyhead motifs versus the 4000 unbound motifs (extended by 300 base pairs each side). The DNA shape data (Helix Twist, Minor Groove Width, Roll and Propeller Twist) were downloaded from ftp://rohslab.usc.edu/dm3/95 and an average score for the bound and unbound regions was calculated.
A similar analysis was performed for the nucleosome prediction data, this time extending 1kb around the motif. The nucleosome prediction data was obtained from https://genie.weizmann.ac.il/software/nucleo_genomes.html55. For the Human nucleosome prediction we used the top 1000 GRHL2 bound regions versus bottom 1000 obtained after orderstatistics.
Drosophila Species and Branch Length Score
We performed ATAC-seq on the eye-antennal discs of 3rd instar larvae from 10 related Drosophila species; Simulans, Sechellia, Yakuba, Erecta, Anasai, Pseudoobscura, Persimilis, Wilistoni, Mojavensis and Virilis.
The ATAC-reads were mapped to their respective genomes and peaks were called using macs277 standard parameters.
We used Kent tools liftover –minMatch=0.1 to obtain the bed coordinates of the Grainyhead enhancers in the other species. Next we used bedtools.2.26.0 intersectBed –wa -wb –f 0.1 –a specie.narrowpPeak –b specie.liftover.bed to obtain the list of conserved Grainyhead enhancers that are also accessible in their respective genomes. For every specie, the sequences of the conserved enhancers were obtained using the bedtools.2.26.0 getfasta command. All sequences were scored with our collection of 18832 motifs, using clusterBuster with the option –m 0 –c 0. For each motif, the highest CRM score per sequence was used to calculate the Branch Length Score according to49. The Newick format was used for the phylogenetic tree data from http://flybase.org/maps/chromosomes/synteny_table.html. Branch length scores were summed over all conserved sequences to obtain a total score for every motif. As control, we shuffled all sequences (keeping same base pair compositions and sequence length) using shuffleseq from the EMBOSS package96 and calculated the BLS score again for all motifs. The corrected Branch Length Score, used to evaluate the co-conservation of motifs, was obtained by subtracting the BLS score from the shuffled sequences from the BLS score calculated on the real sequences.
Co-conserved motif clustering
After calculating the BLS scores for our 18832 motifs, we selected those with a corrected BLS score over 700 (387 motifs or top ~2%). Next, to visualize these motifs we first clustered them using STAMP: a tool for DNA-binding motif similarities97. We obtained 77 motifs clusters, from which we only retained those motifs that were directly annotated for a transcription factor that is expressed in the eye-antennal discs (supplementary table 5).
Gene Set Enrichment Analysis
Publicly available micro array data (Atonal gain of function: GSE1671346, ionizing radiation: GSE3740498 was used to generate the ranked gene list. Differential gene expression was calculated using GEO2R, the log fold change was used to rank the genes.
Regions were assigned to neighboring genes (5kb upstream or intronic) and these gene lists were used in the Gene Set Enrichment Analysis99.
Generation of Transgenic lines
Genomic DNA was extracted from an adult fly of lines 25208, 28123, 28222.
The four enhancer-pairs of interest were obtained by genomic PCR from the specific lines using the primers listed in Supplementary table 6.
The PCR product was purified on an 1.2% agarose gel. The correct band was cut out and further purified using the Qiagen gel extraction kit. The constructs were cloned into an entry vector using the pENTR/D-TOPO cloning kit following the standard online protocol. The plasmids were transformed into chemically competent DH5alpha cells. The right construct was confirmed by sequencing. In a second cloning step, we set up the LR Gateway reaction (Invitrogen) between the entry clone and the modified pHStinger vector46. The final constructs were stably and site specific (VK37(2L)22A3) integrated into the Drosophila Melanogaster genome by injecting it into embryos (done by GenetiVision) using the PhiC31 system.
Activity of Grh enhancers
Drosophila lines from the Janelia-Gal4 FlyLight14 enhancer project were selected that had a Grh-ChIP and ATAC peak + strong Grh motif (Sup table 3). These lines were crossed to a line with UAS-eGFP and eye-antennal and wing imaginal discs from 3rd instar larvae were dissected, fixed and stained.
Immunohistochemistry and image analysis
Imaginal eye and wing discs from third instar larvae were fixed in 5% formaldehyde at RT for 30 min. Next, they were washed in PBT (PBS + 0.3% TritonX-100) and blocked in 5% normal Donkey Serum in PBT (PBNT) for 15 min. For testing the Grainyhead enhancers, the tissues were incubated with the primary antibody mix rabbit anti-GFP (Invitrogen), rat anti-ELAV (DSHB) at 4°C overnight. The secondary antibody Cy3-Donkey-anti rabbit (Molecular probes) and DAPI were added for 2h at RT, and then the samples were washed with PBNT, PBT, and PBS (3x10 min). Samples were postfixed 10 minutes in 4%Formaldehyde and washed (3x) with PBS before mounting the discs in Vectashield (Vector Laboratories). Rabbit-anti-Grh-Cterm (1:500) was stained similarly as for Dcp1 and visualized using Alexa647 conjugated Donkey-anti-Rabbit secondary antibody (Molecular probes).
For imaging the Olympus FV1200 confocal microscope was used (20x oil, z-stack). ImageJ (Bio-formats Importer plug-in) was used to merge and process the images. The quantification of the GFP positive fraction of cells in the discs was also done in ImageJ, manually selecting the threshold for each disc before measurement.
Knock-down of GRH-like transcription factors in human epithelial cell culture
Human epithelial, breast carcinoma MCF7 cells were cultured at 37°C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium (ThermoFisher Scientific), supplemented with 10% fetal bovine serum (Lonza) and penicillin-streptomycin (ThermoFisher Scientific). Combined knockdown of GRHL1, GRHL2 and GRHL3 was performed using a mix of the ON-TARGETplus GRHL1-3 siRNA SMARTpools (Dharmacon) at a final concentration of 50nM for each pool in opti-MEM medium (ThermoFisher Scientific), we used qPCR to confirm the knockdown. Omni-ATAC was performed after 48 hours according the previously-published protocol100.
ATAC-seq reads were mapped to the reference genome (hg19-Gencode v18) using Bowtie273 2.2.6 (with the --sensitive-local parameter). Reads with mapping quality lower 4 were filtered out. Differential peaks were called using MACS277 algorithm (q < 0.01, --nomodel), with the NTC sample as treatment and GRH_kd sample as control.
The 1000 most differential peaks (lost in the GRHL knockdown) were used as input set for i-cisTarget42, a motif enrichment tool that found GRHL motifs as top enriched. We used the Fisher’s Exact Test for Count Data88 in R to calculate for the regions that lost accessibility the overall enrichment between the GRHL target regions (4017) and all other accessible regions (13555) in MCF7 cells. f
Statistics and Reproducibility
caQTLs FDR: see GLM and Chromatin Accessibility QTLs.
The enrichment for caQTLs for each of the 18k motifs was calculated using the Fisher’s exact test (one sided) and pvals were corrected using Benjamini-Hochberg. The top scoring motif (Grh) had a Padj=6.75*10-22 having 34 out of 19804 negatives versus 45 out of 2048 positives.
All comparisons between accessible regions (normalized ATAC-seq reads) were done using the two-sided Welch Two Sample t-test in R. Fig 2a: Grh regions n =3246, Other accessible regions n = 27528, t = 29.057, df = 3477, true difference in means is not equal to 0, p-value = 2.250596e-166. Fig. 6c: n = 3227, t = 5.5326, df = 6390.7, true difference in means is not equal to 0, p-value = 3.281e-08. Fig. 6e: n=3227, t = -6.3823, df = 6428.9, true difference in means is not equal to 0, p-value = 1.866e-10.
Fig. 8d Two-sided Fisher’s Exact Test (MCF7: 13555 accessible regions without GRHL motif of which 265 go down, 4017 with GRHL motif of which 285 go down) pval=2.11*10-47.
Data availability and Accession Code Availability Statements
UCSC Genome Browser hub with ATAC-seq tracks for all DGRP lines, eye/wing injected lines ATAC-seq and ChIPmentation against Grh-GFP in eye-antennal and wing discs:
http://ucsctracks.aertslab.org/users/jjacobs/DGRP_injections/bigWig_norm/hub_norm.txt;
as well as a hub with all cross-species ATAC-seq: Evolution hub:
http://ucsctracks.aertslab.org/papers/evolution/hub.txt. To load these in the UCSC Genome Browser, go to My Data – Track Hubs.
The raw and processed data is available from Gene Expression Omnibus under accession number GSE102441.
Supplementary Material
Acknowledgements
We would like to thank F. Casares for the helpful discussions. We would like to thank L. Vanden Broeck (Laboratory of Behavioural and Developmental Genetics, KULeuven) for sharing the DGRP lines with us. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. Computing was performed on the Flemish Supercomputer Center (VSC). We would like to thank S. Bray (Department of Physiology, Development and Neuroscience, Cambridge) for sharing the UAS::grhN line and for the insightful discussions. We would like to thank M. Harrison (Department of Biomolecular Chemistry, UW School of Medicine and Public Health) for sharing and aliquot of the Grh C-terminal antibody.
Funding: FWO project grants to S.A. (G.0640.13, G.0791.14, and G.0C04.17), to G.Ha. (G.0954.16N), Special Research Fund (BOF) KU Leuven grants to S.A. (PF/10/016 and OT/13/103), Foundation Against Cancer grants to S.A. (2012-F2 and 2016-070), an ERC CoG (724226_cis-CONTROL) to S.A.. J.J., K.D and H.I. have a personal PhD Fellowship from the Flemish Agency for Innovation by Science and Technology. J.W. is funded by a postdoctoral research fellowship from Kom op tegen Kanker (Stand up to Cancer), the Flemish cancer society.
Footnotes
Author contributions
Conceived and designed the experiments: J.J. and S.A.
Performed all bulk ATAC-seq and generated transgenic flies: J.J. and V.C. Performed singe-cell and sorted ATAC-seq: K.D. and V.C. Performed ATAC-seq on the Drosophila Species: D.P. and V.C. Performed Grh-ChIPmentation: V.C. Performed imaginal disc dissections, stainings and imaging: L.R., M.A., V.C. and J.J.
Analysed the data: J.J., with assistance from G.Hu. on evolution part and BLS, I.I.T and G.P. on Random Forest, C.B.G.B and K.D. on single-cell analysis, S.Ai. on DNA-footprinting.
J.W. designed and performed the human GRHL experiments, H.I. analysed the Human GRHL data.
Wrote the paper: J.J. and S.A., gave insightful feedback M.A. and G.Ha.
Competing financial interests Statement
The authors have no competing interests as defined by Nature Research, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Bibliography
- 1.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 3.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 5.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5384. pdb.prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Tollefsbol TO. DNA methylation detection: Bisulfite genomic sequencing analysis. Methods Mol Biol Clifton NJ. 2011;791:11–21. doi: 10.1007/978-1-61779-316-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 10.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 11.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kvon EZ, et al. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512:91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- 14.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- 16.Reiter F, Wienerroither S, Stark A. Combinatorial function of transcription factors and cofactors. Curr Opin Genet Dev. 2017;43:73–81. doi: 10.1016/j.gde.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 18.Soufi A, et al. Pioneer Transcription Factors Target Partial DNA Motifs on Nucleosomes to Initiate Reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barozzi I, et al. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell. 2014;54:844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Younger ST, Rinn JL. p53 regulates enhancer accessibility and activity in response to DNA damage. Nucleic Acids Res. doi: 10.1093/nar/gkx577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Cui W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J Stem Cells. 2014;6:305–311. doi: 10.4252/wjsc.v6.i3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verfaillie A, et al. Multiplex enhancer-reporter assays uncover unsophisticated TP53 enhancer logic. Genome Res. 2016 doi: 10.1101/gr.204149.116. gr.204149.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang H-L, et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foo SM, et al. Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr Biol CB. 2014;24:1341–1346. doi: 10.1016/j.cub.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackay TFC, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W, et al. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014;24:1193–1208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Rahman R, Guo F, Rosbash M. Genome-wide identification of neuronal activity-regulated genes in Drosophila. eLife. 2016;5:e19942. doi: 10.7554/eLife.19942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkatesan K, McManus HR, Mello CC, Smith TF, Hansen U. Functional conservation between members of an ancient duplicated transcription factor family, LSF/Grainyhead. Nucleic Acids Res. 2003;31:4304–4316. doi: 10.1093/nar/gkg644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paré A, Kim M, Juarez MT, Brody S, McGinnis W. The Functions of Grainy Head-Like Proteins in Animals and Fungi and the Evolution of Apical Extracellular Barriers. PLOS ONE. 2012;7:e36254. doi: 10.1371/journal.pone.0036254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narasimha M, Uv A, Krejci A, Brown NH, Bray SJ. Grainy head promotes expression of septate junction proteins and influences epithelial morphogenesis. J Cell Sci. 2008;121:747–752. doi: 10.1242/jcs.019422. [DOI] [PubMed] [Google Scholar]
- 35.Nevil M, Bondra ER, Schulz KN, Kaplan T, Harrison MM. Stable Binding of the Conserved Transcription Factor Grainy Head to its Target Genes Throughout Drosophila melanogaster Development. Genetics. 2017;205:605–620. doi: 10.1534/genetics.116.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varma S, et al. The Transcription Factors Grainyhead-like 2 and NK2-Homeobox 1 Form a Regulatory Loop That Coordinates Lung Epithelial Cell Morphogenesis and Differentiation. J Biol Chem. 2012;287:37282–37295. doi: 10.1074/jbc.M112.408401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyne R, et al. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8:R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods. 2015;12:963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spokony R, White K. Spokony insertions. 2012 [Google Scholar]
- 40.Potier D, et al. Mapping gene regulatory networks in Drosophila eye development by large-scale transcriptome perturbations and motif inference. Cell Rep. 2014;9:2290–2303. doi: 10.1016/j.celrep.2014.11.038. [DOI] [PubMed] [Google Scholar]
- 41.Herrmann C, Van de Sande B, Potier D, Aerts S. i-cisTarget: an integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Res. 2012;40:e114. doi: 10.1093/nar/gks543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imrichová H, Hulselmans G, Kalender Atak Z, Potier D, Aerts S. i-cisTarget 2015 update: generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res. 2015;43:W57–W64. doi: 10.1093/nar/gkv395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mace KA, Pearson JC, McGinnis W. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science. 2005;308:381–385. doi: 10.1126/science.1107573. [DOI] [PubMed] [Google Scholar]
- 44.Wang S, et al. The tyrosine kinase Stitcher activates Grainy head and epidermal wound healing in Drosophila. Nat Cell Biol. 2009;11:890–895. doi: 10.1038/ncb1898. [DOI] [PubMed] [Google Scholar]
- 45.Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aerts S, et al. Robust Target Gene Discovery through Transcriptome Perturbations and Genome-Wide Enhancer Predictions in Drosophila Uncovers a Regulatory Basis for Sensory Specification. PLOS Biol. 2010;8:e1000435. doi: 10.1371/journal.pbio.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, et al. Transcription Factors Bind Thousands of Active and Inactive Regions in the Drosophila Blastoderm. PLOS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostrin EJ, et al. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nüsslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle inDrosophila melanogaster. Wilhelm Rouxs Arch Dev Biol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- 51.Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 52.Svetlichnyy D, Imrichova H, Fiers M, Atak ZK, Aerts S. Identification of High-Impact cis-Regulatory Mutations Using Transcription Factor Specific Random Forest Models. PLOS Comput Biol. 2015;11:e1004590. doi: 10.1371/journal.pcbi.1004590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Propeller-Twisting of Base-pairs and the Conformational Mobility of Dinucleotide Steps in DNA. [Accessed: 26th December 2017]; doi: 10.1006/jmbi.1996.0304. ScienceDirect. Available at: http://www.sciencedirect.com.kuleuven.ezproxy.kuleuven.be/science/article/pii/S0022283696903046?via%3Dihub. [DOI] [PubMed] [Google Scholar]
- 54.Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20:267. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cirillo LA, Zaret KS. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell. 1999;4:961–969. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- 57.Wilanowski T, et al. A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech Dev. 2002;114:37–50. doi: 10.1016/s0925-4773(02)00046-1. [DOI] [PubMed] [Google Scholar]
- 58.Gao X, et al. Evidence for multiple roles for grainyhead-like 2 in the establishment and maintenance of human mucociliary airway epithelium.[corrected] Proc Natl Acad Sci U S A. 2013;110:9356–9361. doi: 10.1073/pnas.1307589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung VY, et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci Rep. 2016;6:19943. doi: 10.1038/srep19943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frisch SM, Farris JC, Pifer PM. Roles of Grainyhead-like transcription factors in cancer. Oncogene. 2017;36:6067. doi: 10.1038/onc.2017.178. [DOI] [PubMed] [Google Scholar]
- 61.Ming Q, et al. Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family. Nucleic Acids Res. 2018 doi: 10.1093/nar/gkx1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumasaka N, Knights AJ, Gaffney DJ. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat Genet. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goltsev Y, Hsiong W, Lanzaro G, Levine M. Different combinations of gap repressors for common stripes in Anopheles and Drosophila embryos. Dev Biol. 2004;275:435–446. doi: 10.1016/j.ydbio.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 66.Varma S, et al. Grainyhead-like 2 (GRHL2) distribution reveals novel pathophysiological differences between human idiopathic pulmonary fibrosis and mouse models of pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2014;306:L405–419. doi: 10.1152/ajplung.00143.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carpinelli MR, de Vries ME, Jane SM, Dworkin S. Grainyhead-like Transcription Factors in Craniofacial Development. J Dent Res. 2017;96:1200–1209. doi: 10.1177/0022034517719264. [DOI] [PubMed] [Google Scholar]
- 68.Harrison MM, Botchan MR, Cline TW. Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev Biol. 2010;345:248–255. doi: 10.1016/j.ydbio.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ye T, et al. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39:e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davie K, et al. Discovery of transcription factors and regulatory regions driving in vivo tumor development by ATAC-seq and FAIRE-seq open chromatin profiling. PLoS Genet. 2015;11:e1004994. doi: 10.1371/journal.pgen.1004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Single-cell chromatin accessibility reveals principles of regulatory variation. [Accessed: 18th July 2017]; doi: 10.1038/nature14590. PubMed - NCBI. Available at: https://www.ncbi.nlm.nih.gov/pubmed/26083756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gramates LS, et al. FlyBase at 25: looking to the future. Nucleic Acids Res. 2017;45:D663–D671. doi: 10.1093/nar/gkw1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H. seqtk: Toolkit for processing sequences in FASTA/Q formats. 2017 [Google Scholar]
- 75.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinforma Oxf Engl. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma Oxf Engl. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 79.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas-Chollier M, et al. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res. 2012;40:e31–e31. doi: 10.1093/nar/gkr1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Picard Tools - By Broad Institute. [Accessed: 24th July 2017]; Available at: https://broadinstitute.github.io/picard/.
- 83.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: Inferring transcription factor variation from single-cell epigenomic data. bioRxiv. 2017 doi: 10.1101/110346. 110346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wei T, et al. corrplot: Visualization of a Correlation Matrix. 2017 [Google Scholar]
- 85.RepeatMasker Home Page. [Accessed: 20th July 2017]; Available at: http://www.repeatmasker.org/.
- 86.bedtools: a powerful toolset for genome arithmetic — bedtools 2.26.0 documentation. [Accessed: 17th July 2017]; Available at: http://bedtools.readthedocs.io/en/latest/.
- 87.R: Fitting Generalized Linear Models. [Accessed: 20th July 2017]; Available at: https://stat.ethz.ch/R-manual/R-devel/library/stats/html/glm.html.
- 88.R: Fisher’s Exact Test for Count Data. [Accessed: 15th July 2017]; Available at: https://stat.ethz.ch/R-manual/R-devel/library/stats/html/fisher.test.html.
- 89.Weirauch MT, et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat Biotechnol. 2013;31 doi: 10.1038/nbt.2486. nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robin X, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frith MC, Li MC, Weng Z. Cluster-Buster: finding dense clusters of motifs in DNA sequences. Nucleic Acids Res. 2003;31:3666–3668. doi: 10.1093/nar/gkg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 93.RSAT Matrix-Clustering: Dynamic Exploration and Redundancy Reduction of Transcription Factor Binding Motif Collections. [Accessed: 12th July 2017]; doi: 10.1093/nar/gkx314. PubMed Journals Available at: https://ncbi.nlm.nih.gov/labs/articles/28591841/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frith MC, Hansen U, Weng Z. Detection of cis -element clusters in higher eukaryotic DNA. Bioinformatics. 2001;17:878–889. doi: 10.1093/bioinformatics/17.10.878. [DOI] [PubMed] [Google Scholar]
- 95.Chiu T-P, et al. GBshape: a genome browser database for DNA shape annotations. Nucleic Acids Res. 2015;43:D103–109. doi: 10.1093/nar/gku977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.EMBOSS: shuffleseq. [Accessed: 20th July 2017]; Available at: http://structure.usc.edu/emboss/shuffleseq.html.
- 97.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Bergeijk P, Heimiller J, Uyetake L, Su TT. Genome-Wide Expression Analysis Identifies a Modulator of Ionizing Radiation-Induced p53-Independent Apoptosis in Drosophila melanogaster. PLOS ONE. 2012;7:e36539. doi: 10.1371/journal.pone.0036539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Corces MR, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UCSC Genome Browser hub with ATAC-seq tracks for all DGRP lines, eye/wing injected lines ATAC-seq and ChIPmentation against Grh-GFP in eye-antennal and wing discs:
http://ucsctracks.aertslab.org/users/jjacobs/DGRP_injections/bigWig_norm/hub_norm.txt;
as well as a hub with all cross-species ATAC-seq: Evolution hub:
http://ucsctracks.aertslab.org/papers/evolution/hub.txt. To load these in the UCSC Genome Browser, go to My Data – Track Hubs.
The raw and processed data is available from Gene Expression Omnibus under accession number GSE102441.