Abstract
New anti-tuberculosis (anti-TB) drugs are urgently needed to battle drug-resistant Mycobacterium tuberculosis strains and to shorten the current 6–12-month treatment regimen. In this work, we have continued the efforts to develop chalcone-based anti-TB compounds by using an in silico design and QSAR-driven approach. Initially, we developed SAR rules and binary QSAR models using literature data for targeted design of new chalcone-like compounds with anti-TB activity. Using these models, we prioritized 33 compounds for synthesis and biological evaluation. As a result, 10 chalcones-like compounds (4, 8, 9, 11, 13, 17–20, and 23) were found to exhibit nanomolar activity against replicating micobacteria, low micromolar activity against nonreplicating bacteria, and nanomolar and micromolar against rifampin (RMP) and isoniazid (INH) monoresistant strains (rRMP and rINH) (<1 µM and <10 µM, respectively). The series also show low activity against commensal bacteria and generally show good selectivity toward M. tuberculosis, with very low cytotoxicity against Vero cells (SI = 11–545). Our results suggest that our designed chalcone-like compounds, due to their high potency and selectivity, are promising anti-TB agents.
Keywords: Tuberculosis, in silico design, QSAR, nitroaromatic compounds, chalcone, anti-TB agents
INTRODUCTION
Tuberculosis (TB) is a chronic infectious disease caused predominantly by Mycobacterium tuberculosis (M. tb). Tuberculosis is reported in every country around the globe and the World Health Organization (WHO) estimates that about a third of the world’s population is infected with M. tb. [1–3]. According to the WHO, in 2014 there were registered almost 10 million of new TB cases and 1.5 million deaths; 400,000 of which were HIV-positive. As a frequent co-infection, TB is aggravated by the spread of HIV and is a major cause of death among HIV/AIDS patients [3–5].
Drug-sensitive TB can be cured by a combination of isoniazid (INH), rifampin (RMP), pyrazinamide (PZA), and ethambutol (EMB) taken under supervision for 4 months, and 2 months of treatment with only two drugs RMP and INH, consisting the basis of the DOTS program (Directly Observed Therapy Short-course). The emergence of multidrug-resistance (MDR-TB) and extensively drug-resistant (XDR-TB) has created substantial new challenges for TB treatment [6,7]. The treatment of resistant strains requires a prolongation of the therapy with drugs that are more toxic, less effective, and more costly [8]. Over the past 16 years, significant investment by academia, funding agencies, and initiatives such as WHO Stop TB Partnership [9] and The Global Alliance for TB Drug Development [10], has led to a renaissance of research in the field of TB and led to the discovery of bedaquiline and delamanid, two new anti-TB drugs approved in 2012 and 2013 respectively for treatment of adults with MDR-TB [11,12].
The development of computer science has found broad application in the drug discovery area [13]. Computer-aided drug design (CADD) has become an integral part of the drug discovery process in both academia and pharma companies [13,14]. Elucidation of quantitative structure-activity relationships (QSAR) is one of the main approaches of CADD [15–18]. QSAR modeling has been widely used for identification of novel anti-TB agents. In many studies, QSAR was used to design new anti-TB agents [2,19–32]. However, in the majority of the cases, QSAR has been used to modify previously discovered congeneric series of chemicals (Table S1, supplementary data).
Chalcones or 1,3-diaryl-2-propen-1-ones represent one class of natural products and essential intermediates in the biosynthesis of flavonoids. Chalcones are low molecular weight compounds possessing a broad spectrum of biological activities [33–46] including antibacterial [47,48] and anti-TB [38,49,50] activities.
The goal of this work was the design, synthesis and discovery of new chalcone and chalcone-like derivatives with potent anti-TB activity. To achieve this goal, we performed the following steps: (i) collection of available data and rigorous data curation; (ii) generation of structure-activity relationships (SAR) using matched molecular pair analysis (MMP) to design new chalcones with potential anti-TB activity by bioisosteric replacement; (iii) development of rigorously validated binary QSAR models; (iv) perform virtual screening of designed compounds; (v), organic synthesis and structure identification (NMR, MS, and IR) of selected VS hits; and (vi) in vitro experimental evaluation of designed hits under normoxic (MABA) and hypoxic (LORA) conditions.
RESULTS AND DISCUSSION
Design of chalcone and chalcone-like compounds
For the initial design of new chalcone derivatives with anti-TB activity, we retrieved 604 chalcones compounds with inhibition data against the M. tb H37Rv strain from PubChem Bioassay [51], ChEMBL [52], SciFinder database [53], and from literature. After collecting and integrating all the data, chemical structures and activity values were rigorously curated following the protocols established by Fourches et al [54–56]. Briefly, structural normalization of specific chemotypes, such as aromatic and nitro groups, was performed using ChemAxon Standardizer (v. 15.10.12.0, ChemAxon, Budapest, Hungary, http://www.chemaxon.com). Inorganic salts, organometallic compounds, and mixtures were also removed. After structural standardization, the duplicates were identified using ISIDA Duplicates[57] and HiT QSAR[58]. Analysis of duplicates also allowed to estimate inter- and intra-lab variability. No suspicious data sources were found. The curated dataset consisted of 571 chalcones, which were the subject for SAR analysis using matched molecular pairs (MMP, Figure 1) of analysis [59] that reveal changes in properties measured between structures with high similarity in this case, evaluated by MACCS keys descriptor [60] and Tanimoto coefficient (>0.7) [61] e.g. lost or gain of activity results of specific changes on structure by comparison between two structures [59,62].
Figure 1.
MMP analysis of molecular pairs of chalcones and chalcone-like compounds with anti-TB activity reported in the literature.
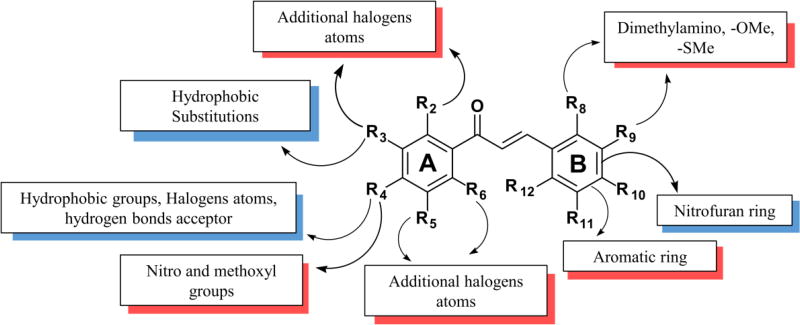
This analysis revealed the following SAR rules (Figure 2): (i) hydrophobic and hydrogen bond acceptor groups, e.g., halogens, phenyl, and heterocyclic amines, in p-position of ring A are favorable to anti-TB activity; (ii) substitution of benzene ring B by nitrofuran, increases the activity; (iii) any substituent in any position of ring B decreases the activity; and (iv) halogen in ortho- or meta-position of the ring A decreases the activity. These rules were used to design new compounds using the bioisosteric replacement using BROOD v.2.0 software [63] and SwissBioisosteres server [64].
Figure 2.
Derived SAR rules for chalcones with anti-TB activity. Modifications in blue shading increase the activity; with red – decrease the activity.
QSAR-DRIVEN design
QSAR modeling
MACCS [65], AtomPairs [66,67], Morgan [67,68], FeatMorgan [69], and Avalon fingerprints [70] combined with support vector machine (SVM) [71], gradient boosting machine (GBM) [72], and random forest (RF) [73] machine learning methods were used for the development of 15 different binary QSAR models. These models were united in a consensus ensemble model (Table 1). The dataset was balanced prior to the modeling to keep the ratio of active to inactive compounds as 1:1. The results of 5-fold external cross-validation demonstrated high predictive power of the developed consensus model (Table 1). Ten rounds of Y-randomization were performed (CCR≈0.5, see Table S2, supplementary Data) and indicated that developed models were not obtained due to chance correlations.
Table 1.
Statistical characteristics of developed QSAR models estimated by 5-fold external CV.
| Models | CCR | Kappa | Se | Sp | Coverage |
|---|---|---|---|---|---|
| MACCS-GBM | 0.73 | 0.46 | 0.76 | 0.70 | 0.71 |
| AtomPairs-GBM | 0.71 | 0.41 | 0.71 | 0.70 | 0.71 |
| Morgan-GBM | 0.76 | 0.51 | 0.77 | 0.74 | 0.68 |
| FeatMorgan-GBM | 0.74 | 0.47 | 0.76 | 0.71 | 0.66 |
| Avalon-GBM | 0.74 | 0.47 | 0.79 | 0.68 | 0.77 |
| MACCS-RF | 0.75 | 0.51 | 0.79 | 0.72 | 0.71 |
| AtomPairs-RF | 0.75 | 0.50 | 0.73 | 0.77 | 0.71 |
| Morgan-RF | 0.76 | 0.52 | 0.79 | 0.73 | 0.68 |
| FeatMorgan-RF | 0.75 | 0.50 | 0.70 | 0.80 | 0.66 |
| Avalon-RF | 0.74 | 0.49 | 0.76 | 0.73 | 0.77 |
| MACCS-SVM | 0.77 | 0.53 | 0.77 | 0.76 | 0.71 |
| AtomPairs-SVM | 0.74 | 0.48 | 0.74 | 0.74 | 0.71 |
| Morgan-SVM | 0.76 | 0.53 | 0.80 | 0.73 | 0.68 |
| FeatMorgan-SVM | 0.76 | 0.51 | 0.75 | 0.76 | 0.66 |
| Avalon-SVM | 0.73 | 0.46 | 0.72 | 0.74 | 0.77 |
| Consensus* | 0.77 | 0.53 | 0.79 | 0.74 | 1.00 |
GBM: Gradient Boosting Machine; SVM: Support Vector Machine; RF: Random Forest; CCR: correct classification rate; Kappa: Cohen’s kappa coefficient; Se: sensitivity; Sp: specificity.
Consensus model was developed by averaging the predictions of all 15 single models.
Then, the developed consensus model was used for virtual screening of the chalcones designed by bioisosteric replacement aiming at prioritize the compounds for synthesis. The chalcones obtained by bioisosteric replacement (Table S3) are drug-like compounds and satisfy Veber [74] and Lipinski [75] rules. In addition, the designed compounds contained no PAINs substructures [76,77].
Chemistry
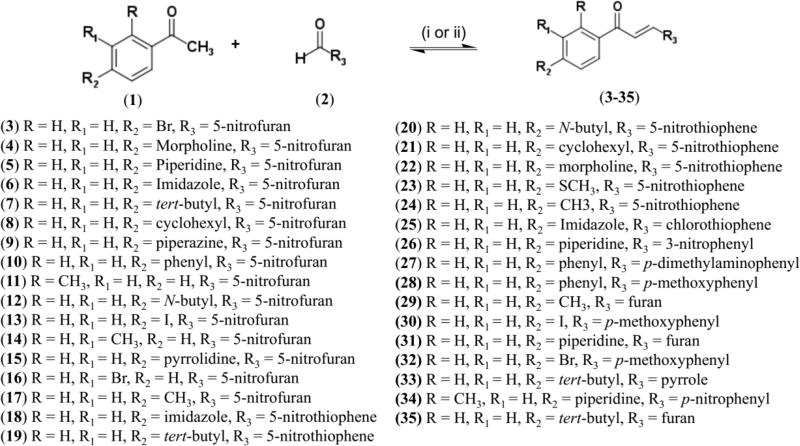
Based on the results of the in silico design, we synthesized the selected nitrofuran- 3–17, nitrothiophene- 18–24 and chlorothiophene 25 containing chalcones (Scheme 1). The standard Claisen-Schmidt condensation [78] under basic condition could not be used because the starting materials (aldehydes, nitrofurans, nitrothiophenes, and chlorothiophenes) are alkali-sensitive. Thus, the modified Claisen-Schmidt condensation was performed using acetic acid as solvent and sulfuric acid as catalyst [79,80]. Compounds 26–35 were synthesized following standard Claisen-Schmidt condensation using 20% NaOH as catalyst [78] (see Experimental Section of the Supplementary Data for details of spectra and purity data).
Scheme 1.
Synthesis of chalcones and chalcone-like derivatives.
Reagents and conditions: (i) H2SO4 conc., AcOH, reflux, 100 °C, 4 – 24 h; (1) acetophenones, (2) nitrofuraldehyde or nitrothiophenecarboxaldehyde, (3–25) analogs nitrofurans or nitrotiophenes. (ii) 20% NaOH, EtOH, room temperature, 10 h; (1) acetophenones; (2) aromatics aldehydes; (26–35) phenyl analogs, furan or pyrrole.
Among designed and synthesized compounds, 17 compounds are new and were not published previously (6–9, 11, 14, 15, 17–23, 25, 31, and 33), and thirty compounds (6–35) were not tested against tuberculosis before.
Antituberculosis activity
The compounds were submitted to biological assays against M. tb H37Rv, under both aerobic (replicating) and anaerobic (non-replicating) conditions using MABA and LORA assays, respectively [81,82]. Minimum inhibitory concentrations (MICs) were defined as the lowest compound concentration effecting ≥90% inhibition of fluorescence or luminescence, respectively. We evaluated 33 chalcones including three known compounds (Table 2) [79]. Twenty-two compounds had low MICs in both the MABA and LORA assays. Compounds containing substituents in the para-position of ring A, and containing nitrofurans and nitrothiophens as ring B (Figure 2) were the most potent. Ortho- and meta-substituted compounds were somewhat less active. Ten designed compounds 4, 8, 9, 11, 13, 17–20, and 23 had MABA MICs of < 1 µM and LORA MICs of < 10 µM (Table 2). And four this compounds 9, 18, and 19 were more potent than (MIC = 0.27, 0.19, and 0.22 µM respectively) of standard drug INH (MIC = 0.41 µM) used on treatment of TB. Already the compound 23 exhibited MIC similar (0.45 µM) to INH.
Table 2.
In vitro antituberculosis activity reported in minimum inhibitory concentration (MIC, µM) (MABA and LORA), MABA MIC of selected compounds against isogenic monodrug-resistant M. tb., rRMP and rINH, spectrum of activity and selectivity index of designed chalcones.
| Code | Compounds | Minimum Inhibitory Concentration (µM) | SI | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||
|
MABA | LORA | rRMP | rINH | C. albicans | E. coli | S. aureus | M. smegmatis | |||||
| R1 | R2 | R3 | R4 | ||||||||||
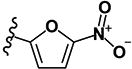
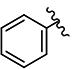
| 3 | H | H | Br |
|
2.50 | 6.76 | 0.76 | 0.58 | 4.93 | >10 | 0.36 | 7.12 | ND |
| 4 | H | H |
|
|
0.81 | 9.85 | 0.55 | 0.11 | >10 | >10 | 1.19 | >10 | 123 |
| 5 | H | H |
|
|
3.42 | 10.91 | 0.07 | <0.03 | >10 | >10 | >10 | >10 | ND |
| 6 | H | H |
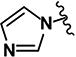
|
|
1.05 | 6.94 | 1.19 | 1.44 | >10 | 3.18 | 0.34 | >10 | 94 |
| 7 | H | H | -C(CH3)3 |
|
0.35 | >10 | 0.29 | 0.22 | >10 | >10 | >10 | >10 | 284 |
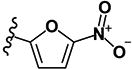
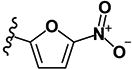
| 8 | H | H |
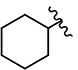
|
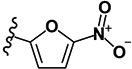
|
0.81 | 3.33 | 1.12 | 1.23 | >10 | >10 | >10 | >10 | 122 |
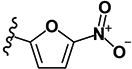
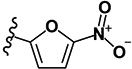
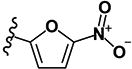
| 9 | H | H |
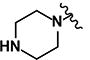
|
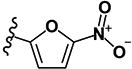
|
0.27 | 9.67 | 0.76 | 0.19 | >10 | >10 | >10 | >10 | 359 |
| 10 | H | H |
|
|
3.91 | 3.18 | 3.45 | 0.29 | >10 | >10 | >10 | >10 | 25 |
| 11 | -CH3 | H | H |
|
0.57 | 4.59 | 1.08 | 1.07 | 6.36 | >10 | 1.09 | 9.50 | 41 |
| 12 | H | H | -(CH2)3CH3 |
|
1.31 | 4.21 | 4.70 | 2.23 | >10 | >10 | 2.23 | >10 | 57 |
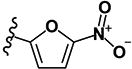
| 13 | H | H | -I |
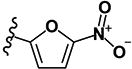
|
0.60 | 4.12 | 1.18 | 1.08 | >10 | >10 | >10 | 4.62 | 166 |
| 14 | H | -CH3 | H |
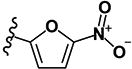
|
1.27 | 4.75 | 3.78 | 1.93 | >10 | >10 | 2.23 | >10 | 43 |
| 15 | H | H |
|
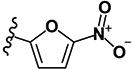
|
>10 | 4.30 | - | - | >10 | >10 | >10 | >10 | ND |
| 16 | H | -Br | H |
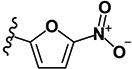
|
1.32 | 5.65 | 1.88 | 1.12 | >10 | >10 | 1.68 | 4.84 | 52 |
| 17 | H | H | -SCH3 |
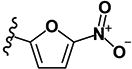
|
0.66 | 5.07 | 0.24 | 0.80 | >10 | >10 | 0.58 | 4.81 | 61 |
| 18 | H | H |
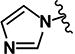
|
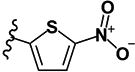
|
0.19 | 1.73 | 0.60 | 0.30 | >10 | >10 | 0.28 | 4.39 | 225 |
| 19 | H | H | -C(CH3)3 |
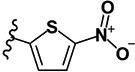
|
0.22 | 4.18 | 0.14 | 0.15 | >10 | >10 | >10 | >10 | 454 |
| 20 | H | H | -(CH2)3CH3 |
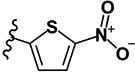
|
0.54 | 5.56 | 0.48 | 0.31 | >10 | >10 | >10 | >10 | 81 |
| 21 | H | H |
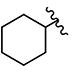
|
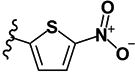
|
>10 | 5.75 | - | - | >10 | >10 | >10 | >10 | ND |
| 22 | H | H |
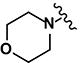
|
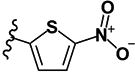
|
>10 | >10 | - | - | >10 | >10 | >10 | >10 | ND |
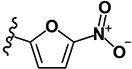
| 23 | H | H | -SCH3 |
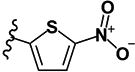
|
0.45 | 5.96 | 0.14 | 0.22 | >10 | >10 | >10 | >10 | 222 |
| 24 | H | H | -CH3 |
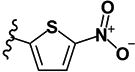
|
1.87 | 2.05 | 1.21 | 0.96 | >10 | >10 | >10 | >10 | ND |
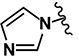
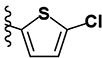
| 25 | H | H |
|
|
9.29 | 9.75 | 8.89 | 9.03 | >10 | >10 | >10 | >10 | ND |
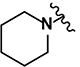
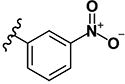
| 26 | H | H |
|
|
>10 | >10 | - | - | >10 | >10 | >10 | >10 | ND |
| 27 | H | H |
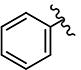
|
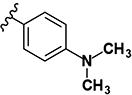
|
>10 | >10 | - | - | >10 | >10 | >10 | >10 | ND |
| 28 | H | H |
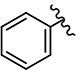
|
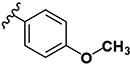
|
>10 | >10 | - | - | >10 | >10 | 1.18 | >10 | ND |
| 29 | H | H | -SCH3 |
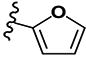
|
>10 | >10 | - | - | >10 | >10 | >10 | >10 | ND |
| 30 | H | -I | H |
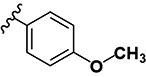
|
4.90 | >10 | 8.31 | 7.75 | >10 | >10 | >10 | >10 | 20 |
| 31 | H | H |
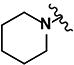
|
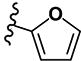
|
>10 | >10 | - | - | >10 | >10 | >10 | >10 | ND |
| 32 | H | -Br | H |
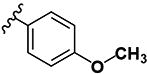
|
8.54 | 8.20 | 9.28 | 4.94 | >10 | >10 | >10 | >10 | 11 |
| 33 | H | H | -C(CH3)3 |
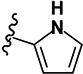
|
>10 | >10 | - | - | >10 | >10 | >10 | >10 | ND |
| 34 | H | H |
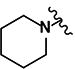
|
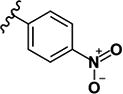
|
>10 | >10 | - | - | >10 | >10 | >10 | >10 | ND |
| 35 | H | H | -C(CH3)3 |
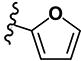
|
>10 | >10 | - | - | >10 | >10 | >10 | >10 | ND |
| RMP | 0.05 | 0.19 | >1 | - | - | - | - | >10 | 2000 | ||||
| INH | 0.41 | >256 | - | >5 | - | - | - | >10 | |||||
| Amph. B | - | - | - | - | <0.004 | - | - | - | |||||
| Amp | - | - | - | - | - | 1.18 | 0.25 | - | |||||
MABA: Microplate Alamar Blue Assay; LORA: Low Oxygen Recovery Assay; rRMP: monoresistant to rifampin; rINH: monoresistant to isoniazid. SI: Selectivity Index (Vero Cell IC50/MABA MIC). RMP: rifampin, INH: isoniazid, Amph. B: amphotericin B, Amp.: ampicillin
The most potent compound was the nitrothiophene analogue 18 with MABA MIC = 0.19 µM and LORA MIC = 1.73 µM. The substitution of furan ring by thiophene or nitro-substituted thiophene (e.g., 6 and 18) led to 5.5-fold increase of the activity in the MABA (1.05 µM to 0.19 µM) and 4-fold for LORA (6.94 µM to 1.73 µM). The compounds 19 and 20 were the most active in MABA, however 20 lost activity in the LORA in comparison to its nitrofuran analogue 12. Nitrothiophenes 21 and 22, unlike their nitrofuran analogues 8 and 4, were inactive (MIC>10 µM) in both MABA and LORA assays.
Cytotoxicity assay
To verify the possibility that the anti-TB activity of the designed compounds arises from general toxicity, Vero cells were used to estimate the in vitro cytotoxicity of the 18 most potent compounds in MABA and LORA assays. These compounds demonstrated modest to high selectivity on this assay, with selectivity indices (SI) ranging between 11 and 454 (Table 2).
Spectrum of activity
We also investigated selectivity of compounds with respect to activity against Candida albicans, Escherichia coli, Staphylococus aureus, and M. smegmatis (Table 2). Most of the tested compounds had MIC >10 µM, except 3, 4, 6, 11–14, 16–18, 20, and 28 that exhibited MICs against S. aureus of 0.28–2.23 µM.
Conversely, these compounds demonstrated broad-spectrum activity against non-tuberculosis mycobacterias (NTMs), i.e., M. abscessus, M. chelonae, M. marinum, M. avium, M. kansasii, and M. bovis (Table S3). Compounds 3, 8–13, 15–25, 30, and 32 had MICs <10 µM against M. avium, M. kansasii, and M. bovis, and compound 10 demonstrated MICs of 0.14 µM and 0.08 µM against M. kansasii and M. bovis, respectively.
Evaluation in M. tb. resistant strains
We evaluated the subset of most potent compounds (3–14, 16–21, 24 and 25) against rifampin- and isoniazid-resistant strains of M. tb. H37Rv (Table 2). All the compounds were potent against resistant strains (MIC < 10 µM), and compounds 3–5, 7, 9, and 17–21 exhibiting MIC < 1 µM. Compound 5 was the most potent compound with MIC of 0.07 µM against rRMP and < 0.03 µM against rINH strains. These results suggest that our designed compounds do not share the same mode of action as these two first line drugs, INH and RMP.
CONCLUSIONS
The integration of in silico design, QSAR-driven virtual screening, synthesis, and experimental evaluation in a single pipeline led to discovery of new and promising anti-TB compounds. After the compilation of the initial dataset and its rigorous curation, the specific SAR rules were developed and used for designing of new chalcones by bioisosteric replacement. For instance, hydrophobic groups and H-bond acceptors are preferred in the para-position of ring A combined with nitrofuran or nitrothiophene serving as ring B. Then, the developed consensus QSAR model of antimicrobial activity and applied it for virtual screening and prioritization of designed compounds. Thirty-three chalcone derivatives were synthesized, structures were confirmed by spectroscopic methods and tested against normoxic, replicating (MABA), and hypoxic, non-replicating (LORA) cultures of M. tb. We identified 20 compounds with MIC <10 µM in MABA including 10 compounds (4, 8, 9, 11, 13, 17–20, and 23) with MIC < 1 µM in MABA and < 10 µM in LORA. All tested compounds were active against M. tb strains mono-resistant to isoniazid or rifampicin. The compounds were not cytotoxic against mammalian (VERO) cells and appeared selective for mycobacteria with moderate activity against S. aureus. Our compounds satisfy the criteria for new anti-TB hits published by Katsuno and coauthors[83] and due to their high potency and activity against resistant strains, they can be considered as perspective anti-TB agents.
EXPERIMENTAL SECTION
Computational Design
Dataset
We retrieved 604 chalcone and chalcones-like compounds with experimental data tested against M. tb H37Rv from the PubChem (AID: 1626 and AID: 1949) [51], ChEMBL [52], SciFinder [53], and from the literature. Compounds that had inconclusive IC50 values were considered unreliable and were not included in the modeling.
Data curation
The compiled dataset of 604 compounds was carefully curated following the protocols proposed by Fourches et al.[54–56] Briefly, explicit hydrogens were added, whereas specifics chemotypes such as aromatic and nitro groups were normalized using ChemAxon Standardizer (v.15.1.26.0, ChemAxon, Budapest, Hungary, http://www.chemaxon.com). Polymers, inorganic salts, organometallic compounds, mixtures, and duplicates were removed. Modeling-ready curated dataset contained 571 compounds.
SAR analysis
SAR analysis was performed using the MMP (Matched Molecular Pairs) approach [84], Structural similarity was calculated using Tanimoto coefficient obtained on MACCS keys.
SAR analysis and bioisosteric replacement
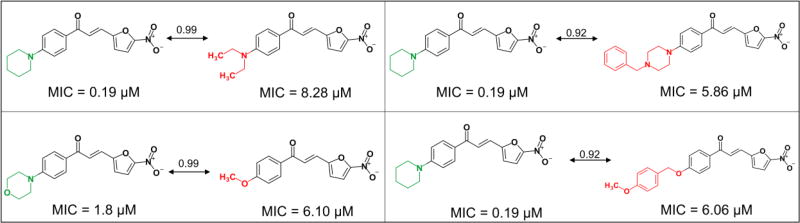
SAR analysis was performed using the MMP (Matched Molecular Pairs) approach [84]. Structural similarity was calculated using Tanimoto coefficient [61] obtained on MACCS keys. Bioisosteric replacement was performed in the p-substituents on the ring A (Figure 1), i.e., piperidin of the most active chalcone (MIC = 0.19 µM), (2E)-3-(5-nitrofuran-2-yl)-1-[4-(piperidin-1-yl)phenyl]prop-2-en-1-one, described in literature [79]. Design of these bioisosters were performed using BROOD v.2.0 software [63] and SwissBioisosteres webserver (http://www.swissbioisostere.ch) [64].
Molecular fingerprints
Five different types of fingerprints were used: molecular access system (MACCS) structural key fingerprints [65], AtomPair [66,67], Morgan, [67,68] FeatMorgan, [69] and Avalon.[70] All fingerprints were calculated using the open-source cheminformatics toolkit RDKit v.2.4.0 [85].
Dataset analysis and under-sampling
The curated dataset was unbalanced (148 active and 423 inactive compounds), which is not recommended to build binary QSAR models. Therefore, we decided to balance the dataset using linear under-sampling strategy developed by Braga, R.C. (Neves et al, 2016) [86]. Unlike the traditional under-sampling methods which randomly balance the dataset, this strategy retains the most representative inactive compounds in the balanced dataset, thus assuring as high as possible coverage of original chemical space. As a result, balanced dataset containing 148 active and 148 inactive compounds was used for the modeling.
Machine learning techniques
SVM[71], GBM [72], and Random Forest (RF) [73], approaches implemented in R v.3.0.3 [87] were used for the building and optimization of statistically acceptable QSAR models. All machine learning classifiers were implemented using the R v.3.0.3 [87]. More details about these machine modeling techniques are given in Supplementary Information.
External validation of developed QSAR models
5-fold external cross-validation is the standard approach for the estimation of predictive power of QSAR models [88]. In this procedure, the dataset is randomly divided in five subsets of equal size (20% of compounds each). One of these subsets serve as an external validation fold and the other four subsets are used building of the model. The same procedure is repeated five times to place each compound once in the corresponding external fold. Then, the predictivity of the models is estimated based on these external folds. Description of statistical characteristics used for estimation of robustness and external predictivity of developed models is provided in supplementary data.
Consensus modeling
The underlying idea of consensus predictions is that an implicit SAR for a given dataset can be formally manifested by a variety of QSAR models built with different types of molecular descriptors and diverse machine learning approaches. Rigorously built individual models form an ensemble that allows for consensus bioactivity prediction using all models at once. The development of consensus models is generally recommended because usually they result in better predictivity and better coverage of chemical space during virtual screening [89]. To obtain consensus prediction, we have averaged the predictions of all individual models.
Chemical synthesis
All the chemicals and solvents were purchased from Sigma Aldrich®. The progress of all reactions was monitored on Merck KGaA precoated silica gel plates 0.25 mm (with fluorescence indicator UV254) using ethyl acetate/n-hexane as solvent system. Spots were visualized by irradiation with ultraviolet light (254 nm). Melting points (mp) were determined using open capillary method on Melting Point III Marte® apparatus. Proton (1H) and (13C) NMR spectra were recorded on Bruker Avance 400 spectrometer at 400 MHz for 1H and 100 MHz for 13C using DMSO-d6 and CDCl3 as solvents referenced. Chemical shifts are given in parts per million (ppm) (δ relative to residual solvent peak for 1H and 13C). Spectra Mass was performed on a LCMS-2020 Liquid Chromatograph Mass Spectrometer Shimadzu, the column was Agilent XDB-C18, 35µM, 21×20 nm. IR spectra were recorded on a PerkinElmer model Spectrum 400 (medium, sweep of 4000 to 400 cm−1). Synthesized compounds were ≥96% pure as determined by high performance liquid chromatography (HPLC) Shimadzu with PDA detector, Nucleodur 100-5 CN-RP column 205×4.6mm, mobile phase water/0.1% TFA and acetonitrile with flow of 1 mL/min.
For the synthesis of 3–25, substituted acetophenones (0.5 equiv, 0.5 mmol) and nitroaromatics (0.5 equiv., 0.5 mmol) were dissolved in acetic acid (1 mL) and concentrated sulfuric acid (0.05 mL) and were stirred at 100° C until completion of the reaction (4–24 h). The cooled mixture and the solid was washed with iced methanol (200 mL) for purification. For the synthesis of 26–35, 0.4 mL of aqueous NaOH (20% w/v) was added to the solution of the acetophenones substituted in 4’ position (1 mmol) in EtOH. The resulting mixture was stirred at the room temperature for 10 hours. The formed precipitate was filtered and washed with cold water. If no precipitation occurred, the resulting mixture was neutralized with 5% HCl filtered and dried. The crude was then subjected to chromatography column with EtOAc/Hexane (7:3, v/v) as eluent.
(2E)-1-(4-bromophenyl)-3-(5-nitrofuran-2-yl)prop-2-en-1-one (LabMol-63) 3
Yellow solid; yield 33% (107 mg, 0.33 mmol); mp 182°C; HPLC purity 98.13%. 1H NMR (CDCl3): δ = 7.95 (d, 2H, J = 8.0 Hz), 7.70 (d, 2H J = 8.0 Hz), 7.72 (d, 1H, J = 15.0 Hz), 7.58 (d, 1H, J = 15.0 Hz), 7.39 (d, 1H, J = 4.0 Hz), 6.87 (d, 1H, J = 4.0 Hz). 13C NMR (CDCl3): δ = 187.1, 152.4, 135.4, 131.8 (2 C), 130.4, 129.7 (2 C), 128.5, 128.1, 123.9, 116.4, 112.7. IR (KBr): ν = 1663 (s; ν(C=O)), 1607 (s; ν(C=Cαβ)), 1475, 1301 (s; ν(Ar-NO2).
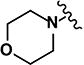
(2E)-1-[4-(morpholin-4-yl)phenyl]-3-(5-nitrofuran-2-yl)prop-2-en-1-one (LabMol-64) 4
Red solid, yield 13% (42 mg, 0.12 mmol); mp 86°C; HPLC purity 98.19%. 1H NMR ([D6] DMSO): δ = 8.02 (d, 2H, J = 8.0 Hz), 7.88 (d, 1H, J = 15.0 Hz), 7.80 (d, 1H, J = 3.6 Hz), 7.51 (d, 1H, J = 15.0 Hz), 7.42 (d, 1H, J = 3.6 Hz), 7.04 (d, 2H, J = 9.2 Hz), 3.73 (m, 4H), 3.41 (m, 4H). 13C NMR ([D6] DMSO): δ = 185.2, 154.1, 153.8, 130.8 (2 C), 127.1, 126.6, 125.6, 116.9, 114.9, 113.1 (2 C), 65.8 (2 C), 46.6 (2 C). IR (KBr): ν = 1660 (s; ν(C=O)), 1601 (s; ν(C=Cαβ)), 1515, 1355 (s; ν(Ar-NO2)), 1238 (s; ν(C-N)), 1119 (s; ν(C-O)). ESI (+)-MS (MeOH): m/z = 329 [M+H]+
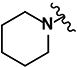
(2E)-3-(5-nitrofuran-2-yl)-1-[4-(piperidin-1-yl)phenyl]prop-2-en-1-one (LabMol-65) 5
Red solid, yield 17% (56 mg, 0.17 mmol); mp 220°C; HPLC purity 98.41%. 1H NMR ([D6] DMSO): δ = 7.97 (d, 2H, J = 9.2 Hz), 7.86 (d, 1H, J = 15.0 Hz), 7.80 (d, 1H, J = 4.0 Hz), 7.44 (d, 1H, J = 15.0 Hz), 7.40 (d, 1H, J = 4,0 Hz), 6.99 (d, 2H, J = 9.2 Hz), 4.43 (s, 4H), 1.60 (s, 6H). 13C NMR ([D6] DMSO): δ = 184.9, 154.1, 153.9, 131.0 (2 C), 126.7, 125.8, 125.3, 116.8, 115.0, 112.9 (2 C), 47.6 (2 C), 24.9 (2 C), 24.0. IR (KBr): ν = 1642 (s, ν(C=O)), 1607 (s; ν(C=Cαβ)), 1578, 1354 (s, ν(Ar-NO2)), 1235 (s; ν(C-N)). ESI (+)-MS (MeOH): m/z = 327 [M+H]+
(2E)-1-[4-(1H-imidazol-1-yl)phenyl]-3-(5-nitrofuran-2-yl)prop-2-en-1-one (LabMol-66) 6
Brown solid; yield 12% (36 mg, 0.12 mmol); mp 232°C; HPLC purity 99.08%. 1H NMR ([D6] DMSO): δ = 9.07 (s, 1H); 8.32 (d, 2H, J = 8.0 Hz), 8.15 (s, 1H), 7.97 (d, 2H, J = 8.0 Hz), 7.94 (d, 1H, J = 16.0 Hz), 7.83 (d, 1H, J = 4.0 Hz), 7.63 (d, 1H, J = 16.0 Hz), 7.52 (s, 1H), 7.48 (d, 1H, J = 4.0 Hz). 13C NMR ([D6] DMSO): δ = 187.2, 153.2, 152.1, 139.6, 135.9, 135.8, 130.7 (2 C), 129.0, 126.3, 121.0 (2 C), 119.2, 118.0, 114.9. IR (KBr): ν = 1662 (s; ν(C=O)), 1609 (s; ν(C=Cαβ)), 1566, 1352 (s, ν(Ar-NO2)). ESI (+)-MS (MeOH): m/z = 310 [M+H]+
(2E)-1-(4-tert-butylphenyl)-3-(5-nitrofuran-2-yl)prop-2-en-1-one (LabMol-68) 7
Yellow solid; yield 22% (67 mg, 0.22 mmol); mp 180°C; HPLC purity 99.89%. 1H NMR (CDCl3): δ = 1.37 (s, 9H), 6.84 (d, 1H, J = 4.0 Hz), 7.39 (d, 1H, J = 4.0 Hz), 7.55 (d, 1H, J = 15.0 Hz), 7.55 (d, 2H, J = 8.4Hz), 7.77 (d, 1H J = 15.0Hz), 8.01 (d, 2H, J = 8.4Hz). 13C NMR (CDCl3): δ = 188.3, 157.2, 152.8, 151.8, 134.2, 128.2 (2 C), 127.3, 125.4 (2 C), 124.8, 115.8, 112.8, 34.8, 30.6 (3 C). IR (KBr): ν = 1651 (s; ν(C=O)), 1596 (s; ν(C=Cαβ)), 1527, 1354 (s, ν(Ar-NO2)), 1391 (m, ν(CH3)). ESI (+)-MS (MeOH): m/z = 300 [M+H]+
(2E)-1-(4-cyclohexylphenyl)-3-(5-nitrofuran-2-yl)prop-en-2-one (LabMol-72) 8
Yellow solid; yield 44% (72 mg, 0.22 mmol); mp 162° C; HPLC purity 98.59%. 1H NMR (CDCl3): δ = 8.00 (d, 2H, J = 8.0 Hz), 7.77 (d, 1H, J = 15.6 Hz), 7.54 (d, 1H, J = 15.6 Hz), 7.38 (d, 1H, J = 3.6 Hz), 7.37 (d, 2H, J = 8.0 Hz), 6.84 (d, 1H, J = 3.6 Hz), 2.61 (s, 1H); 2.18 (s, 2H); 1.89 (s, 2H), 1.78 (s, 1H), 1.39 (m, 6H). 13C NMR (CDCl3): 187.7, 154.1 (2 C), 152.9, 134.6, 128.5 (2 C), 127.3, 127.0 (2 C), 124.8, 115.8, 112.8, 44.3, 33.6 (2 C), 26.3 (2 C), 25.6. IR (KBr): ν = 2925 (s; ν(C-H)) 1651 (s; ν(C=O)), 1593 (s; ν(C=Cαβ)), 1526, 1353 (s, ν(Ar-NO2)), 1481 (m, ν(CH2)). ESI (+)-MS (MeOH): m/z = 326 [M+H]+.
(2E)-3-(5-nitrofuran-2-yl)-1-[4-(piperazin-1-yl)phenyl]prop-2-en-1-one (LabMol-73) 9
Red solid; yield 42% (59 mg, 0.12 mmol); mp 221° C; HPLC purity 98.07%. 1H NMR (CDCl3): δ = 8.00 (d, 2H, J = 8.8 Hz), 7.79 (d, 1H, J = 15.2 Hz), 7.51 (d, 1H, J = 15.2 Hz), 7.38 (d, 1H, J = 4.0 Hz), 6.90 (d, 2H, J = 8.8 Hz), 6.78 (d, 1H J = 4.0 Hz), 3.44 (s, 4H), 1.69 (s, 5H). 13C NMR (CDCl3): δ 185.2, 154.2 (2 C), 153.5, 130.8 (2 C), 125.9, 125.7, 125.3, 115.1, 112.9, 112.7 (2 C), 47.9 (2 C), 24.9 (2 C), 23.9. IR (KB:r) ν = 1618 (s; ν(C=O)), 1609 (s; ν(C=Cαβ)), 1580, 1354 (s, ν(Ar-NO2)), 1513 (m, ν(N-H)), HR-MS (m/z) (ESI): calcd for C17H18N3O4 [M + H+]: 328.1291; found: 328.1289.
(2E)-3-(5-nitrofuran-2-yl)-1-(4-phenylphenyl)prop-2-en-1-one (LabMol-74) 10
Yellow solid; yield 62% (100 mg, 0.31 mmol); mp 200°C; HPLC purity 99.29%. 1H NMR (CDCl3): δ = 8.16 (d, 2H, J = 8.4 Hz), 7.83 (d, 1H, J = 15.5 Hz), 7.77 (d, 2H, J = 8.4 Hz), 7.67 (d, 2H, J = 7.2 Hz), 7.59 (d, J = 15.5 Hz, CH α, 1H), 7.50 (s, 2H); 7.44 (d, 1H J = 7.2 Hz), 7.40 (s, 1H), 6.86 (s, 1H). 13C NMR (CDCl3): δ = 187.6, 152.7 (2 C), 145.9, 139.2, 135.4, 128.0 (2 C), 128.6 (2 C), 128.0, 127.6, 127.1 (2 C), 126.9 (2 C), 124.6, 116.1, 112.8. IR (KBr): ν = 1660 (s; ν(C=O)), 1598 (s; ν(C=Cαβ)), 1597, 1352 (s, ν(Ar-NO2)), 1513, 1474 (s, ν(ArC=C)). ESI (+)-MS (MeOH): m/z = 320 [M+H]+.
(2E)-1-(2-methylphenyl)-3-(5-nitrofuran-2-yl)prop-2-en-1-ona (LabMol-75) 11
Yellow solid; yield 9% (12 mg, 0.04 mmol); mp 114° C; HPLC purity 99.20%. 1H NMR (CDCl3): δ = 7.61 (s, 1H), 7.44 (s, 1H), 7.41 (s, 1H), 7.35 (s, 1H), 7.37 (d, 1H, J = 3.6 Hz), 7.31 (s, 2H), 6.83 (d, 1H, J = 3.6 Hz), 2.50 (s, 3H). 13C NMR (CDCl3): δ = 193.0, 152.5, 137.6, 137.3, 131.4, 131.1, 128.7, 128.2, 127.9, 125.3, 115.7, 112.7 (2 C), 20.2. IR (KBr): ν = 1663 (s; ν(C=O)), 1608 (s; ν(C=Cαβ)), 1483, 1348 (s, ν(Ar-NO2)), 1476 (s, ν(CH3)). ESI (+)-MS (MeOH): m/z = 258 [M+H]+, HR-MS (m/z) (ESI): calcd for C14H12NO4 [M + H+]: 258.0760; found: 258.0770.
(2E)-1-(4-butylphenyl)-3-(5-nitrofuran-2-yl)prop-2-en-1-one (LabMol-77) 12
Yellow solid, yield 10% (16 mg, 0.05 mmol); mp 100°C; HPLC purity 97.93%. 1H NMR (MHz CDCl3): δ = 8.00 (d, 2H, J = 8.4 Hz), 7.78 (d, 1H, J = 15.6 Hz), 7.55 (d, 1H, J = 15.6 Hz), 7.39 (d, 1H, J = 4.0 Hz), 7.35 (d, 2H, J = 8.4 Hz), 6.84 (d, 1H, J = 4.0 Hz), 2.71 (t, 2H, J = 8.0 Hz), 1.65 (q, 2H, J = 8.0 Hz), 1.39 (s, 2H J = 8.0 Hz), 0.95 (t, 3H, J = 8.0 Hz). 13C NMR (CDCl3): δ = 188.5, 161.5, 145.1, 140.8, 139.9, 136.9, 130.0 (2 C), 129.8, 127.1, 126.9, 118.6, 114.1 (2 C), 94.0, 55.0. IR (KBr): ν = 2934 (s; ν(C-H), 1652 (s; ν(C=O)), 1607 (s; ν(C=Cαβ)), 1594, 1351 (s; ν(Ar-NO2)), 1481 (s; ν(CH3)), 810 (s; ν(CH2)). ESI (+)-MS (MeOH): m/z = 300 [M+H]+, HR-MS (m/z) (ESI): calcd for C17H18NO4 [M + H+]: 300.1230; found: 300.1235.
(2E)-1-(4-iodophenyl)-3-(5-nitrofuran-2-yl)prop-2-en-1-one (LabMol-78) 13
Brown solid; yield 33% (61 mg, 0.16 mmol); mp 194°C; HPLC purity 99.49%. 1H NMR (CDCl3): δ = 7.91 (d, 2H, J = 8.4 Hz), 7.78 (d, 2H, J = 8.4 Hz), 7.70 (d, 1H, J = 15.6 Hz), 7.57 (d, 1H, J = 15.6 Hz), 7.39 (d, 1H, J = 4.0 Hz), 6.87 (d, 1H, J = 4.0 Hz). 13C NMR (CDCl3): δ = 187.4, 152.4 (2 C), 137.8 (2 C), 136.0, 129.5 (2 C), 128.1, 123.9, 116.4, 112.7, 101.4. IR (KBr): ν = 1658 (s; ν(C=O)), 1606 (s; ν(C=Cαβ)), 1579, 1351 (s; ν(Ar-NO2)).
(2E)-1-(3-methylphenyl)-3-(5-nitrofuran-2-yl)prop-2-en-1-one (LabMol-79) 14
Yellow solid; yield 17% (22 mg, 0.08 mmol); mp 141° C; HPLC purity 97.92%. 1H NMR (CDCl3): δ = 7.86 (s, 2H); 7.76 (d, 1H, J = 15.6 Hz), 7.55 (d, 1H, J = 15.6 Hz), 7.45 (s, 2H), 7.39 (d, 1H, J = 4.0 Hz), 6.85 (d, 1H, J = 4,0 Hz), 2,48 (s, 3H). 13C NMR (CDCl3): δ = 188.3, 152.8, 138.4, 138.8, 134.0, 128.7, 128.3, 127.5, 125.5, 124.8, 116.0, 112.8 (2 C), 20.2. IR (KBr): ν = 3131 (s; ν(C-H)) 1661 (s; ν(C=O)), 1606 (s; ν(C=Cαβ)), 1579, 1349 (s; ν(Ar-NO2)). ESI (+)-MS (MeOH): m/z = 258 [M+H]+, HR-MS (m/z) (ESI): calcd for C14H12NO4 [M + H+]: 258.0760; found: 258.0764.
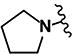
(2E)-3-(5-nitrofuran-2-yl)-1-[4-(pyrrolidin-1-yl)phenyl]prop-2-en-1-one (LabMol-81) 15
Red solid; yield 36% (57 mg, 0.18 mmol); mp 242° C; HPLC purity 96.87%. 1H NMR (CDCl3): δ = 8.03 (d, 2H, J = 8.8 Hz), 7.82 (d, 1H, J = 15.6 Hz), 7.52 (d, 1H, J = 15.6 Hz), 7.38 (d, 1H, J = 4.0 Hz), 6.77 (d, 1H, J = 3.6 Hz), 6.59 (d, 2H, J = 8.8 Hz), 3.42 (s, 4H); 2.07 (s, 4H). 13C NMR (CDCl3): δ = 185.0, 153.6, 151.1, 130.9 (2 C), 125.6, 125.5, 124.3, 114.9, 113.0, 110.7 (2 C), 42.2 (2 C), 24.0 (2 C). IR (KBr): ν = 2854 (s; ν(C-H)), 1643 (s; ν(C=O)), 1610 (s; ν(C=Cαβ)), 1578, 1354 (s; ν(Ar-NO2)) 1199 (s; ν(C-N)). ESI (+)-MS (MeOH): m/z = 313 [M+H]+
(2E)-1-(3-bromophenyl)-3-(5-nitrofuran-2-yl)prop-2-en-1-one (LabMol-82) 16
Brown solid, yield 32% (62 mg, 0.19 mmol); mp 158°C; HPLC purity 99.98%. 1H NMR (CDCl3): δ = 8.18 (s, 1H); 7.98 (d, 1H, J = 7.6 Hz), 7.76 (d, 1H, J = 8.0 Hz), 7.69 (d, 1H, J = 15.6 Hz), 7.57 (d, 1H, J = 15.6 Hz), 7.44 (t, 1H, J1 = 7.6Hz, J2 = 8.0 Hz), 7.39 (d, 1H, J = 4.0 Hz), 6.88 (d, 1H, J = 4.0 Hz). 13C NMR (CDCl3): δ = 186.9, 152.3, 151.9, 138.5, 136.0, 131.2, 130.0, 128.4, 126.7, 123.9, 122.8. 116.5, 112.7. IR (KBr): ν = 1664 (s; ν(C=O)), 1607 (s; ν(C=Cαβ)), 1566, 1354 (s; ν(Ar-NO2)). ESI (+)-MS (MeOH): m/z = 321 [M+H]+; HR-MS (m/z) (ESI): calcd for C13H9BrNO4 [M + H+]: 321.9709; found: 321.9701
(2E)-1-[4-(methylsulfanyl)phenyl]-3-(5-nitrofuran-2-yl)prop-2-en-1-one (LabMol-92) 17
Brown solid; yield 24% (35 mg, 0.12 mmol); mp 160° C; HPLC purity 99.15%. 1H NMR (CDCl3): δ = 8.00 (d, 2H, J = 8.0 Hz), 7.76 (d, 1H, J = 16.0 Hz), 7.56 (d, 1H, J = 16.0 Hz), 7.39 (d, 1H, J = 4.0 Hz), 7.34 (d, 2H, J = 8.0 Hz), 6.84 (d, 1H, J = 4.0 Hz), 2.56 (s, 3H). 13C NMR (CDCl3): δ = 186.6, 152.8 (2 C), 146.6, 132.9, 128.7 (2 C), 127.4, 124.7 (2 C), 124.5, 115.9, 112.8, 14.3. IR (KBr): ν = 3117 (m, ν(C-H)), 1658 (s; ν(C=O)), 1607 (s; ν(C=Cαβ)), 1589, 1354 (s; ν(Ar-NO2)), 1394 (s; ν(CH3)). ESI (+)-MS (MeOH): m/z = 290 [M+H]+, HR-MS (m/z) (ESI): calcd for C14H12NO4 [M + H+]: 258.0760; found: 258.0770
(2E)-1-[4-(1H-imidazol-1-yl)phenyl]-3-(5-nitrothiofen-2-yl)prop-2-en-1-one (LabMol-84) 18
Green solid; yield 55% (180 mg, 0.55 mmol); mp 223° C; HPLC purity 98.99%. 1H NMR ([D6] DMSO): δ = 9.80 (s, 1H), 8.42 (d, 3H, J = 8.0 Hz), 8.17 (d, 1H, J = 4.0 Hz), 8.05 (d, 1H, J = 16.0 Hz), 8.05 (d, 2H, J = 8.0 Hz), 7.95 (d, 1H, J = 8.0 Hz), 7.94 (d, 1H, J = 16.0 Hz), 7.84 (d, 1H, J = 4.0 Hz). 13C NMR ([D6] DMSO): δ = 187.2, 151.9, 146.4, 138.5, 137.1, 135.5, 135.2, 131.5, 130.6 (2 C), 130.6, 125.1, 122.0 (2 C), 121.7, 120.6. IR (KBr): ν = 1663 (s; ν(C=O)), 1605 (s; ν(C=Cαβ)), 1595, 1339 (s; ν(Ar-NO2)), 1284 (m; ν(C-NAr)). ESI (+)-MS (MeOH): m/z = 326 [M+H]+. HR-MS (m/z) (ESI): calcd for C16H12N3O3S [M+H+]: 326.0593; found: 326.0607.
(2E)-1-(4-tert-butylphenyl)-3-(5-nitrothiophen-2-yl)prop-2-en-1-one (LabMol-86) 19
Green solid; yield 63% (99 mg, 0.31 mmol); mp 192° C; HPLC purity 99.55%. 1H NMR (CDCl3): δ = 7.96 (d, 2H, J = 8.4 Hz), 7.88 (d, 1H, J = 4.4 Hz), 7.80 (d, 1H, J = 15.6 Hz), 7.55 (d, 2H, J = 8.4 Hz), 7.52 (d, 1H, J = 15.6 Hz), 7.27 (d, 1H, J = 4.4 Hz), 1.37 (s, 9H). 13C NMR (CDCl3): δ = 187.8, 157.1, 151.6, 146.1, 134.2, 134.09, 129.1, 128.7, 128.1 (2 C), 125.4 (2 C), 124.6, 34.8, 30.6 (3 C). IR (KBr): ν = 2962 (s; ν(C-H)), 1657 (s; ν(C=O)), 1691 (s; ν(C=Cαβ)), 1586, 1334 (s; ν(Ar-NO2)), 1366 (m; ν(CH3)). ESI (+)-MS (MeOH): m/z = 316 [M+H]+, HR-MS (m/z) (ESI): calcd for C17H18NO3S [M + H+]: 316.1001; found: 316.1006.
(2E)-1-(4-butylphenyl)-3-(5-nitrothiophen-2-yl)prop-2-en-1-one (LabMol-87) 20
Green solid; yield 38% (61 mg, 0.19 mmol); mp 145° C; HPLC purity 99.76%. 1H NMR (CDCl3): δ = 7.95 (d, 2H, J = 8.0 Hz), 7.89 (d, 1H, J = 4.4 Hz), 7.80 (d, 1H, J = 15.6 Hz), 7.51 (d, 1H, J = 15.6 Hz), 7.34 (d, 2H, J = 8.0 Hz), 7.27 (d, 1H, J = 4.0 Hz), 2.71 (t, 2H, J = 8.0 Hz), 1.65 (q, 2H, J = 8.0 Hz), 1.38 (s, 2H, J = 8.0 Hz), 0.95 (t, 3H, J = 8.0 Hz) 3H). 13C NMR (CDCl3): δ = 187.7, 151.5, 149.1, 146.1, 134.5, 134.0, 129.1, 128.7, 128.5 (2 C), 128.3 (2 C), 124.6, 35.3, 32.8, 21.9, 13.4. IR (KBr): ν = 2928 (s; ν(C-H)), 1657 (s; ν(C=O)), 1598 (s; ν(C=Cαβ)), 1593, 1330 (s; ν(Ar-NO2)), 1366 (m; ν(CH3)), 816 (m; ν(CH2)). ESI (+)-MS (MeOH): m/z = 316 [M+H]+, HR-MS (m/z) (ESI): calcd for C17H18NO3S [M + H+]: 316.1001; found: 316.1005
(2E)-1-(4-cyclohexylphenyl)-3-(5-nitrothiofen-2-yl)prop-2-en-1-one (LabMol-88) 21
Green solid; yield 64% (110 mg, 0.32 mmol); mp 182° C; HPLC purity 99.79%. 1H NMR (CDCl3): δ = 7.95 (d, 2H, J = 8.0 Hz), 7.88 (d, 1H, J = 4.0 Hz), 7.79 (d, 1H, J = 15.2 Hz), 7.52 (d, 1H, J = 15.2 Hz), 7.37 (d, 2H, J = 8.0 Hz), 7.27 (d, 1H, J = 4.0 Hz), 2.61 (s, 1H), 1.89 (s, 4H), 1.79 (s, 1H), 1.47 (s, 4H), 1.30 (s, 1H). 13C NMR (CDCl3): δ = 187.7, 154.0, 151.5, 146.1, 134.6, 134.0, 129.1, 128.7, 128.4 (2 C), 126.9 (2 C), 124.7, 44.3, 33.6 (2 C), 26.3 (2 C), 25.6. IR (KBr): ν = 2926 (s; ν(C-H)), 1656 (s; ν(C=O)), 1606 (s; ν(C=Cαβ)), 1589, 1334 (s; ν(Ar-NO2)), 1427 (m; ν(CH2)).
(2E)-1-[4-morpholin-4-yl)phenyl]-3-(nitrothiofen-2-yl)prop-2-en-1-one (LabMol-89) 22
Yellow solid; yield 9% (17 mg, 0.04 mmol); mp 240° C; HPLC purity 98.62%. 1H NMR (CDCl3): δ = 7.99 (d, 2H, J = 9.2 Hz), 7.89 (d, 1H, J = 4.0 Hz), 7.79 (d, 1H, J = 15.2 Hz), 7.53 (d, 1H, J = 15.6 Hz), 7.25 (d, 1H, J = 4.4 Hz), 6.93 (d, 2H, J = 9.2 Hz), 3.88 (t, 4H, J = 4.0 Hz), 3.38 (t, 4H, J = 4.0 Hz). 13C NMR (CDCl3): δ = 185.6, 154.1, 146.6 (2 C), 133.1, 130.4 (2 C), 128.7, 127.3, 124.8, 112.9 (2 C), 66.1 (2 C), 46.8 (2 C). IR (KBr): ν = 1648 (s; ν(C=O)), 1604 (s; ν(C=Cαβ)), 1584, 1335 (s; ν(Ar-NO2)), 1428 (m; ν(CH2)), 1119 (w; ν(C-O)). ESI (+)-MS (MeOH): m/z = 345 [M+H]+.
(2E)-1-[4-(methylsulfanyl)phenyl]-3-(5-nitrotiophen-2-yl)prop-2-en-1-one (LabMol-93) 23
Green solid; yield 61% (189 mg, 0.61 mmol); mp 204° C; HPLC purity 99.21%. 1H NMR (CDCl3): δ = 7.95 (d, 2H, J = 8.0 Hz), 7.89 (d, 1H, J = 4.0 Hz), 7.81 (d, 1H, J = 16.0 Hz), 7.50 (d, 1H, J = 16.0 Hz), 7.34 (d, 2H, J = 8.0 Hz), 7.29 (d, 1H, J = 4.0 Hz), 2.57 (s, 3H). 13C NMR (CDCl3): δ = 186.9, 146.6, 146.0, 134.2, 133.0, 129.2, 128.7, 128.5 (2 C), 124.7 (2 C), 124.3, 14.3. IR (KBr): ν = 1654 (s; ν(C=O)), 1604 (s; ν(C=Cαβ)), 1589, 1331 (s; ν(Ar-NO2)), 1428 (m; ν(CH3)).
(2E)-1-(4-methylphenyl)-3-(5-nitrothiophen-2-yl)prop-2-en-1-one (LabMol-95) 24
Green solid; yield 68% (186 mg, 0.68 mmol); mp 194° C; HPLC purity 99.4%. 1H NMR (CDCl3): 7.94 (d, 2H, J = 8.0 Hz), 7.90 (d, 1H, J = 4.0 Hz), 7.81 (d, 1H, J = 16.0 Hz), 7.53 (d, 1H, J = 16.0 Hz), 7.35 (d, 2H, J = 8.0 Hz), 7.28 (d, 1H, J = 4.0 Hz), 2.47 (s, 3H). 13C NMR (CDCl3): 188.1, 146.4, 144.6, 134.7, 134.5, 129.6 (2C), 129.5, 129.1, 128.7 (2C), 125.0, 21.7. IR (KBr): ν = 3077 (m; ν(CH3)), 1659 (s; ν(C=O)), 1609 (s; ν(C=Cαβ)), 1594, 1336 (s; ν(Ar-NO2)). ESI (+)-MS (MeOH): m/z = 274 [M+H]+.
(2E)-3-(5-chlorothiophen-2-yl)-1-[4-(1H-imidazol-1-yl)phenyl]prop-2-en-1-one (LabMol-94) 25
Green solid; yield 17% (54 mg, 0.17 mmol); mp 178° C; HPLC purity 99.2%. 1H NMR (CDCl3): δ = 8.13 (d, 2H, J = 8.0 Hz), 7.98 (s, 1H), 7.84 (d, 1H, J = 16.0 Hz), 7.54 (d, 2H, J = 8.0 Hz), 7.38 (s, 1H), 7.24 (d, 2H, J = 16.0 Hz), 7.19 (s, 1H), 7.17 (d, 1H, J = 4.0 Hz), 6.94 (d, 1H, J = 4.0 Hz). 13C NMR (CDCl3): δ = 188.6, 140.5, 138.8, 137.1, 136.5, 135.3, 134.0, 132.1, 131.1, 130.3 (2C), 127.7, 120.8 (2C), 119.9, 117.7. IR (KBr): ν = 1645 (s; ν(C=O)), 1608 (s; ν(C=Cαβ)), 810 (s; ν(C-Cl)). ESI (+)-MS (MeOH): m/z = 315 [M+H]+.
(2E)-3-(3-nitrophenyl)-1-[4-(piperidin-1-yl)phenyl]prop-2-en-1-one (LabMol-67) 26
Yellow solid, yield 84% (84 mg, 0.25 mmol); mp 181°C; HPLC purity 99.97%. 1H NMR (CDCl3): 8.51 (s, 1H), 8.23 (d, 2H, J = 8.0 Hz), 8.01 (d, 2H, J = 8.0 Hz), 7.91 (d, 1H, J = 8.0 Hz), 7.80 (d, 1H, J = 16.0 Hz), 7.69 (d, 1H, J = 16.0 Hz), 7.60 (t, 1H, J = 8.0 Hz), 6.91 (d, 2H, J = 8.0 Hz), 3.43 (s, 4H), 1.69 (s, 6H). 13C NMR (CDCl3): 186.5, 154.1, 148.3, 139.1, 136.8, 133.8, 130.6 (2 C), 129.5, 126.1, 124.4, 123.7, 121.6, 112.6 (2 C), 48.0 (2 C), 25.0 (2 C), 24.2. IR (KBr): ν = 2933 (m, ν(C-H)), 1651 (s; ν(C=O)), 1610 (s; ν(C=Cαβ)), 1588, 1349 (s; ν(Ar-NO2)), 1227 (s; ν(C-N)). ESI (+)-MS (MeOH): m/z = 337 [M+H]+
(2E)-3-[4-(dimethylamino)phenyl]-1-(4-phenylphenyl)prop-2-en-1-one (LabMol-69) 27
Yellow solid; yield 53% (143 mg, 0.43 mmol); mp 164°C; HPLC purity 99.36%. 1H NMR (CDCl3): 8.07 (d, 2H, J = 4.0 Hz), 7.81 (s, 1H), 7.69 (d, 2H, J = 4.0 Hz), 7.63 (d, 2H, J = 4.0 Hz), 7.56 (d, 2H, J = 8.0 Hz), 7.45 (d, 2H, J = 8.0 Hz), 7.38 (d, 2H, J = 4.0 Hz), 6.68 (d, 2H, J = 4.0 Hz), 3.02 (s, 6H). 13C NMR (CDCl3): 190.3, 152.3, 146.0, 145.1, 140.4, 138.0, 130.7 (2 C), 129.1 (3 C), 128.3 (2 C), 127.5 (2 C), 127.3 (2 C), 122.9, 112.1 (2 C), 40.3 (2 C). IR (KBr): ν = 1647 (s; ν(C=O)), 1603 (s; ν(C=Cαβ)), 1228 (s; ν(C-N)) ESI (+)-MS (MeOH): m/z = 328 [M+H]+
(2E)-3-(4-methoxyphenyl)-1-(4-phenylphenyl)prop-2-en-1-one (LabMol-70) 28
Yellow solid; yield 25% (79 mg, 0.25 mmol); mp 152°C; HPLC purity 100.00%. 1H NMR (CDCl3): 7.81 (d, 1H, J = 15.0Hz), 8.09 (d, 2H, J = 10 Hz), 7.71 (d, 2H, J = 5.0 Hz), 7,64 (d, 2H, J = 10 Hz), 7.61 (d, 2H, J = 10 Hz); 7,45 (d, 1H, J = 15.0 Hz), 6.93 (d, 2H, J = 5.0 Hz). 3.84 (s, 3H). 13C NMR (CDCl3): 190.2, 161.9, 145.5, 144.8, 140.2, 137.4, 130.5 (2 C), 129.3 (3 C), 128.4 (2 C), 127.9, 127.5 (2 C), 127.4 (2 C), 120.0, 114.7 (2 C), 55.6. IR (KBr): ν = 1647 (s; ν(C=O)), 1597 (s; ν(C=Cαβ)), 1303, 1037 (s; ν(C-O)). ESI (+)-MS (MeOH): m/z = 315 [M+H]+
(2E)-3-(furan-2-yl)-1-[4-(methylsulfanyl)phenyl]prop-2-en-1-one (LabMol-71) 29
Yellow solid; yield 19% (49 mg, 0.20 mmol); mp 114°C; HPLC purity 99.05%. 1H NMR (CDCl3): 7.95 (d, 2H, J = 6.8 Hz), 7.58 (d, 1H, J = 12.4 Hz), 7.51 (s, 1H), 7.43 (d, 1H, J = 12.4 Hz), 7.29 (d, 2H, J = 6.8 Hz), 6.70 (d, 1H, J = 2.4 Hz), 6.50 (q, 1H, J = 2.4 Hz), 2.52 (s, 3H). 13C NMR (CDCl3): 188.6, 151.9, 145.7, 145.0, 134.6, 130.5, 129.0 (2 C), 125.2 (2 C), 119.2, 116.2, 112.8, 14.9. IR (KBr): ν = 1656 (s; ν(C=O)), 1596 (s; ν(C=Cαβ)), 1549, 1476 (ArC=C), 1336 (s; ν(CH3)), 1297, 1094 (s; ν(C-O)). ESI (+)-MS (MeOH): m/z = 245 [M+H]+.
(2E)-1-(3-iodophenyl)-3-(4-methoxyphenyl)prop-2-en-1-one (LabMol-76) 30
White solid, yield 6% (22 mg, 0.06 mmol); mp 110°C; HPLC purity 99.64%. 1H NMR (CDCl3): δ = 8.32 (s, 1H), 7.96 (d, 1H, J = 8.0 Hz), 7.89 (d, 1H, J = 8.0 Hz), 7.79 (d, 1H, J = 15.6 Hz), 7.61 (d, 2H, J = 8.4 Hz), 7.33 (d, 1H, J = 15.6 Hz), 7.24 (t, 1H, J = 8.0 Hz), 6.95 (d, 2H, J = 8.0 Hz), 3.86 (s, 3H). 13C NMR (CDCl3): δ = 188.5, 161.5, 145.1, 140.8, 139.9, 136.9, 130.0 (2 C), 129.8, 127.1, 126.9, 118.6, 114.1 (2 C), 94.0, 55.0. IR (KBr): ν = 1657 (s; ν(C=O)), 1600 (s; ν(C=Cαβ)), 1559, 1510 (s, ν((ArC=C)), 1323 (s; ν(CH3)). ESI (+)-MS (MeOH): m/z = 365 [M+H]+.
(2E)-3-(furan-2-yl)-1-[4-(piperidin-1-yl)fenil]prop-2-en-1-one (LabMol-80) 31
Yellow solid; yield 30% (86 mg, 0.30 mmol); mp 182° C; HPLC purity 99.32%. 1H NMR (CDCl3): δ = 8.00 (d, 2H, J = 8.8 Hz), 7.58 (d, 1H, J = 15.2 Hz), 7.54 (m, 1H), 7.50 (d, 1H, J = 15.2 Hz), 6.90 (d, 2H, J = 8,8 Hz), 6.67 (d, 1H, J = 3.2 Hz), 6.50 (dd, 1H, J = 3.2 Hz), 3.40 (s, 4H), 1.68 (s, 6H). 13C NMR (CDCl3): δ = 186.8, 153.9, 151.7, 143.9, 130.3 (2 C), 128.6, 126.6, 119.2, 114.6, 112.9 (2 C), 112.0, 48.1 (2 C), 24.9 (2 C), 23.9. IR (KBr): ν = 2941 (m; ν(C-H), 1648 (s; ν(C=O)), 1604 (s; ν(C=Cαβ)), 1597, 1559 (s, ν(ArC=C)), 1390 (s; ν(C-N)). ESI (+)-MS (MeOH): m/z = 282 [M+H]+.
(2E)-1-(3-bromophenyl)-3-(4-methoxyphenyl)prop-2-en-1-one (LabMol-83) 32
Brown solid; yield 41% (130 mg, 0.41 mmol); mp 90°C; HPLC purity 99.66%. 1H NMR (CDCl3): δ = 8.13 (s, 1H), 7.93 (d, 1H, J = 8.0 Hz), 7.80 (d, 1H, J = 15.6 Hz), 7.70 (d, 1H, J = 8.0 Hz), 7.62 (d, 2H, J = 8.0 Hz), 7.39 (d, 1H, J = 8.0 Hz), 7.35 (d, 1H, J = 15.6 Hz), 6.95 (d, 2H J = 8.0 Hz), 3.87 (s, 3H). 13C NMR (CDCl3): δ = 188.6, 161.5, 145.1, 139.9, 134.9, 131.0, 130.0 (2 C), 129.7, 126.9, 126.5, 122.5, 118.6, 114.1 (2 C), 55.0. IR (KBr): ν = 1662 (s; ν(C=O)), 1594 (s; ν(C=Cαβ)), 1570, 1513 (s, ν(ArC=C)), 1260, 1042 (s; ν(C-O)), 556 (m, ν(C-Br)).
(2E)-1-(4-tert-butylphenyl)-3-(1H-pyrrol-2-yl)prop-2-en-1-one (LabMol-85) 33
White solid; yield 7% (20 mg, 0.07 mmol); mp 158° C; HPLC purity 99.81%. 1H NMR (CDCl3): δ = 9.07 (s, 1H), 7.95 (d, 2H, J = 8.4 Hz), 7.77 (d, 1H, J = 15.6 Hz), 7.50 (d, 2H, J = 8.4 Hz), 7.19 (d, 1H, J = 15.6 Hz), 7.00 (s, 1H), 6.72 (s, 1H), 6.34 (s, 1H), 1.36 (s, 9H). 13C NMR (CDCl3): δ = 189.6, 155.7, 135.5, 133.9, 128.9, 127.8 (2 C), 125.1 (2 C), 122.6, 115.4, 114.6, 111.0, 34.6, 30.7 (3 C). ESI (+)-MS (MeOH): m/z = 254 [M+H]+.
(2E)-3-(4-nitrophenyl)-1-[4-piperidin-1-yl)phenyl]prop-2-en-1-one (LabMol-90) 34
Yellow solid, yield 89% (300 mg, 0.89 mmol); mp 198°C; HPLC purity 99.50%. 1H NMR (CDCl3): δ = 8.25 (d, 2H, J = 8.0 Hz), 7.99 (d, 2H, J = 8.0 Hz), 7.78 (d, 1H, J = 16.0 Hz), 7.77 (d, 2H, J = 8.0 Hz), 7.68 (d, J = 16.0 Hz), 6.90 (d, 2H, J = 8.0 Hz), 3.41 (s, 4H), 1.69 (s, 6H). 13C NMR (CDCl3): δ = 186.2, 154.1, 147.8, 141.3, 139.0, 130.6 (2 C), 130.0, 128.2 (2 C), 126.0, 125.6, 123.7 (2 C), 112.7 (2 C), 48.0 (2 C), 24.9 (2 C), 23.9. IR (KBr): ν = 2942 (m, ν(C-H)), 1655 (s; ν(C=O)), 1609 (s; ν(C=Cαβ)), 1595, 1514 (s, ν(ArC=C)), 1593, 1336 (s, ν(Ar-NO2)), 1196 (s; ν(C-N)), 556 (m, ν(C-Br)). ESI (+)-MS (MeOH): m/z = 337 [M+H]+.
(2E)-1-(4-tert-butylphenyl)-3-(furan-2-yl)prop-2-en-1-one (LabMol-91) 35
Yellow solid; yield 7.8% (20 mg, 257 mmol); mp 84°C; HPLC purity 99.8%. 1H NMR (CDCl3): δ = 7.99 (d, 2H, J = 8.4 Hz), 7.60 (d, 1H, J = 15.2 Hz), 7.52 (d, 3H, J = 8.4 Hz), 7.48 (d, 1H, J = 15.2 Hz), 6.72 (s, 1H), 6.52 (s, 1H), 1.37 (s, 9H). 13C NMR (CDCl3): δ = 188.7, 156.1, 151.3, 144.3, 135.1, 129.9, 128.0 (2 C), 125.1 (2 C), 119.0, 115.5, 112.2, 34.7, 30.7 (3 C). IR (KBr): ν = 2962 (m, ν(C-H)), 1655 (s; ν(C=O)), 1605 (s; ν(C=Cαβ)), 1285 (s, ν(C-O)). ESI (+)-MS (MeOH): m/z = 255 [M+H]+.
Biological Evaluation
Anti-TB activity
MICs against M. tb H37Rv (ATCC 27294) as well as the rifampin (rRMP, ATCC 35838) and isoniazid (rINH, ATCC 35822) mono-resistant strains under normoxic, replicating conditions were determined using the Microplate Assay Blue Alamar (MABA) as previously described [90–92]. Briefly, cultures were incubated in 200 µL Middlebrook 7H12 medium together with test compound in 96-well plates for 7d and 37° C. Resazurin and Tween 80 were added and incubation continued for 24h at 37° C. Fluorescence was determined at excitation/emission wavelengths of 530/590 nm, respectively. The MIC was defined as the lowest concentration effecting a reduction in fluorescence of 90% relative to controls.61 MICs against M. tb. H37Rv under hypoxic, non-replicating conditions were determined using the Low Oxygen Recovery Assay as previously described [82,92] except that the luxABCDE reporter[94] was used instead of the luxAB reporter gene. The MIC was defined as the lowest concentration of compound which reduced luminescence by 90% after 10 days exposure to compound under hypoxic conditions followed by 28 hours of normoxic recovery and comparison to untreated controls.
Cytotoxicity in mammalian cells
Vero cells (ATCC CRL-1586) were cultured in 10% Fetal Bovine Serum (FBS) in Eagle minimum essential medium plus penicillin and streptomycin. Cells were prepared and washed in HBSS (1× pH = 7.4) and Trypsin-EDTA 0.25%, and then morphology was verified by microscopy. After adjusting the density to 3–5×105 cells/mL in MEM media, 100 µL of the cell suspension were incubated with test compounds at 37° C for 72 hours; visual inspection was performed each 24 hours. Then, 20 µL of 0.6 mM resazurin were added into each well and incubated for 4 hours. The fluorescence was determined by excitation/emission wavelengths of 530/590 nm. The concentration of test compound effecting a reduction in fluorescence of 50% relative to untreated cells was calculated as the IC50.
Spectrum of activity
Mycobacterium abscessus (ATCC 19977), M. chelonae (ATCC 35752), M. marinum (ATCC 927), M. avium (ATCC 15769), M. kansasii (ATCC 12478), and M. bovis (ATCC 35734) were cultured in Middlebrook 7H9 Broth with 0.2% (v/v) glycerol, 0.05% Tween 80, and 10% (v/v) albumin-dextrose-catalase (BBL™ OADC Enrichment, Cat. N°. 212352). M. smegmatis (ATCC MC2155) was cultured in 7H12 medium. Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 29213) were cultured in cation-adjusted Mueller Hinton (CAMH) broth and Candida albicans (ATCC 90028) in RPMI media until an absorbance at 570 nm of 0.2–0.5 was achieved. Cultures were diluted 1:5000 to 1:10,000 into fresh media in 96-well plates and incubated at 37° C with test compounds. Incubation times were 3 days for M. smegmatis, 3–7 days for other mycobacteria, 36–48 hours for C. albicans and 16–20 hours for S. aureus and E. coli. The MIC for C. albicans, S. aureus and E. coli was defined as the lowest concentration effecting a reduction of ≥90% in A570 relative to untreated cultures. The MABA MICs for mycobacteria are defined as described above.
Supplementary Material
Acknowledgments
Funding Sources
M.N.G. was supported by a sandwich fellowship from the Coordination for the Improvement of Higher Education Personnel (CAPES-PDSE) during his stay in ITR-UIC. M.N.G. is also supported by a Ph.D. fellowship from the State of Goiás Research Foundation (FAPEG). This work has been funded by the National Counsel of Technological and Scientific Development (CNPq), the State of Goiás Research Foundation (FAPEG). C.H.A. is CNPq productivity fellow. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors would like to thank Brazilian funding agencies, CNPq, CAPES and FAPEG for financial support and fellowships. We are grateful to OpenEye Scientific Software, Inc. and ChemAxon for providing academic license of their software.
ABBREVIATIONS
- TB
tuberculosis
- M. tb
Mycobacterium tuberculosis
- WHO
World Health Organization
- DOTS
Directly Observed Therapy Short-course
- RMP
rifampin
- INH
isoniazid
- PZA
pyrazinamide
- EMB
ethambutol
- DS-TB
drug sensitive TB
- MDR-TB
multidrug-resistance
- XDR-TB
extensively drug-resistance
- STOP-TB
STOP tuberculosis strategy
- CADD
computer assisted drug design
- QSAR
Quantitative Structure Activity Relationship
- MMPA
Matched Molecular Pairs of Analysis
- SAR
Structure Activity Relationship
- MACCS
Molecular ACCess System keys
- SVM
Support Vector Machine
- RF
Random Forest
- CCR
correct classification rate
- Se
sensitivity
- Sp
Specificity
- NMR
Nuclear Magnetic Resonance
- MS
Mass Spectrometry
- HPLC
High-Performance Liquid Chromatography
- MABA
microplate alamar blue assay
- LORA
low oxygen recovery assay
- MIC
minimum inhibitory concentration
- SI
selectivity index
- NTM
non-tuberculosis mycobacterias
- AD
applicability domain
- DMSO
dimethylsulfoxide
- ATCC
American Type Culture Collection
- CAMH
Mueller Hinton Media
- RPMI
Roswell Park Memorial Institute
- HBSS
Hank’s Balanced Salt Solution
- rRMP
resistant isogenic strain rifampin
- rINH
resistant isogenic isoniazid
Footnotes
ASSOCIATED CONTENT
Supporting Information. More computational details regarding molecular fingerprints calculation and QSAR model development are available in the Supporting Information, as well as additional tables of results and structural characterization for all synthetized compounds. This material is available free of charge via the Internet at http://ejmedch.
The authors declare no competing financial interest.
References
- 1.Palmer BD, Sutherland HS, Blaser A, Kmentova I, Franzblau SG, Wan B, Wang Y, Ma Z, Denny Wa, Thompson AM. Synthesis and Structure-Activity Relationships for Extended Side Chain Analogues of the Antitubercular Drug (6 S)-2-Nitro-6-{[4-(trifluoromethoxy)benzyl]oxy}-6,7-dihydro-5 H-imidazo[2,1-b][1,3]oxazine (PA-824) J. Med. Chem. 2015;58:3036–3059. doi: 10.1021/jm501608q. [DOI] [PubMed] [Google Scholar]
- 2.Martins F, Santos S, Ventura C, Elvas-Leitão R, Santos L, Vitorino S, Reis M, Miranda V, Correia HF, Aires-de-Sousa J, Kovalishyn V, Latino DaRS, Ramos J, Viveiros M. Design, synthesis and biological evaluation of novel isoniazid derivatives with potent antitubercular activity. Eur. J. Med. Chem. 2014;81:119–138. doi: 10.1016/j.ejmech.2014.04.077. [DOI] [PubMed] [Google Scholar]
- 3.WHO. [accessed October 28, 2015];Global Tuberculosis 2015. 2015 http://www.who.int/tb/publications/global_report/en/
- 4.Tang J, Yam W-C, Chen Z. Mycobacterium tuberculosis infection and vaccine development. Tuberculosis. 2016;98:30–41. doi: 10.1016/j.tube.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Cole ST, Riccardi G. New tuberculosis drugs on the horizon. Curr. Opin. Microbiol. 2011;14:570–576. doi: 10.1016/j.mib.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 6.Andrade CH, Salum LDB, Castilho MS, Pasqualoto KFM, Ferreira EI, Andricopulo AD. Fragment-based and classical quantitative structure–activity relationships for a series of hydrazides as antituberculosis agents. Mol. Divers. 2008;12:47–59. doi: 10.1007/s11030-008-9074-z. [DOI] [PubMed] [Google Scholar]
- 7.Lilienkampf A, Pieroni M, Wan B, Wang Y, Franzblau SG, Kozikowski AP. Rational design of 5-phenyl-3-isoxazolecarboxylic acid ethyl esters as growth inhibitors of Mycobacterium tuberculosis. a potent and selective series for further drug development. J. Med. Chem. 2010;53:678–88. doi: 10.1021/jm901273n. [DOI] [PubMed] [Google Scholar]
- 8.Pieroni M, Lilienkampf A, Baojie W, Yuehong W, Franzblau SG, Kozikowski AP. Synthesis, biological evaluation, and structure-activity relationships for 5-[(E)-2-arylethenyl]-3-isoxazolecarboxylic acid alkyl ester derivatives as valuable antitubercular chemotypes. J. Med. Chem. 2009;52:6287–6296. doi: 10.1021/jm900513a. [DOI] [PubMed] [Google Scholar]
- 9.WHO. [accessed October 8, 2015];The Stop TB Strategy. 2015 http://www.who.int/tb/strategy/stop_tb_strategy/en/
- 10.TBALLIANCE, TB ALLIANCE. [accessed September 22, 2015];Putting science to work for a faster TB cure. 2015 Www.tballiance.org. www.tballiance.org.
- 11.Mahajan R. Bedaquiline: First FDA-approved tuberculosis drug in 40 years. Int. J. Appl. Basic Med. Res. 2013;2:124–128. doi: 10.4103/2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xavier aS, Lakshmanan M. Delamanid: A new armor in combating drug-resistant tuberculosis. J Pharmacol Pharmacother. 2014;5:222–224. doi: 10.4103/0976-500X.136121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobchev D, Pillai G, Karelson M. In silico machine learning methods in drug development. Curr. Top. Med. Chem. 2014;14:1913–1922. doi: 10.2174/1568026614666140929124203. [DOI] [PubMed] [Google Scholar]
- 14.Song CM, Lim SJ, Tong JC. Recent advances in computer-aided drug design. Brief. Bioinform. 2009;10:579–591. doi: 10.1093/bib/bbp023. [DOI] [PubMed] [Google Scholar]
- 15.Melo-Filho CC, Braga RC, Andrade CH. 3D-QSAR Approaches in Drug Design: Perspectives to Generate Reliable CoMFA Models. Curr. Comput. Aided. Drug Des. 2014;10:148–59. doi: 10.2174/1573409910666140410111043. [DOI] [PubMed] [Google Scholar]
- 16.Braga RC, Alves VM, Silva AC, Nascimento MN, Silva FC, Liao LM, Andrade CH. Virtual screening strategies in medicinal chemistry: the state of the art and current challenges. Curr. Top. Med. Chem. 2014;14:1899–1912. doi: 10.2174/1568026614666140929120749. [DOI] [PubMed] [Google Scholar]
- 17.Fort PO, Silva CHTP, De Alencastro RB, Augusto O, Antunes C, Castro HC, Rodrigues CR, De Janeiro R, Biologia D, Fluminense UF. Multidimensional-QSAR: Beyond the third-dimension in drug design. 2007;1:91–100. [Google Scholar]
- 18.Guido RVC, Oliva G, Andricopulo AD. Modern Drug Discovery Technologies: Opportunities and Challenges in Lead Discovery. Comb. Chem. High Throughput Screen. 2011;14:830–839. doi: 10.2174/138620711797537067. [DOI] [PubMed] [Google Scholar]
- 19.Kovalishyn V, Aires-de-Sousa J, Ventura C, Elvas Leitao R, Martins F. QSAR modeling of antitubercular activity of diverse organic compounds. Chemom. Intell. Lab. Syst. 2011;107:69–74. doi: 10.1016/j.chemolab.2011.01.011. [DOI] [Google Scholar]
- 20.Klopman G, Fercu D, Jacob J. Computer-aided study of the relationship between structure and antituberculosis activity of a series of isoniazid derivatives. Chem. Phys. 1996;204:181–193. doi: 10.1016/0301-0104(95)00415-7. [DOI] [Google Scholar]
- 21.Mehra R, Rani C, Mahajan P, Vishwakarma RA, Khan IA, Nargotra A. Computationally Guided Identification of Novel Mycobacterium tuberculosis GlmU Inhibitory Leads, Their Optimization, and in Vitro Validation. ACS Comb. Sci. 2016;18:100–116. doi: 10.1021/acscombsci.5b00019. [DOI] [PubMed] [Google Scholar]
- 22.Khunt RC, Khedkar VM, Chawda RS, Chauhan NA, Parikh AR, Coutinho EC. Synthesis, antitubercular evaluation and 3D-QSAR study of N-phenyl-3-(4-fluorophenyl)-4-substituted pyrazole derivatives. Bioorg. Med. Chem. Lett. 2012;22:666–678. doi: 10.1016/j.bmcl.2011.10.059. [DOI] [PubMed] [Google Scholar]
- 23.Nayyar A, Malde A, Jain R, Coutinho E. 3D-QSAR study of ring-substituted quinoline class of anti-tuberculosis agents. Bioorganic Med. Chem. 2006;14:847–856. doi: 10.1016/j.bmc.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Virsodia V, Pissurlenkar RRS, Manvar D, Dholakia C, Adlakha P, Shah A, Coutinho EC. Synthesis, screening for antitubercular activity and 3D-QSAR studies of substituted N-phenyl-6-methyl-2-oxo-4-phenyl-1,2,3,4-tetrahydro-pyrimidine-5-carboxamides. Eur. J. Med. Chem. 2008;43:2103–2115. doi: 10.1016/j.ejmech.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Ragno R, Ballante F. Pharmacophore Assessment Through 3-D QSAR:evaluation of the predictive ability on new derivatives by the application on a serie of antitubercularagents. J. Chem. Inf. Model. 2013 doi: 10.1021/ci400132q. [DOI] [PubMed] [Google Scholar]
- 26.Gupta RA, Kaskhedikar SG. Synthesis, antitubercular activity, and QSAR analysis of substituted nitroaryl analogs: Chalcone, pyrazole, isoxazole, and pyrimidines. Med. Chem. Res. 2013;22:3863–3880. doi: 10.1007/s00044-012-0385-3. [DOI] [Google Scholar]
- 27.Ventura C, Latino DARS, Martins F. Comparison of multiple linear regressions and neural networks based QSAR models for the design of new antitubercular compounds. Eur. J. Med. Chem. 2013;70:831–845. doi: 10.1016/j.ejmech.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 28.Joshi SD, More UA, Aminabhavi TM, Badiger AM. Two- and three-dimensional QSAR studies on a set of antimycobacterial pyrroles: CoMFA, Topomer CoMFA, and HQSAR. Med. Chem. Res. 2014;23:107–126. doi: 10.1007/s00044-013-0607-3. [DOI] [Google Scholar]
- 29.Khunt RC, Khedkar VM, Coutinho EC. Synthesis and 3D-QSAR analysis of 2-chloroquinoline derivatives as H 37 RV MTB inhibitors. Chem. Biol. Drug Des. 2013;82:669–684. doi: 10.1111/cbdd.12178. [DOI] [PubMed] [Google Scholar]
- 30.Stavrakov G, Valcheva V, Philipova I, Doytchinova I. Design of novel camphane-based derivatives with antimycobacterial activity. J. Mol. Graph. Model. 2014;51:7–12. doi: 10.1016/j.jmgm.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Frecer V, Seneci P, Miertus S. Computer-assisted combinatorial design of bicyclic thymidine analogs as inhibitors of Mycobacterium tuberculosis thymidine monophosphate kinase. J. Comput. Aided. Mol. Des. 2011;25:31–49. doi: 10.1007/s10822-010-9399-4. [DOI] [PubMed] [Google Scholar]
- 32.Kumar A, Siddiqi MI. CoMFA based de novo design of Pyrrolidine Carboxamides as Inhibitors of Enoyl Acyl Carrier Protein Reductase from Mycobacterium tuberculosis. J. Mol. Model. 2008;14:923–935. doi: 10.1007/s00894-008-0326-8. [DOI] [PubMed] [Google Scholar]
- 33.Syam S, Abdelwahab SI, Al-Mamary MA, Mohan S. Synthesis of chalcones with anticancer activities. Molecules. 2012;17:6179–95. doi: 10.3390/molecules17066179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoki N, Muko M, Ohta E, Ohta S. C-geranylated chalcones from the stems of Angelica keiskei with superoxide-scavenging activity. J. Nat. Prod. 2008;71:1308–1310. doi: 10.1021/np800187f. [DOI] [PubMed] [Google Scholar]
- 35.Chen YH, Wang WH, Wang YH, Lin ZY, Wen CC, Chern CY. Evaluation of the anti-inflammatory effect of chalcone and chalcone analogues in a Zebrafish model. Molecules. 2013;18:2052–2060. doi: 10.3390/molecules18022052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahapatra DK, Bharti SK, Asati V. Anti-cancer chalcones: Structural and molecular target perspectives. Eur. J. Med. Chem. 2015;98:69–114. doi: 10.1016/j.ejmech.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Rizvi SUF, Siddiqui HL, Johns M, Detorio M, Schinazi RF. Anti-HIV-1 and cytotoxicity studies of piperidyl-thienyl chalcones and their 2-pyrazoline derivatives. Med. Chem. Res. 2012;21:3741–3749. doi: 10.1007/s00044-011-9912-x. [DOI] [Google Scholar]
- 38.Hans RH, Guantai EM, Lategan C, Smith PJ, Wan B, Franzblau SG, Gut J, Rosenthal PJ, Chibale K. Synthesis, antimalarial and antitubercular activity of acetylenic chalcones. Bioorg. Med. Chem. Lett. 2010;20:942–4. doi: 10.1016/j.bmcl.2009.12.062. [DOI] [PubMed] [Google Scholar]
- 39.Chen M, Zhai L, Christensen SB, Thor G, Zhai LIN. Inhibition of Fumarate Reductase in Leishmania major and L. donovani by Chalcones. Antimicrob. Agents Chemother. 2001;45:2023–2029. doi: 10.1128/AAC.45.7.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouattara M, Sissouma D, Koné MW, Hervé E, Touré SA, Ouattara L. Synthesis and anthelmintic activity of some hybrid Benzimidazolyl-chalcone derivatives. 2011;10:767–775. [Google Scholar]
- 41.López SN, Castelli MV, Zacchino Sa, Domínguez JN, Lobo G, Charris-Charris J, Cortés JC, Ribas JC, Devia C, Rodríguez aM, Enriz RD. In vitro antifungal evaluation and structure-activity relationships of a new series of chalcone derivatives and synthetic analogues, with inhibitory properties against polymers of the fungal cell wall. Bioorg. Med. Chem. 2001;9:1999–2013. doi: 10.1016/s0968-0896(01)00116-x. http://www.ncbi.nlm.nih.gov/pubmed/11504637. [DOI] [PubMed] [Google Scholar]
- 42.Avila-Villarreal G, Hernández-Abreu O, Hidalgo-Figueroa S, Navarrete-Vázquez G, Escalante-Erosa F, Peña-Rodríguez LM, Villalobos-Molina R, Estrada-Soto S. Antihypertensive and vasorelaxant effects of dihydrospinochalcone-A isolated from Lonchocarpus xuul Lundell by NO production: Computational and ex vivo approaches. Phytomedicine. 2013 doi: 10.1016/j.phymed.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto T, Yoshimura M, Yamaguchi F, Kouchi T, Tsuji R, Saito M, Obata A, Kikuchi M. Anti-allergic activity of naringenin chalcone from a tomato skin extract. Biosci. Biotechnol. Biochem. 2004;68:1706–1711. doi: 10.1271/bbb.68.1706. [DOI] [PubMed] [Google Scholar]
- 44.Jamal H, Ansari WH, Rizvi SJ. Evaluation of chalcones - A flavonoid subclass, for, their anxiolytic effects in rats using elevated plus maze and open field behaviour tests. Fundam. Clin. Pharmacol. 2008;22:673–681. doi: 10.1111/j.1472-8206.2008.00639.x. [DOI] [PubMed] [Google Scholar]
- 45.Lam KW, Uddin R, Liew CY, Tham CL, Israf Da, Syahida A, Rahman MBA, Ul-Haq Z, Lajis NH. Synthesis and QSAR analysis of chalcone derivatives as nitric oxide inhibitory agent. Med. Chem. Res. 2011;21:1953–1966. doi: 10.1007/s00044-011-9706-1. [DOI] [Google Scholar]
- 46.Sato Y, He J-X, Nagai H, Tani T, Akao T. Isoliquiritigenin, one of the antispasmodic principles of Glycyrrhiza ularensis roots, acts in the lower part of intestine. Biol. Pharm. Bull. 2007;30:145–149. doi: 10.1248/bpb.30.145. [DOI] [PubMed] [Google Scholar]
- 47.Batovska D, Parushev S, Stamboliyska B, Tsvetkova I, Ninova M, Najdenski H. Examination of growth inhibitory properties of synthetic chalcones for which antibacterial activity was predicted. Eur. J. Med. Chem. 2009;44:2211–8. doi: 10.1016/j.ejmech.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 48.Nowakowska Z. A review of anti-infective and anti-inflammatory chalcones. Eur. J. Med. Chem. 2007;42:125–37. doi: 10.1016/j.ejmech.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 49.Chiaradia LD, Mascarello A, Purificação M, Vernal J, Cordeiro MNS, Zenteno ME, Villarino A, Nunes RJ, Yunes RA, Terenzi H. Synthetic chalcones as efficient inhibitors of Mycobacterium tuberculosis protein tyrosine phosphatase PtpA. Bioorg. Med. Chem. Lett. 2008;18:6227–30. doi: 10.1016/j.bmcl.2008.09.105. [DOI] [PubMed] [Google Scholar]
- 50.García A, Bocanegra-García V, Palma-Nicolás JP, Rivera G. Recent advances in antitubercular natural products. Eur. J. Med. Chem. 2012;49:1–23. doi: 10.1016/j.ejmech.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 51.PubChem. PubChem Bioassay. (n.d.). https://pubchem.ncbi.nlm.nih.gov.
- 52.ChEMBL. ChEMBL-European Bioinformatics Institute. 2014 https://www.ebi.ac.uk/chembl/
- 53.Scifinder. SciFinder-Chemical Abstracts Service. 2014 https://scifinder.cas.org/
- 54.Fourches D, Muratov EN, Tropsha A. Trust, But Verify II. A Practical Guide to Chemogenomics Data Curation. J. Chem. Inf. Model. 2016 doi: 10.1021/acs.jcim.6b00129. doi:10.1021/acs.jcim.6b00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fourches D, Muratov E, Tropsha A. Trust, but verify: On the importance of chemical structure curation in cheminformatics and QSAR modeling research. J. Chem. Inf. Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fourches D, Muratov E, Tropsha A. Curation of chemogenomics data. Nat. Chem. Biol. 2015;11:535–535. doi: 10.1038/nchembio.1881. [DOI] [PubMed] [Google Scholar]
- 57.Varnek A, Fourches D, Horvath D, Klimchuk O, Gaudin C, Vayer P, Solov’ev V, Hoonakker F, Tetko I, Marcou G. ISIDA - Platform for Virtual Screening Based on Fragment and Pharmacophoric Descriptors. Curr. Comput. Aided-Drug Des. 2008;4:191–198. doi: 10.2174/157340908785747465. [DOI] [Google Scholar]
- 58.Kuz’min VE, Artemenko AG, Muratov EN. Hierarchical QSAR technology based on the Simplex representation of molecular structure. J. Comput. Aided. Mol. Des. 2008;22:403–21. doi: 10.1007/s10822-008-9179-6. [DOI] [PubMed] [Google Scholar]
- 59.Dossetter AG, Griffen EJ, Leach AG. Matched molecular pair analysis in drug discovery. Drug Discov. Today. 2013;18:724–731. doi: 10.1016/j.drudis.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 60.MACCS structural keys. Accelrys; San Diego, CA: 2013. (n.d.) [Google Scholar]
- 61.Bajusz D, Rácz A, Héberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform. 2015;7:1–13. doi: 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kramer C, Fuchs JE, Whitebread S, Gedeck P, Liedl KR. Matched molecular pair analysis: Significance and the impact of experimental uncertainty. J. Med. Chem. 2014;57:3786–3802. doi: 10.1021/jm500317a. [DOI] [PubMed] [Google Scholar]
- 63.Wang L, Evers A, Monecke P, Naumann T. Ligand based lead generation - considering chemical accessibility in rescaffolding approaches via BROOD. J. Cheminform. 2012;4:O20. doi: 10.1186/1758-2946-4-S1-O20. [DOI] [Google Scholar]
- 64.Wirth M, Zoete V, Michielin O, Sauer WHB. SwissBioisostere: A database of molecular replacements for ligand design. Nucleic Acids Res. 2013;41:1–7. doi: 10.1093/nar/gks1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Boyle NM, Morley C, Hutchison GR. Pybel: a Python wrapper for the OpenBabel cheminformatics toolkit. Chem. Cent. J. 2008;2:5. doi: 10.1186/1752-153X-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carhart RE, Smith DH, Venkataraghavan R. Atom pairs as molecular features in structure-activity studies: definition and applications. J Chem. Inf. Comput. Sci. 1985;25:64–73. [Google Scholar]
- 67.Riniker S, Landrum Ga. Open-source platform to benchmark fingerprints for ligand-based virtual screening. J. Cheminform. 2013;5:26. doi: 10.1186/1758-2946-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rogers D, Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–54. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 69.Morgan HL. The Generation of a Unique Machine Description for Chemical Structures-A Technique Developed at Chemical Abstracts Services. J. Chem. Doc. 1965;5:107–113. [Google Scholar]
- 70.Gedeck P, Rohde B, Bartels C. Article QSAR – How Good Is It in Practice? Comparison of Descriptor Sets on an Unbiased Cross Section of Corporate Data Sets QSAR - How Good Is It in Practice? Comparison of Descriptor Sets on an Unbiased Cross Section of Corporate Data Sets. 2006:1924–1936. doi: 10.1021/ci050413p. [DOI] [PubMed] [Google Scholar]
- 71.Vapnik V. The Nature of Statistical Learning Theory. 2. Springer; New York: 2000. [Google Scholar]
- 72.Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front. Neurorobot. 2013;7 doi: 10.3389/fnbot.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Breiman L. Random Forest. Mach. Learn. 2001:1–33. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 74.Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–23. doi: 10.1021/jm020017n. http://www.ncbi.nlm.nih.gov/pubmed/12036371. [DOI] [PubMed] [Google Scholar]
- 75.Franc I, Lipinski a, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. 1997;23 doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- 76.Baell J, Walters MA. Chemistry: Chemical con artists foil drug discovery. Nature. 2014;513:481–483. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 77.Baell JB, Holloway Ga. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 78.Nasir Abbas Bukhari S, Jasamai M, Jantan I, Ahmad W. Review of Methods and Various Catalysts Used for Chalcone Synthesis. Mini. Rev. Org. Chem. 2013;10:73–83. doi: 10.2174/1570193X11310010006. [DOI] [Google Scholar]
- 79.Tawari NR, Bairwa R, Ray MK, Rajan MGR, Degani MS. Design, synthesis, and biological evaluation of 4-(5-nitrofuran-2-yl)prop-2-en-1-one derivatives as potent antitubercular agents. Bioorg. Med. Chem. Lett. 2010;20:6175–8. doi: 10.1016/j.bmcl.2010.08.127. [DOI] [PubMed] [Google Scholar]
- 80.Cisak A, Rzeszowska-Modzelewska, Brzezinska E. Reactivity of 5-nitro-2-furaldehyde in alkaline and acidic solutions. 2001:427–434. [PubMed] [Google Scholar]
- 81.Collins La, Franzblau Scott G. Microplate Almar Blue Assay versus BACTEC 460 System for High-throughput Screeing of Compunds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cho SH, Warit S, Wan B, Hwang CH, Pauli GF, Franzblau SG. Low-Oxygen-Recovery Assay for High-Throughput Screening of Compounds against Nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2007;51:1380–1385. doi: 10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katsuno K, Burrows JN, Duncan K, van Huijsduijnen RH, Kaneko T, Kita K, Mowbray CE, Schmatz D, Warner P, Slingsby BT. Hit and lead criteria in drug discovery for infectious diseases of the developing world. Nat. Rev. Drug Discov. 2015:1–8. doi: 10.1038/nrd4683. [DOI] [PubMed] [Google Scholar]
- 84.Hu X, Hu Y, Vogt M, Stumpfe D. MMP-Cliffs: Systematic Identification of Activity Cliffs on the Basis of Matched Molecular Pairs. 2012:1–8. doi: 10.1021/ci3001138. [DOI] [PubMed] [Google Scholar]
- 85.Landrum G. RDKit: Open-Source Cheminformatics. 2014 http://www.rdkit.org/
- 86.Neves BJ, Dantas RF, Senger MR, Melo-Filho CC, Valente WCG, Almeida ACM, Rezende-Neto JM, Lima EFC, Paveley R, Furnham N, Muratov E, Kamentsky L, Carpenter AE, Braga RC, Silva-Júnior FP, Andrade CH. Discovery of New Anti-Schistosomal Hits by Integration of QSAR-Based Virtual Screening and High Content Screening. J. Med. Chem. 2016 doi: 10.1021/acs.jmedchem.5b02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- 88.Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, Dearden J, Gramatica P, Martin YC, Todeschini R, Consonni V, Kuz’Min VE, Cramer R, Benigni R, Yang C, Rathman J, Terfloth L, Gasteiger J, Richard A, Tropsha A. QSAR modeling: Where have you been? Where are you going to? J. Med. Chem. 2014;57:4977–5010. doi: 10.1021/jm4004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tropsha A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 90.Falzari K, Zhu Z, Pan D, Liu H, Hongmanee P, Franzblau SG. In Vitro and In Vivo Activities of Macrolide Derivatives against. Society. 2005;49:1447–1454. doi: 10.1128/AAC.49.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Franzblau SG, Degroote MA, Cho SH, Andries K, Nuermberger E, Orme IM, Mdluli K, Angulo-Barturen I, Dick T, Dartois V, Lenaerts AJ. Comprehensive analysis of methods used for the evaluation of compounds against Mycobacterium tuberculosis. Tuberculosis. 2012;92:453–488. doi: 10.1016/j.tube.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 92.Cho SH, Lee HS, Franzblau S. Microplate Alamar Blue Assay (MABA) and Low Oxygen Revovery Assay (LORA) for Mycobacterium tuberculosis. In: Parish T, Roberts DM, editors. Mycobact. Protoc. Vol. 3. New York: 2015. pp. 281–291. [DOI] [PubMed] [Google Scholar]
- 93.Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, Degnan MT, Cook MB, Quenzer VK, Ferguson RM, Gilman RH. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 1998;36:362–6. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andreu N, Zelmer A, Fletcher T, Elkington PT, Ward TH, Ripoll J, Parish T, Bancroft GJ, Schaible U, Robertson BD, Wiles S. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.