ABSTRACT
All animals must coordinate growth rate and timing of maturation to reach the appropriate final size. Here, we describe hobbit, a novel and conserved gene identified in a forward genetic screen for Drosophila animals with small body size. hobbit is highly conserved throughout eukaryotes, but its function remains unknown. We demonstrate that hobbit mutant animals have systemic growth defects because they fail to secrete insulin. Other regulated secretion events also fail in hobbit mutant animals, including mucin-like ‘glue’ protein secretion from the larval salivary glands. hobbit mutant salivary glands produce glue-containing secretory granules that are reduced in size. Importantly, secretory granules in hobbit mutant cells lack essential membrane fusion machinery required for exocytosis, including Synaptotagmin 1 and the SNARE SNAP-24. These membrane fusion proteins instead accumulate inside enlarged late endosomes. Surprisingly, however, the Hobbit protein localizes to the endoplasmic reticulum. Our results suggest that Hobbit regulates a novel step in intracellular trafficking of membrane fusion proteins. Our studies also suggest that genetic control of body size, as a measure of insulin secretion, is a sensitive functional readout of the secretory machinery.
KEY WORDS: Body size, Growth, Insulin secretion, Regulated exocytosis, Intracellular trafficking, Drosophila
Summary: Phenotypic analysis of hobbit mutants reveals that Hobbit is a novel and conserved regulator of body size in Drosophila that plays a specific role in secretory granule membrane protein trafficking.
INTRODUCTION
Body size is exquisitely regulated during metazoan development. In Drosophila, an explosive period of growth occurs during larval development; however, this systemic growth ceases and final body size is specified at the onset of metamorphosis (Nijhout et al., 2014). One of the crucial regulators of body size in Drosophila is the insulin signaling pathway. In this pathway, Drosophila insulin-like peptides (Dilps) are secreted from the insulin-producing cells (IPCs), which are neuroendocrine cells located in the larval central nervous system. The IPCs function in a manner that is analogous to mammalian pancreatic β-cells (Geminard et al., 2006). Dilps then circulate systemically and bind to the insulin receptor (InR) on the membrane of target cells, triggering a downstream signaling cascade that regulates both metabolism and cellular growth (Brogiolo et al., 2001; Rulifson et al., 2002). Disruption of the insulin signaling pathway has a profound effect on Drosophila body size. Ablation of the IPCs (Rulifson et al., 2002), genetic deletion of Dilps (Grönke et al., 2010), or mutation of targets downstream of InR (Böhni et al., 1999; Brogiolo et al., 2001; Murillo-Maldonado et al., 2011) all result in a dramatic reduction in body size. However, despite the importance of the insulin signaling pathway for systemic growth in flies and for disease in humans, the factors regulating insulin secretion remain largely unknown.
Insulin secretion in both flies and mammals occurs via regulated exocytosis. This crucial cellular process begins with the biogenesis of cargo protein-containing secretory granules. Cargo proteins targeted to the regulated exocytosis pathway, such as insulin, are translated by ribosomes bound to the rough endoplasmic reticulum (RER) and translocated into the ER lumen co-translationally. These cargo proteins are then transported to ER exit sites (ERES), specialized ER subdomains characterized by a coating of coat protein complex II (COPII), and packaged into COPII-coated vesicles (Brandizzi and Barlowe, 2013). COPII-coated vesicles mediate trafficking of cargo proteins from the ER to the Golgi cisternae; in the Golgi, cargo proteins undergo additional processing and post-translational modification. Cargo proteins are then selectively packaged into secretory granules, specialized organelles that bud from the trans-Golgi Network (TGN) as immature secretory granules (Kögel and Gerdes, 2010). The immature granules undergo a maturation process to refine their content and membrane composition, rendering them competent for exocytosis (Kögel and Gerdes, 2010). Mature secretory granules traffic to the plasma membrane, where they undergo tethering, docking and priming steps, attaching them to the membrane so that cargo proteins can be quickly released to the extracellular environment in a stimulus-dependent manner (Burgoyne and Morgan, 2003).
The membrane fusion events leading to extracellular release are mediated by membrane fusion proteins, such as synaptotagmins and soluble N-ethylmaleimide-sensitive factor (NSF) activating protein receptors (SNAREs) (Burgoyne and Morgan, 2003; Hong, 2005). To drive membrane fusion during regulated exocytosis, these proteins must be properly trafficked to secretory granule membranes. Like cargo proteins, secretory granule membrane proteins must first enter the ER. Synaptotagmins contain a single-pass transmembrane domain at their N terminus (Südhof, 2002) that acts as a signal, directing the ribosome to the RER and allowing co-translational translocation of the nascent protein into the ER (Hegde and Keenan, 2011). In contrast, most SNAREs contain a single-pass transmembrane domain at their C terminus and are therefore translated by free ribosomes in the cytosol. SNAREs must then be post-translationally translocated into the ER, a process that requires chaperones to prevent the nascent SNARE from complexing with SNAREs on other organelles and to shelter the hydrophobic transmembrane domain, preventing aggregation (Hegde and Keenan, 2011; Kutay et al., 1995). Once in the ER, secretory granule membrane proteins are thought to follow a similar path to that of cargo proteins: they leave the ER via ERESs, traffic through the Golgi cisternae, and are eventually loaded onto secretory granules produced in the TGN. However, despite the importance of regulated exocytosis for both normal development and disease, many of the trafficking steps during this process remain largely unknown.
Here, we report the identification and characterization of hobbit, a novel and conserved protein that plays a crucial role in the trafficking of secretory granule membrane proteins. We have identified hobbit in a chemical mutagenesis screen for Drosophila mutant animals with a small body size. hobbit is highly conserved throughout eukaryotes, but the protein has not been functionally characterized. We find that hobbit mutant animals have a small body size because they fail to secrete insulin and demonstrate that hobbit is required for maintenance of the major endocrine signaling axis that regulates insulin release. Furthermore, other regulated exocytosis events are also disrupted in hobbit mutant animals, including the secretion of mucin-like ‘glue’ proteins from the larval salivary glands at the onset of metamorphosis. hobbit mutant cells produce secretory granules that contain cargo proteins; however, these granules lack SNAREs and synaptotagmins and therefore are not competent for release, providing an explanation for why secretion fails upon loss of hobbit function. We find that the Hobbit protein itself localizes to the ER, suggesting that Hobbit may play a new role in trafficking of secretory granule membrane proteins through the secretory pathway.
RESULTS
A chemical mutagenesis screen for new regulators of body size
To find new regulators of body size in Drosophila, we have conducted a large-scale ethyl methanesulfonate (EMS) mutagenesis screen on the third chromosome and identified metamorphosis-specific lethal mutations. We screened over 26,000 mutagenized chromosomes and identified 8636 lethal mutations; of these, 566 were pupal lethal mutations (Wang et al., 2008). Among the 566, we identified 26 mutant strains in 19 complementation groups that displayed a dramatic small pupa (sP) phenotype. We first focused on mapping one of the mutants with a dramatic small body size: sP1. Recombination mapping with pairs of dominant markers (Sapiro et al., 2013) followed by complementation tests with chromosomal deficiencies and known lethal mutations identified sP1 as an allele of the insulin receptor (InR) (Fig. S1A-C), a gene that has previously been shown to play a crucial role in body size determination (Chen et al., 1996; Fernandez et al., 1995). InRsP1 does not contain any sequence changes within the InR coding sequence; however, InR mRNA transcript levels were significantly reduced in the mutant animals (Fig. S1D), suggesting that this allele contains a lesion disrupting critical regulatory sequences. The identification of a novel allele of InR confirms that our screening approach has uncovered bona fide regulators of systemic growth.
Identification of hobbit, a novel and conserved regulator of body size
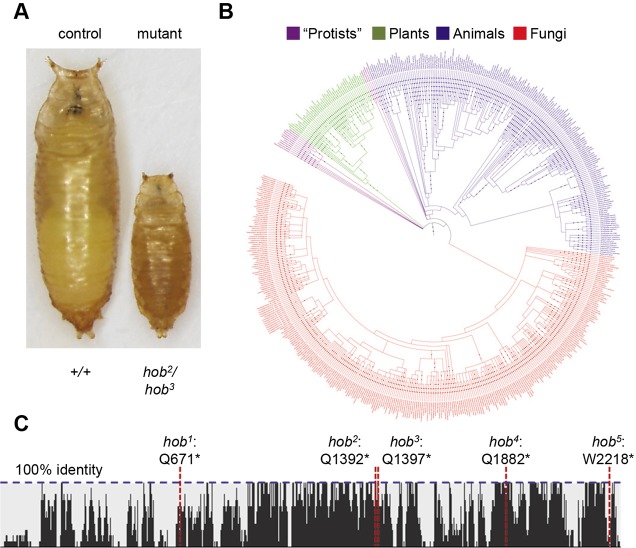
We next mapped the most frequently mutated locus (sP2), which contained five alleles. Recombination mapping with dominant markers placed this mutation between Roughened (R) and Dichaete (D); however, the mutant animals complemented all chromosomal deficiencies in this region, indicating that the disrupted gene was located in a gap between chromosomal deletions (Fig. S2A). Subsequent Sanger sequencing of candidate genes not covered by available deletions revealed lesions in a previously uncharacterized gene, CG14967 (Fig. S2A). We named this gene hobbit because of the dramatic small pupa phenotype displayed by the mutant animals (Fig. 1A and Fig. 2). All five hobbit alleles contained nonsense mutations (Fig. 1C, Fig. S2A), and all exhibited the same small body size and lethal phase during metamorphosis (Fig. S2B), indicating that these are likely loss-of-function alleles. Although the majority of hobbit mutant animals arrested during metamorphosis, some animals appeared to die earlier in development, as only 80% of the expected progeny were observed as pupae on the side of vials (hob2/hob3, total n=274). Importantly, hobbit is conserved across eukaryotes, with orthologs in fungi, green plants, protists and animals (Fig. 1B). Additionally, a protein sequence identity plot among 13 animal model organisms (from worms to humans) revealed a high degree of primary sequence conservation across the length of the protein (Fig. 1C). However, no functionally characterized domains are predicted beyond a possible transmembrane domain at the N terminus. Additionally, sequence homology searches did not reveal any hits outside of hobbit and its orthologs. These results indicate that hobbit encodes a truly novel protein.
Fig. 1.
Identification of hobbit, a novel and conserved regulator of systemic growth. (A) hobbit mutant animals exhibit a dramatic small pupa phenotype. (B) ‘Tree of life’ phylogeny showing hobbit orthologs throughout Eukaryota, in green plants (green), animals (blue), fungi (red) and some protists (purple). (C) Protein identity plot showing primary sequence conservation between D. melanogaster hobbit and 12 metazoan orthologs (Caenorhabditis elegans, Bombyx mori, Anopheles gambiae, Danio rerio, Xenopus tropicalis, Gallus gallus, Oryctolagus cuniculus, Rattus norvegicus, Mus musculus, Macaca mulatta, Pan troglodytes and Homo sapiens); horizontal blue dashed line shows 100% identity. Vertical red dashed lines indicate the position of Drosophila hobbit mutant alleles, which are all nonsense mutations. hob: hobbit.
Fig. 2.
hobbit mutant animals can be rescued by fly and human proteins. Size quantification and lethal phase analysis for control, ubiquitous hobbit-RNAi [UAS-Dcr/+; tub>/UAS-hob(i) 1 and 2], hobbit mutants (hob2/hob3), hobbit mutant controls for rescue experiments (act>,hob2/hob3, hob2/UAS-hob,hob3 and UAS-Hs-hob, hob2/hob3), fly hobbit mutant rescue (act>,hob2/UAS-hob,hob3), and human ortholog rescue (UAS-Hs-hob,hob2/act>,hob3). Ubiquitous expression of hobbit-RNAi reduces body size and results in lethality during metamorphosis. Controls for rescue experiments do not significantly alter body size or lethality compared with hob2/hob3 mutant animals. Expression of fly hobbit or the human ortholog (Hs-hob) both rescue body size and lethality. Body size quantified by pupa volume is expressed as a percentage relative to wild type (100%). Data are shown as mean±s.e.m. Volume quantification and lethal phase analysis were performed on the same animals; n=100 animals per genotype. > in genotypes denotes GAL4; for example, ‘act>’ is shorthand for act-GAL4. A, adult; PA, pharate adult; P/iP, pupa/incomplete pupa; PP, prepupa.
Without any obvious molecular signatures or protein domains to guide our studies, we relied on phenotypic clues to characterize the function of hobbit. We began these phenotypic studies by validating our hobbit mutations using RNAi knockdown and rescue experiments. Publicly available RNAi lines did not affect hobbit expression levels; therefore, we generated two new RNAi constructs that significantly reduced hobbit levels (Fig. S3C). Importantly, ubiquitous hobbit RNAi knockdown phenocopied both the small body size and lethality of our mutant animals (Fig. 2). To conduct a rescue experiment, we cloned the wild-type fly Hobbit protein and ubiquitously overexpressed it in the hobbit mutant background. This treatment rescued both the small body size and metamorphosis lethality of hobbit mutant animals (Fig. 2). Additionally, we cloned the human ortholog of hobbit, Hs hob (KIAA0100), and ubiquitously overexpressed it in the hobbit mutant background; this treatment also rescued both body size and lethality (Fig. 2), suggesting that the function of hobbit is evolutionarily conserved.
hobbit regulates body size in a non-cell-autonomous manner
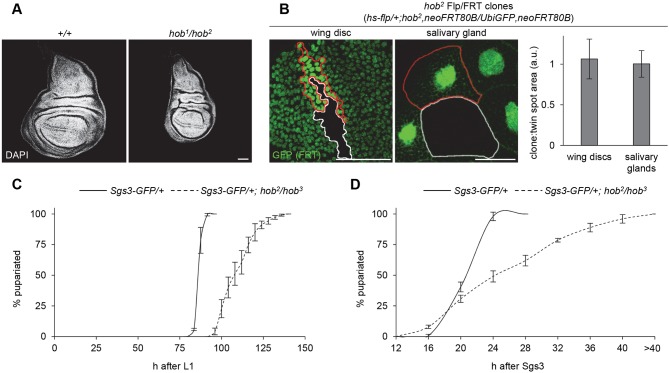
As a first step toward understanding the function of hobbit, we sought to determine why the mutant animals are small. We first wanted to determine whether hobbit had a cell-autonomous effect on growth. To do this, we generated somatic clones in both mitotic (wing disc) and endocycling (salivary gland) cells and measured the area of the mutant clones and twin spots. Although hobbit mutant tissues were small (Fig. 3A), the ratio of clone:twin spot area in both wing disc and salivary gland cells was 1:1 (Fig. 3B), suggesting that hobbit has a non-cell-autonomous effect on growth. We next used qPCR to determine whether hobbit mRNA expression levels were regulated in a stage- or tissue-specific manner. We found that hobbit appeared to be ubiquitously expressed across wandering L3 (wL3) tissues (Fig. S3B). Additionally, during development, we observed that expression levels were highest in early embryos (Fig. S3A), suggesting that hobbit mRNA may be maternally loaded. hobbit then appears to be expressed at relatively steady levels in whole animals, with only a modest increase in expression from early larval stages to the onset of metamorphosis (Fig. S3A). However, levels of hobbit do not appear to dictate body size, as ubiquitous overexpression of hobbit had no effect on final size (Fig. S3D). Altogether, these results indicate that hobbit does not have a cell-autonomous effect on growth and therefore must regulate body size in a non-cell-autonomous manner.
Fig. 3.
hobbit has a non-cell-autonomous effect on growth. (A) Wing imaginal discs dissected from wandering L3 (wL3) larvae of control and hobbit mutant (hob1/hob2) animals show that tissues are small in hobbit mutant animals. Nuclei stained with DAPI (white). (B) hob2 mutant Flp/FRT clones (marked by loss of GFP and outlined in white) are the same size as paired twin spots (marked by 2× GFP and outlined in red) in both wing discs and salivary glands. Quantification of the ratio of clone:twin spot area (in arbitrary units, a.u.) confirms a 1:1 ratio between clone area:twin spot area. Data are shown as mean±s.d. n=10 clones/twin spots per tissue. (C) Analysis of developmental timing throughout larval development in control and hobbit mutant animals. Animals were synchronized at 0-4 h after hatching from the embryo (0-4 h after L1), then allowed to age until the onset of metamorphosis (puparium formation). hobbit mutant animals (dashed line) are delayed and asynchronous compared with control animals (solid line). y-axis shows percentage of animals pupariated, x-axis shows developmental time point in h after L1. Three replicates of n>50 animals (control total n=172, hob2/hob3 total n=214) were analyzed; data are shown as mean±s.d. (D) Analysis of developmental timing during the third larval instar in control and hobbit mutant animals. Animals were synchronized at 0-4 h after the start of the mid-third larval instar transition (0-4 h after Sgs3-GFP expression), then allowed to age until puparium formation. All control animals pupariate within 24 h of Sgs3-GFP expression (solid line); in contrast, hobbit mutant animals take considerably longer to pupariate (dashed line). y-axis shows percentage of animals pupariated, x-axis shows developmental time point in h after Sgs3-GFP expression. Three replicates of n>40 animals (control total n=146, hob2/hob3 total n=137) were analyzed; data are shown as mean±s.d. Scale bars: 50 µm.
We next began to investigate known causes of non-cell-autonomous growth defects in Drosophila. Developmental timing abnormalities leading to a shortened larval growth phase and precocious entry into metamorphosis can cause reduced body size through non-cell-autonomous mechanisms (King-Jones et al., 2005). Larvae with this defect develop normally until the third larval instar, but then prematurely enter metamorphosis. We used a marker, called Sgs3-GFP, which is induced in the larval salivary glands at the mid-third larval instar transition, to synchronize animals at this developmental time point and then measure the duration of the final stage of the third larval instar. All control animals entered metamorphosis within 24 h of Sgs3-GFP expression (Fig. 3D, solid line). hobbit mutant animals induced expression of Sgs3-GFP, indicating that the animals reached the mid-third instar transition. Importantly, hobbit mutant animals did not pupariate precociously; instead, hobbit mutant animals were developmentally delayed following the mid-third instar transition, with some animals taking >40 h to pupariate after induction of Sgs3-GFP (Fig. 3D, dashed line). To analyze this developmental delay phenotype further, we also measured timing throughout the larval growth phase [from L1 to puparium formation (PF)]. We found that hobbit mutant animals were developmentally delayed and developed asynchronously throughout larval development (Fig. 3C; compare the slopes of the solid and dashed lines). Taken together, these results indicate that hobbit regulates growth non-cell-autonomously, but a precocious developmental timing defect is not responsible for the small body size of hobbit mutant animals.
Insulin secretion fails in hobbit mutant animals
The insulin signaling pathway plays a central role in the regulation of Drosophila body size. Interestingly, hobbit mutant animals exhibited a dominant genetic interaction with our mutant allele of InR; double heterozygous animals were significantly smaller than either single heterozygote (Fig. S4A), suggesting that hobbit may play a role in the insulin pathway. Like loss of insulin itself, hobbit has a non-cell-autonomous effect on growth, leading us to test whether hobbit mutant animals had defects in insulin secretion. Consistent with previous reports (Géminard et al., 2009), control (fed) animals secreted insulin and therefore exhibited little staining for Dilp2 (Ilp2 – FlyBase) or Dilp5 (Ilp5 – FlyBase) in the IPCs; however, starved animals retained insulin, resulting in bright Dilp2/5 signal that returned to steady-state levels within 2 h of re-feeding (Fig. 4A, Fig. S5A). In contrast, hobbit mutant animals exhibited bright Dilp2 and Dilp5 signal in fed, starved, and re-fed states (Fig. 4B, Fig. S5B), indicating that these animals do not properly secrete insulin and suggesting that the insulin secretion defect may underlie the small body size of hobbit mutant animals.
Fig. 4.
hobbit is required for insulin secretion. (A) Immunofluorescence staining for Drosophila insulin-like peptide Dilp2 (red) in IPCs (marked with GFP in green) of control animals shows that Dilp2 is secreted under fed conditions, retained under starved conditions, and secreted within 2 h of re-feeding. (B) Dilp2 staining in hobbit mutant IPCs in fed, starved, and re-fed states shows that Dilp2 is not secreted under any condition. (C) IPC-specific hobbit RNAi knockdown (with Dilp2-GAL4) or fat body-specific hobbit RNAi knockdown (with Cg-GAL4) reduces body size. However, IPC- or fat body-specific (with ppl-GAL4 or Cg-GAL4) hobbit expression does not rescue hobbit mutant small pupae, whereas expression in both IPCs and fat body does rescue size. None of these treatments rescues hobbit mutant lethality. Size was quantified by pupa volume and expressed relative to wild type (100%); data are shown as mean±s.e.m. Volume and lethal phase data shown for hob2/UAS-hob,hob3 are the same as that shown in Fig. 2 but are included here for comparison. Volume measurements and lethal phase analysis were performed on the same animals; n=100 per genotype. ***P<0.0001 calculated by two-tailed t-test. (D) Representative image of body size rescue in hobbit mutant animals when hobbit is expressed in the IPCs only, fat body only, or IPCs and fat body. (E) Dilp2 (red) is not secreted in fed hobbit mutant animals upon expression of hobbit in the IPCs only (with Dilp2-GAL4) or fat body only (with ppl-GAL4). (F) Dilp2 (red) secretion is rescued in hobbit mutant animals when hobbit is expressed in both the IPCs (marked with GFP in green) and fat body (with Cg-GAL4). IPC images in all panels representative of n≥20 analyzed per condition/genotype. > in genotypes denotes GAL4; for example, ‘Dilp2>’ is shorthand for Dilp2-GAL4. A, adult; FB, fat body; n.s., not significant; PA, pharate adult; P/iP, pupa/incomplete pupa; PP, prepupa. Scale bars: 50 µm.
To confirm this observed insulin secretion defect, we analyzed insulin/insulin-like growth factor (IGF) signaling (IIS) responses in peripheral tissues. When insulin is secreted, it binds to InR, initiating a downstream kinase signaling cascade that includes the activation of phosphatidylinositol 3-kinase (PI3K), a lipid kinase that phosphorylates phosphatidylinositol 4,5-bisphosphate (PIP2) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3) (Britton et al., 2002). We used a ubiquitously expressed GFP-tagged PIP3 reporter (tGPH) (Britton et al., 2002) to determine plasma membrane levels of PIP3 in the fat body of control and hobbit mutant animals, and saw a reduction in plasma membrane signal in hobbit mutant fat body cells (Fig. S4B). This result indicates that hobbit mutant tissues contain less PIP3, indicative of reduced IIS signaling. Additionally, activation of IIS inhibits activity of the transcription factor foxo; therefore, reduced IIS should result in increased expression of foxo target genes, such as InR and the translational regulator 4E-BP (Thor – FlyBase) (Jünger et al., 2003; Puig et al., 2003). Using qPCR, we observed significant upregulation of both InR and 4E-BP in hobbit mutant whole animals (Fig. S4C). Taken together, these results indicate that IIS is reduced in hobbit mutant peripheral tissues, as would be expected with a defect in insulin secretion.
We next wanted to determine which tissues require hobbit function for insulin secretion. IPC-specific hobbit RNAi knockdown significantly reduced body size (Fig. 4C), suggesting that hobbit is required in the IPCs for insulin secretion. Consistent with this result, pan-neuronal RNAi knockdown of hobbit using elav-GAL4 also reduced animal body size (Fig. S4D). However, IPC-specific expression of hobbit in the mutant background did not rescue body size (Fig. 4C,D) or insulin secretion (Fig. 4E, Fig. S5C), suggesting that additional tissues may require hobbit function for insulin release. Previous studies have shown that the Drosophila fat body functions as a nutrient sensor, releasing secreted signals that circulate systemically and promote insulin release in response to dietary inputs (Delanoue et al., 2016; Géminard et al., 2009; Rajan and Perrimon, 2012), prompting us to test whether hobbit was also required in the fat body. Fat body-specific RNAi knockdown of hobbit significantly reduced body size (Fig. 4C). In contrast, fat body-specific expression of hobbit in mutant animals did not rescue body size (Fig. 4C,D) or insulin secretion (Fig. 4E, Fig. S5C). However, expression of hobbit in both the IPCs and fat body significantly rescued the small body size of hobbit mutant animals and restored the ability to secrete insulin (Fig. 4C,D,F, Fig. S5D). Other tissues, including the midgut (Song et al., 2017), prothoracic gland (Colombani et al., 2005) and muscles (Demontis et al., 2014; Langerak et al., 2018) also secrete signals that influence systemic growth and regulation; however, RNAi knockdown of hobbit in these tissues had no significant effect on body size (Fig. S4D), suggesting that hobbit is primarily required in the IPCs and fat body to regulate systemic growth. Taken together, these results indicate that hobbit regulates body size by maintaining the major endocrine axis required for insulin secretion.
Secretion of mucin-like glue proteins is disrupted in hobbit mutant salivary glands
Although hobbit expression in both the IPCs and fat body rescued body size, it did not rescue lethality (Fig. 4C), suggesting that hobbit has essential functions in other tissues. Because the fat body and IPCs both utilize regulated exocytosis, we investigated whether other regulated secretion events were disrupted in hobbit mutant animals. First, hobbit mutant pupae have a soft cuticle, suggesting that hobbit function may be required for deposition of chitin. Another conspicuous regulated exocytosis event during Drosophila development is the secretion of mucin-like ‘glue’ proteins from the larval salivary glands at the onset of metamorphosis (Biyasheva et al., 2001). Glue proteins are synthesized in the acinar cells of the salivary glands during the final day of larval development, secreted into the lumen via regulated exocytosis just prior to puparium formation, and expelled onto the surface of the animal at puparium formation, allowing it to adhere to a solid surface during metamorphosis (Biyasheva et al., 2001; Kang et al., 2017; Rousso et al., 2016; Tran et al., 2015). Interestingly, glue secretion, assayed by imaging GFP-tagged glue protein (Sgs3-GFP), failed in hobbit mutant animals (Fig. S6A), and this effect was cell-autonomous, as hobbit mutant Flp/FRT somatic clones also failed to secrete glue (Fig. 5A). Taken together, these results demonstrate that hobbit plays a crucial, cell-autonomous role in regulated exocytosis.
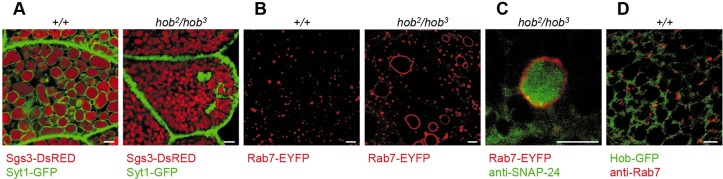
Fig. 5.
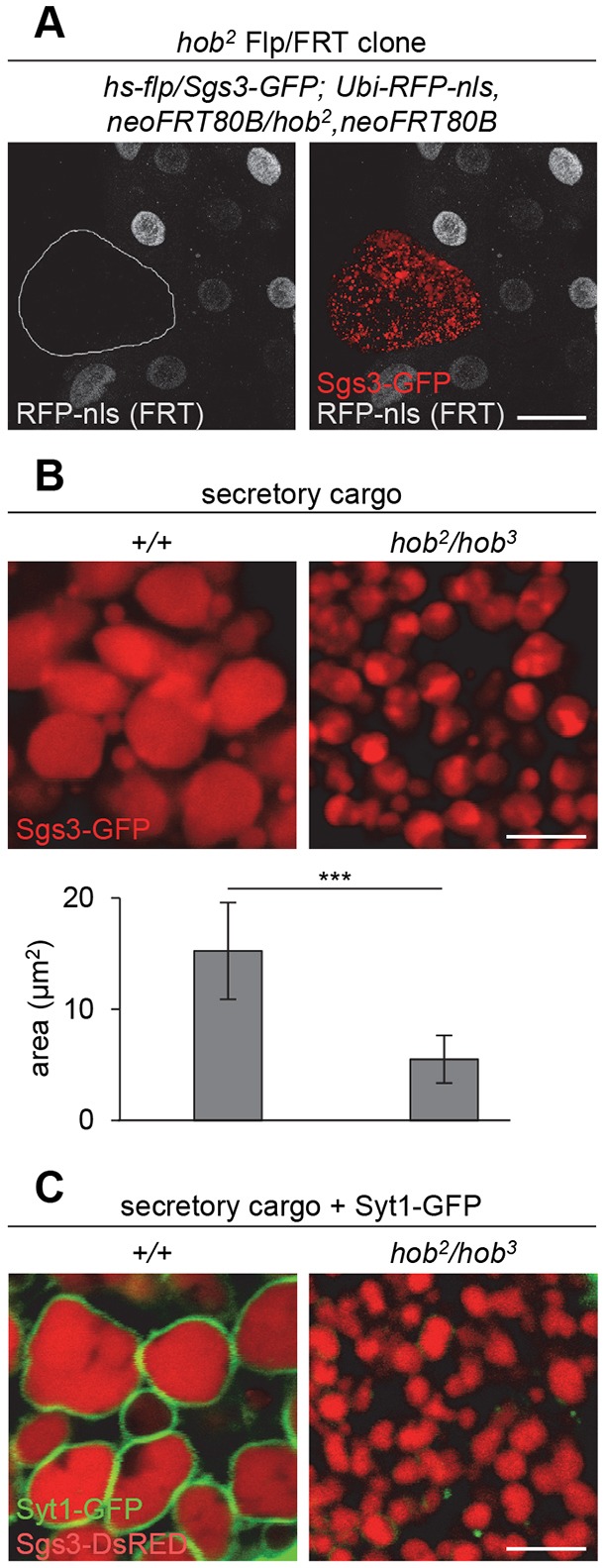
Secretory granules in hobbit mutant cells are not competent for exocytosis. (A) Flp/FRT hob2 clone, marked by loss of RFP-nls (gray) and outlined in white, shows that Sgs3-GFP glue proteins (red) are not secreted in hobbit mutant salivary gland cells dissected from prepupal animals. (B) Sgs3-GFP glue granules in hobbit mutant salivary glands are significantly smaller than those in controls. Graph shows quantification by granule area; data are shown as mean±s.d. n≥60 granules per genotype. ***P<0.001 calculated by two-tailed t-test. (C) Synaptotagmin-1-GFP (Syt1-GFP, in green, expression driven in salivary glands by hs-GAL4), is loaded onto glue granule membranes in control glands but not in hobbit mutant glands. Glue protein shown in red. All images were acquired from live, unfixed tissue. Scale bars: 50 µm (A); 5 µm (B,C).
Secretory granules in hobbit mutant tissues lack membrane proteins required for maturation and exocytosis
Our next goal was to identify the mechanism by which hobbit regulates secretion. We first analyzed the morphology of the glue-containing secretory granules in hobbit mutant salivary glands to determine whether there were any defects. We found that these granules were significantly smaller compared with controls (Fig. 5B, Fig. S6B), suggesting that the granules do not mature properly, as glue granules are known in increase in size prior to release (Niemeyer and Schwarz, 2000). Exocytosis and granule-granule fusion during maturation are mediated by the presence and activity of membrane fusion proteins, such as SNAREs and synaptotagmins, which are present on both secretory granules and target membranes. Strikingly, secretory granules in hobbit mutant salivary glands do not contain these membrane fusion proteins. Salivary gland-specific expression of GFP-tagged Synaptotagmin 1 (Syt1-GFP) showed that this protein was present around secretory granules in control glands but absent from granule membranes in hobbit mutant glands (Fig. 5C). Immunofluorescence staining with an anti-Syt1 antibody confirmed these results (Fig. S6C). Furthermore, immunofluorescence staining for the SNARE protein SNAP-24 also showed a similar mislocalization in hobbit mutant glands (Fig. S6D). Because secretory granules in hobbit mutant cells do not contain membrane fusion proteins, these granules are not competent to undergo maturation or exocytosis, explaining why regulated exocytosis fails in hobbit mutant cells.
hobbit is required for normal trafficking of secretory granule membrane proteins
We next wanted to determine what happens to secretory granule membrane proteins in hobbit mutant cells. We observed that these proteins accumulated in large structures that were distinct from secretory granules (Fig. 6A, Fig. S6C,D). This result suggests that granule membrane proteins may aberrantly accumulate in one or more specific subcellular compartments. To determine where Syt1 and SNAP-24 accumulate, we used a collection of endogenously regulated, EYFP-tagged Rab proteins (Dunst et al., 2015), because Rab proteins are markers for many subcellular compartments and organelles. We first compared Rab protein localization and morphology in control and hobbit mutant salivary glands and observed striking defects with Rab7; hobbit mutant salivary glands contained numerous enlarged Rab7-positive late endosomes (Fig. 6B). Furthermore, secretory granule membrane proteins accumulated inside the enlarged late endosomes (Fig. 6C). Late endosomes are essential sorting sites, traditionally thought to direct proteins to either recycling or degradative pathways (Maxfield and McGraw, 2004). These results indicate that trafficking of secretory granule membrane proteins is severely disrupted in hobbit mutant secretory cells.
Fig. 6.
Trafficking of membrane fusion proteins on secretory granules is disrupted in hobbit mutant cells. (A) Syt1-GFP (green) localizes in large structures that are distinct from glue granules (red) in hobbit mutant cells. UAS-Syt1-GFP was expressed in salivary glands using hs-GAL4. Images were acquired from live, unfixed tissue. (B) Rab7-positive late endosomes (red) are dramatically enlarged in hobbit mutant salivary glands. Rab7-EYFP contains an EYFP tag at the endogenous Rab7 locus (Dunst et al., 2015). Images were acquired from live, unfixed tissue. (C) Rab7-positive endosomes (red) contain accumulations of the SNARE protein SNAP-24 (green) in hobbit mutant glands. (D) Hobbit-GFP (green, expression driven by Sgs3-GAL4) does not colocalize with Rab7 (red). Scale bars: 5 µm.
To begin to understand how hobbit regulates trafficking of secretory granule membrane proteins, we looked at where the Hobbit protein localized within the cell. To do this, we generated a GFP-tagged hobbit overexpression construct. Ubiquitous overexpression of this Hobbit-GFP construct rescued hobbit mutant animals (Fig. S7A), indicating that the construct produced functional Hobbit protein. Given that Syt1 and SNAP-24 are trapped inside Rab7-positive endosomes in hobbit mutant cells, we first tested whether Hobbit-GFP colocalized with Rab7. However, we did not observe any significant overlap between Hobbit-GFP and Rab7 (Fig. 6D). We next tested the possibility that Hobbit localizes to secretory granules to facilitate loading of secretory granule membrane proteins. Instead, we observed that Hobbit-GFP was present between secretory granules and did not appear to colocalize with SNAP-24 (Fig. 7B), indicating that Hobbit is not enriched on secretory granule membranes. Given that secretory granules are generated in the Golgi body, we also tested whether Hobbit-GFP colocalized with a pan-Golgi marker, but we did not observe any colocalization between Hobbit-GFP and Golgi-RFP (Fig. S7B). In contrast, we found that Hobbit-GFP strongly colocalized with the ER marker KDEL-RFP (Fig. 7A), suggesting that Hobbit localizes to the ER. The presence of many secretory granules dramatically condenses the cytoplasm; therefore, to confirm that Hobbit-GFP localizes to the ER, we analyzed Hobbit-GFP localization in salivary glands at puparium formation, the stage immediately following the completion of glue secretion. Hobbit-GFP did not appear to colocalize with a cytoplasmic/soluble mCherry at puparium formation (Fig. 7C), nor did KDEL-RFP appear to colocalize with a cytoplasmic/soluble GFP (Fig. 7D). However, Hobbit-GFP and KDEL-RFP strongly colocalized at puparium formation (Fig. 7E, Fig. S7C), demonstrating that Hobbit-GFP is strongly enriched within the ER. Taken together, our results indicate that hobbit is a novel and conserved regulator of intracellular trafficking; furthermore, hobbit appears to play a specific role in trafficking secretory granule membrane proteins through the secretory pathway.
Fig. 7.

Hobbit localizes to the endoplasmic reticulum. (A) Hobbit-GFP (green) strongly colocalizes with the endoplasmic reticulum (ER) marker KDEL-RFP (red) in wandering L3 salivary glands. Expression of Hobbit-GFP and KDEL-RFP was driven by Sgs3-GAL4; images were acquired from live, unfixed tissue. (B) Hobbit-GFP (expressed using Sgs3-GAL4; green) does not appear to colocalize with the secretory granule membrane protein SNAP-24 (red). (C) Hobbit-GFP (green) does not colocalize with cytoplasmic mCherry (red) in glands at puparium formation (0 h PF). Expression was driven by Sgs3-GAL4; images were acquired from live, unfixed tissue. (D) KDEL-RFP (red) does not colocalize with cytoplasmic GFP (green) in glands at 0 h PF. Expression was driven by Sgs3-GAL4; images were acquired from live, unfixed tissue. (E) Hobbit-GFP, in green, strongly colocalizes with KDEL-RFP (red) in glands at 0 h PF. Expression was driven by Sgs3-GAL4; images were acquired from live, unfixed tissue. Scale bars: 50 µm (A); 5 µm (B-E).
DISCUSSION
Regulated exocytosis is a crucial process for both systemic signaling and homeostatic mechanisms in multicellular organisms. Secretory granule membrane proteins, such as SNAREs and synaptotagmins, are indispensable for regulated exocytosis; without these proteins, secretory granules cannot mature properly, nor can they fuse with the plasma membrane. Therefore, proper trafficking and loading of these proteins onto secretory granules is an essential step in the production of mature granules that are competent for release. In this study, we have identified that hobbit, a novel and conserved protein, is required for proper trafficking of secretory granule membrane proteins during regulated exocytosis.
We first identified hobbit in a screen for Drosophila mutant animals that arrest during metamorphosis with a small pupa phenotype. Other mutant animals with reduced body size have been identified in flies; however, most of these mutant alleles are viable and specifically disrupt insulin secretion or insulin signaling (Böhni et al., 1999; Brogiolo et al., 2001; Chen et al., 1996; Rajan and Perrimon, 2012). Additionally, ablation of the IPCs results in viable animals that are reduced in size (Rulifson et al., 2002), demonstrating that insulin secretion is not essential for development. As a result, screening efforts to identify animals with reduced body size have primarily focused on mutations that result in viable animals. In contrast, our characterization of hobbit suggests that lethal mutations with a small body size represent a new, uncharacterized class of systemic growth regulators. Mutant alleles in this class, such as hobbit, are required for insulin secretion from the IPCs but are also required for secretory events in other tissues. These crucial functions in other tissues underlie the lethality observed in the mutant animals. Further analysis and additional screening for these hobbit-like mutations, including those from our small pupa mutant collection, may reveal other important regulators of insulin secretion and intracellular trafficking.
Despite the striking evolutionary conservation of the Hobbit protein, it does not contain any functionally characterized domains or obvious signatures that provide clues to its molecular function. Our identity plot analysis reveals several highly conserved regions within the Hobbit protein; however, these conserved regions do not exhibit homology with any other proteins outside of hobbit orthologs. Databases and prediction programs, such as Pfam, do annotate potential protein domains within Hobbit; however, the location of these domains within the protein sequence does not correspond with the regions of high sequence conservation that we have identified. Furthermore, these automated prediction algorithms annotate domains containing sequence motifs, such as ‘GFWDK’, that are not present in the Hobbit protein, suggesting that the domains have been incorrectly assigned. Some bioinformatic programs predict a transmembrane domain at the N terminus of the Hobbit protein. If present, a transmembrane domain would likely play a crucial role in hobbit localization and function. However, detailed structure-function studies will be required to validate and assign a function to each of the uncharacterized conserved regions within the Hobbit protein. These studies will reveal new insights into the molecular function of hobbit and may also identify new sequence motifs that are essential for the regulation of intracellular trafficking.
Our results demonstrate that hobbit plays a crucial role in regulated exocytosis by regulating trafficking of secretory granule membrane proteins. Secretory granules in hobbit mutant cells lack SNAREs and synaptotagmins, membrane fusion proteins that are required for exocytosis. These secretory granule membrane proteins accumulate inside enlarged late endosomes in hobbit mutant cells. However, how hobbit regulates trafficking of SNAREs and synaptotagmins remains unclear. Integral membrane proteins, such as synaptotagmins, are synthesized via co-translational translocation into the ER. These proteins are still produced in hobbit mutant cells, suggesting that hobbit may function post-ER entry, such as in ER-to-Golgi traffic. However, disruption of ER exit disrupts all secretory traffic (Ward et al., 2001) and would therefore be expected to inhibit trafficking of secretory granule cargo proteins, but this process is unaffected in hobbit mutant cells. Therefore, our data could suggest that there is a selective pathway required for ER-to-Golgi transport of secretory granule membrane proteins. We do not, however, observe accumulation of secretory granule membrane proteins in the ER; instead, these proteins accumulate inside Rab7-positive late endosomes. A similar accumulation phenotype has been described in mutant alleles of PI4KII, a lipid kinase that appears to play an important role in retrograde trafficking (Burgess et al., 2012). Retrograde trafficking recycles many proteins from endosomes to the TGN (Bonifacino and Hurley, 2008), although it is not known whether secretory granule membrane proteins undergo such recycling. A retrograde trafficking function would be consistent with the phenotypes we observe in hobbit mutant cells. However, given that known regulators of retrograde trafficking, including PI4KII, localize to endosomes (Burgess et al., 2012), it is unclear how hobbit, an ER-localized protein, could regulate this process. Our data raises the possibility that a new trafficking route from the ER, to endosomes, to secretory granules may be used for loading of secretory granule membrane proteins. Direct sorting of cargo proteins from endosomes to secretory granules has been described (Topalidou et al., 2016), raising the possibility that a similar trafficking route could be used for secretory granule membrane proteins. Although further studies will be required to resolve these questions, it appears that hobbit does not function in any of the characterized trafficking pathways that are known to play a role in regulated exocytosis.
In conclusion, here we have described hobbit, a novel and highly conserved regulator of intracellular trafficking and regulated exocytosis. Our studies of hobbit mutant phenotypes reveal that this protein regulates a new trafficking step that is required to deliver membrane proteins to secretory granules, rendering them competent for release. Additionally, our data suggests that hobbit, and other metamorphosis-lethal mutants with growth defects, define a new class of body size regulators that drive growth by playing crucial roles in intracellular trafficking and secretion of insulin. These results also highlight the power of unbiased chemical mutagenesis screens to uncover new genes and new biology.
MATERIALS AND METHODS
Fly stocks, food and developmental staging
The following stocks were obtained from the Bloomington Drosophila Stock Center: w1118, Df(3R)BSC677, Df(3R)ED6058, Df(3R)BSC678, Df(3R)Exel6186, InRE19, InR93Dj−4, InR05545, tGPH, UAS-Dcr-2, tub-GAL4, act-GAL4, elav-GAL4, Myo1A-GAL4, repo-GAL4, Mhc-GAL4, hs-flp, neoFRT80B, Ubi-GFP 61EF, Sgs3-GFP, Dilp2-GAL4, UAS-GFP, UAS-mCherry, ppl-GAL4, Cg-GAL4, Ubi-mRFP-nls 3L, UAS-Syt1-GFP, hs-GAL4, Sgs3-GAL4, Rab7-EYFP, UAS-Golgi-RFP, UAS-KDEL-RFP. Sgs3-DsRED was kindly provided by A. Andres (University of Nevada, Las Vegas, NV, USA), and phm-GAL4 was kindly provided by Carl Thummel (University of Utah, UT, USA). The five hobbit mutations and InRsP1 were generated in an EMS mutagenesis screen (Wang et al., 2008); the methods used to map these mutations are described elsewhere (Sapiro et al., 2013). Experiments using hs-GAL4 were carried out without heat-shock treatment, as hs-GAL4 ‘leaks’ constitutively in the salivary glands. All experiments were performed in temperature-controlled incubators at 25°C in uncrowded bottles or vials. For insulin secretion and tGPH assays, larvae were picked at 0-4 h after hatching from the embryo (0-4 h after L1) and grown on food containing 34 g inactivated yeast, 83 g corn flour, 60 g sucrose and 10 g agar per liter. Forty-eight hours later, at the start of the third larval instar, these larvae were starved on agar plates containing PBS/1% sucrose for 24 h. Larvae were then re-fed for the appropriate time on the food described above. To generate somatic clones in the salivary glands, embryos were heat-shocked at 37°C for 30 min at 0-4 h after egg lay (AEL); for wing discs, the same heat-shock treatment was given to larvae at 48-52 h AEL. For developmental timing from L1, larvae containing the Sgs3-GFP transgene were collected at 0-4 after hatching from the embryo (0-4 h after L1) and allowed to age at 25°C until expression of Sgs3-GFP was detected. Animals were then scored for puparium formation beginning 12 h after induction of Sgs3-GFP. For developmental experiments from Sgs3-GFP, larvae containing the Sgs3-GFP transgene were collected at 88 h AEL and scored for the absence of Sgs3-GFP expression. These larvae were then checked every 4 h for induction of Sgs3-GFP; once expressed, larvae were allowed to age at 25°C on grape agar plates with yeast paste until puparium formation.
Phylogenetic tree and identity plot analysis
hobbit orthologs were identified using the HMMER web server (Finn et al., 2015) with the CG14967 protein sequence as the query sequence. Significant hits (P≤0.0001) were submitted to phyloT (http://phylot.biobyte.de/) to generate a phylogenetic tree based on NCBI taxonomy information. We used Interactive Tree of Life (Letunic and Bork, 2016) to visualize, edit and annotate the final tree. To assay amino acid conservation, we used ClustalOmega (Sievers et al., 2011) to generate a protein alignment between fly hobbit and its orthologs in C. elegans, B. mori, A. gambiae, D. rerio, X. tropicalis, G. gallus, O. cuniculus, R. norvegicus, M. musculus, M. mulatta, P. troglodytes and H. sapiens. The resulting alignment was visualized in JalView (Waterhouse et al., 2009), and identity/similarity scores at each amino acid position were extracted to generate the final identity plot.
Molecular cloning and transgenic animal generation
To generate the UAS-hobbit transgene, the full-length hobbit coding sequence was amplified from w1118 cDNA using the following primers: 5′-AAAAGGTACCATGATGCTACAGCTACTTCTATTCTGCCTGG-3′ and 5′-AAAAAAGCGGCCGCTCAATCATTTCCCGATCTCTTTCCG-3′. The resulting ∼6.9 kb product was digested with KpnI and NotI, ligated into the Gateway entry vector pENTR 1A (Invitrogen), and sequence verified. LR Clonase (Invitrogen) was used to recombine the entry clone into the pTW (Drosophila Genomics Resource Center) destination vector. Successful recombination was confirmed by sequencing. The resulting UAS-hobbit plasmid was then injected into w1118 flies using standard methods (Genetic Services). A similar strategy was used to generate the UAS-KIAA0100 transgene. The KIAA0100 coding sequence was amplified from oligo(dT)-primed qPCR human reference cDNA (Clontech) using the following primers: 5′-CCCCCCTTTAAAACCATGCCTCTGTTCTTCTCCGC-3′ and 5′-AAAAAAGCGGCCGCTTTCATTTGCGCCTGCCAAA-3′. The resulting ∼6.2 kb product was digested with DraI and NotI, and entry and destination clones were generated as described above. KIAA0100 has many predicted isoforms; therefore, only the full-length product matching the NCBI consensus coding sequence (CCDS) was used. Sequence-verified plasmids were injected into w1118 flies using standard protocols (Genetic Services). To generate UAS-hobbit-GFP, we first amplified the GFP coding sequence from pTGW (Drosophila Genomics Resource Center) using the following primers: 5′-AAAAAAGGTACCAGTGAGCAAGGGCGAGGAGCT-3′ and 5′-AAAAAATCTAGACTTGTACAGCTCGTCCATGC-3′. The resulting PCR product was digested with KpnI and XbaI and ligated into pBID-UASC-G (Wang et al., 2012). The plasmid was sequence verified, and the hobbit entry clone was recombined into this destination vector as described above. Sequence-verified plasmids were injected into VK00027 flies for phiC31-mediated site-directed integration (Rainbow Transgenic Flies). To generate the UAS-hobbit-RNAi lines, we followed the strategy outlined by the Transgenic RNAi Project (TRiP) (Ni et al., 2011). The primers for hobbit-RNAi 1 were: 5′-CTAGCAGTATGATGCTACAGCTACTTCTATAGTTATATTCAAGCATATAGAAGTAGCTGTAGCATCATGCG-3′ and 5′-AATTCGCATGATGCTACAGCTACTTCTATATGCTTGAATATAACTATAGAAGTAGCTGTAGCATCATAC-3′. The primers for hobbit-RNAi 2 were: 5′-CTAGCAGTATGCAGCGAATTGTTGTTAAATAGTTATATTCAAGCATATTTAACAACAATTCGCTGCATGCG-3′ and 5′-AATTCGCATGCAGCGAATTGTTGTTAAATATGCTTGAATATAACTATTTAACAACAATTCGCTGCATACTG-3′. The primers were annealed, phosphorylated, and ligated into pVALIUM20 (TRiP) that had been linearized by digestion with NheI and EcoRI. The resulting plasmids were sequence verified and injected for phiC31-mediated site-directed integration into attP2-containing flies (Rainbow Transgenic Flies). (Note: A newly generated RNAi construct from TRiP contains the same shRNA hairpin as hobbit-RNAi 2 inserted on chromosome 2; hobbit-RNAi 2 is inserted on chromosome 3.)
Pupa volume quantification and lethal phase analysis
All light microscope images of pupae were captured on an Olympus SXZ16 stereomicroscope coupled to an Olympus DP72 digital camera with DP2-BSW software. All pupae pictured together were originally captured in the same image but were individually rotated and aligned post-acquisition to improve image aesthetics. To quantify body size, we used Photoshop CS6 to measure the length and width of each pupa, and calculated pupa volume using a published formula (Delanoue et al., 2016). No statistical methods were used to determine sample size. Animals were grown at controlled density and on a controlled diet to avoid any nutritional effects on body size. After image capture, the pupae were transferred to a grape agar plate and aged at 25°C for one week for lethal phase analysis. Lethal phases were determined using Bainbridge and Bownes staging criteria (Bainbridge and Bownes, 1981).
Quantitative real-time PCR (qPCR)
qPCR was performed as previously described (Ihry et al., 2012). Total RNA was extracted from whole animals or tissues from the appropriate developmental stage and genotype using the RNeasy Plus Mini Kit (Qiagen), and cDNA was synthesized from 100-400 ng of total RNA using SuperScript III First-Strand Synthesis System (Invitrogen). For wL3 paired tissue samples, the wing discs, salivary glands, central nervous system, and the rest of the carcass were dissected from the same pool of animals to enable a direct comparison of expression levels across tissues. qPCR was performed on a Roche LightCycler 480 using LightCycler 480 SYBR Green I Master Mix (Roche). In all experiments, samples were run simultaneously in biological triplicate, and amplification efficiencies were calculated for each primer pair. The Relative Expression Software Tool (REST) (Pfaffl et al., 2002) was used to calculate relative expression. REST calculates standard error by using a confidence interval centered on the median, which generates error bars that reflect asymmetric tendencies in the data. Primer sequences for rp49 (RpL32 – FlyBase) (Ihry et al., 2012) and 4E-BP (Demontis and Perrimon, 2010) were published previously. New primers were designed using ApE for hobbit or FlyPrimerBank (Hu et al., 2013) for InR. hob F: 5′-TTTGGTGAAGAGGTTTCGGGTC-3′; hob R: 5′-TCTTTGTTGATGCGAATGTCACG-3′. InR F: 5′-GAAGTGGAGACGACGGGTAAA-3′; InR R: 5′-TCGCGCTGTTGTCGATTGTT-3′.
Immunofluorescence staining and confocal microscopy
Tissues were dissected from the appropriate animals, fixed for 30 min in PBS with 0.1% Triton X-100 (PBST) and 4% formaldehyde, blocked overnight in PBST/4% bovine serum albumin (BSA) at 4°C, and stained using the appropriate primary and secondary antibodies diluted in PBST/4% BSA. Primary antibodies used were rat anti-Dilp2 (1:200; Géminard et al., 2009), rabbit anti-Dilp5 (1:800; Géminard et al., 2009) (both a gift from P. Leopold, Institute of Biology Valrose, France), rabbit anti-SNAP-24 (1:200; Niemeyer and Schwarz, 2000) (gift from T. Schwarz, Harvard Medical School, MA, USA), mouse anti-Syt1 (1:50; Developmental Studies Hybridoma Bank, 3H2 2D7), mouse anti-Rab7 (1:50; Developmental Studies Hybridoma Bank, Rab7). The Rab7 antibody was generated and validated previously (Riedel et al., 2016). Secondary antibodies were used at 1:200 and included anti-rat Alexa Fluor 568 (Invitrogen, A11077), anti-rabbit Alexa Fluor 633 (Invitrogen, A21071), anti-rabbit Cy3 (Jackson ImmunoResearch Laboratories, 711-165-152), anti-mouse Cy3 (Jackson ImmunoResearch Laboratories, 715-165-150). DAPI was used to label nuclei (1:1000; Invitrogen). Stained tissues were mounted in Vectashield (Vector Laboratories). Images were captured on an Olympus FluoView FV1000 confocal microscope and optimized with FV10-ASW software. For live tissue imaging, tissues were dissected and mounted in PBS and imaged for no longer than 5 min after dissection. IPC images were acquired from a z-series comprising 40 slices at a 0.59 µm step size. Identical laser settings and optimization parameters were used for all samples; images shown as sum-intensity projections generated by FV10-ASW software. Quantification of clone:twin spot area ratios and glue granule areas was performed using ImageJ. Although true blinding was not possible given the experimental conditions and genotypes analyzed, whenever possible, results were independently confirmed by another lab member who was blinded to the genotype or experimental condition of the samples being analyzed.
Supplementary Material
Acknowledgements
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and reagents obtained from the Drosophila Genomics Resource Center (NIH 2P40OD010949) were used in this study. We are grateful to Jessica Smoko for technical assistance and Yunsik Kang for helpful discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.D.N., A.B.; Methodology: S.D.N., A.B.; Validation: S.D.N.; Formal analysis: S.D.N., A.B.; Investigation: S.D.N.; Data curation: S.D.N.; Writing - original draft: S.D.N.; Writing - review & editing: S.D.N., A.B.; Visualization: S.D.N., A.B.; Supervision: A.B.; Project administration: A.B.; Funding acquisition: A.B.
Funding
This work was supported in part by the National Institutes of Health (GM123204 to A.B.) and the National Science Foundation (GRFP DGE-1256259 to S.D.N.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.161356.supplemental
References
- Bainbridge B. S. P. and Bownes M. (1981). Staging the metamorphosis of Drosophila melanogaster. Development 66, 57-80. [PubMed] [Google Scholar]
- Biyasheva A., Do T.-V., Lu Y., Vaskova M. and Andres A. J. (2001). Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Dev. Biol. 231, 234-251. 10.1006/dbio.2000.0126 [DOI] [PubMed] [Google Scholar]
- Böhni R., Riesgo-Escovar J., Oldham S., Brogiolo W., Stocker H., Andruss B. F., Beckingham K. and Hafen E. (1999). Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 97, 865-875. 10.1016/S0092-8674(00)80799-0 [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S. and Hurley J. H. (2008). Retromer. Curr. Opin. Cell Biol. 20, 427-436. 10.1016/j.ceb.2008.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F. and Barlowe C. (2013). Organization of the ER–Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14, 382-392. 10.1038/nrm3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton J. S., Lockwood W. K., Li L., Cohen S. M. and Edgar B. A. (2002). Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2, 239-249. 10.1016/S1534-5807(02)00117-X [DOI] [PubMed] [Google Scholar]
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R. and Hafen E. (2001). An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213-221. 10.1016/S0960-9822(01)00068-9 [DOI] [PubMed] [Google Scholar]
- Burgess J., Del Bel L. M., Ma C.-I. J., Barylko B., Polevoy G., Rollins J., Albanesi J. P., Kramer H. and Brill J. A. (2012). Type II phosphatidylinositol 4-kinase regulates trafficking of secretory granule proteins in Drosophila. J. Cell Sci. 125, 3040-3050. 10.1242/jcs.117010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne R. D. and Morgan A. (2003). Secretory granule exocytosis. Physiol. Rev. 83, 581-632. 10.1152/physrev.00031.2002 [DOI] [PubMed] [Google Scholar]
- Chen C., Jack J. and Garofalo R. S. (1996). The Drosophila insulin receptor is required for normal growth. Endocrinology 137, 846-856. 10.1210/endo.137.3.8603594 [DOI] [PubMed] [Google Scholar]
- Colombani J., Bianchini L., Layalle S., Pondeville E., Dauphin-Villemant C., Antoniewski C., Carré C., Noselli S. and Léopold P. (2005). Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310, 667-670. 10.1126/science.1119432 [DOI] [PubMed] [Google Scholar]
- Delanoue R., Meschi E., Agrawal N., Mauri A., Tsatskis Y., McNeill H. and Leopold P. (2016). Drosophila insulin release is triggered by adipose Stunted ligand to brain Methuselah receptor. Science 353, 1553-1556. 10.1126/science.aaf8430 [DOI] [PubMed] [Google Scholar]
- Demontis F. and Perrimon N. (2010). FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813-825. 10.1016/j.cell.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F., Patel V. K., Swindell W. R. and Perrimon N. (2014). Intertissue control of the nucleolus via a myokine-dependent longevity pathway. Cell Rep. 7, 1481-1494. 10.1016/j.celrep.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunst S., Kazimiers T., von Zadow F., Jambor H., Sagner A., Brankatschk B., Mahmoud A., Spannl S., Tomancak P., Eaton S. et al. (2015). Endogenously tagged rab proteins: a resource to study membrane trafficking in Drosophila. Dev. Cell 33, 351-365. 10.1016/j.devcel.2015.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez R., Tabarini D., Azpiazu N., Frasch M. and Schlessinger J. (1995). The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 14, 3373-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., Bateman A. and Eddy S. R. (2015). HMMER web server: 2015 Update. Nucleic Acids Res. 43, W30-W38. 10.1093/nar/gkv397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geminard C., Arquier N., Layalle S., Bourouis M., Slaidina M., Delanoue R., Bjordal M., Ohanna M., Ma M., Colombani J. et al. (2006). Control of metabolism and growth through insulin-like peptides in Drosophila. Diabetes 55, S5-S8. 10.2337/db06-S001 [DOI] [Google Scholar]
- Géminard C., Rulifson E. J. and Léopold P. (2009). Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10, 199-207. 10.1016/j.cmet.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Grönke S., Clarke D.-F., Broughton S., Andrews T. D. and Partridge L. (2010). Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6, e1000857 10.1371/journal.pgen.1000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R. S. and Keenan R. J. (2011). Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 12, 787-798. 10.1038/nrm3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. (2005). SNAREs and traffic. Biochim. Biophys. Acta 1744, 120-144. 10.1016/j.bbamcr.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Hu Y., Sopko R., Foos M., Kelley C., Flockhart I., Ammeux N., Wang X., Perkins L., Perrimon N. and Mohr S. E. (2013). FlyPrimerBank: an online database for drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3 (Bethesda) 3, 1607-1616. 10.1534/g3.113.007021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihry R. J., Sapiro A. L., Bashirullah A., Akagi K. and Takai M. (2012). Translational control by the DEAD box RNA helicase belle regulates ecdysone-triggered transcriptional cascades. PLoS Genet. 8, e1003085 10.1371/journal.pgen.1003085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jünger M. A., Rintelen F., Stocker H., Wasserman J. D., Végh M., Radimerski T., Greenberg M. E. and Hafen E. (2003). The Drosophila Forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2, 20 10.1186/1475-4924-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Neuman S. D. and Bashirullah A. (2017). Tango7 regulates cortical activity of caspases during reaper-triggered changes in tissue elasticity. Nat. Commun. 8, 603 10.1038/s41467-017-00693-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K., Charles J.-P., Lam G. and Thummel C. S. (2005). The ecdysone-induced DHR4 orphan nuclear receptor coordinates growth and maturation in Drosophila. Cell 121, 773-784. 10.1016/j.cell.2005.03.030 [DOI] [PubMed] [Google Scholar]
- Kögel T. and Gerdes H.-H. (2010). Maturation of secretory granules. Results Probl. Cell Differ. 50, 1-20. [DOI] [PubMed] [Google Scholar]
- Kutay U., Ahnert-Hilger G., Hartmann E., Wiedenmann B. and Rapoport T. A. (1995). Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 14, 217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak S., Kim M.-J., Lamberg H., Godinez M., Main M., Winslow L., O'Connor M. B. and Zhu C. C. (2018). The Drosophila TGF-beta/Activin-like ligands Dawdle and Myoglianin appear to modulate adult lifespan through regulation of 26S proteasome function in adult muscle. Biol. Open 7, bio.029454 10.1242/bio.029454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I. and Bork P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242-W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F. R. and McGraw T. E. (2004). Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5, 121-132. 10.1038/nrm1315 [DOI] [PubMed] [Google Scholar]
- Murillo-Maldonado J. M., Sánchez-Chávez G., Salgado L. M., Salceda R. and Riesgo-Escovar J. R. (2011). Drosophila insulin pathway mutants affect visual physiology and brain function besides growth, lipid, and carbohydrate metabolism. Diabetes 60, 1632-1636. 10.2337/db10-1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., Yang-Zhou D., Shim H.-S., Handler D., Karpowicz P., Binari R. et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405-407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer B. A. and Schwarz T. L. (2000). SNAP-24, a Drosophila SNAP-25 homologue on granule membranes, is a putative mediator of secretion and granule-granule fusion in salivary glands. J. Cell Sci. 113, 4055-4064. [DOI] [PubMed] [Google Scholar]
- Nijhout H. F., Riddiford L. M., Mirth C., Shingleton A. W., Suzuki Y. and Callier V. (2014). The developmental control of size in insects. Wiley Interdiscip. Rev. Dev. Biol. 3, 113-134. 10.1002/wdev.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W. and Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Marr M. T., Ruhf M. L. and Tjian R. (2003). Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006-2020. 10.1101/gad.1098703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A. and Perrimon N. (2012). Drosophila Cytokine Unpaired 2 Regulates Physiological Homeostasis by Remotely Controlling Insulin Secretion. Cell 151, 123-137. 10.1016/j.cell.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel F., Gillingham A. K., Rosa-Ferreira C., Galindo A. and Munro S. (2016). An antibody toolkit for the study of membrane traffic in Drosophila melanogaster. Biol. Open 84, bio.018937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso T., Schejter E. D. and Shilo B.-Z. (2016). Orchestrated content release from Drosophila glue-protein vesicles by a contractile actomyosin network. Nat. Cell Biol. 18, 181-190. 10.1038/ncb3288 [DOI] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K. and Nusse R. (2002). Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296, 1118-1120. 10.1126/science.1070058 [DOI] [PubMed] [Google Scholar]
- Sapiro A. L., Ihry R. J., Buhr D. L., Konieczko K. M., Ives S. M., Engstrom A. K., Wleklinski N. P., Kopish K. J. and Bashirullah A. (2013). Rapid recombination mapping for high-throughput genetic screens in Drosophila. G3 (Bethesda) 3, 2313-2319. 10.1534/g3.113.008615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Cheng D., Hong S., Sappe B., Hu Y., Wei N., Zhu C., O'Connor M. B., Pissios P. and Perrimon N. (2017). Midgut-derived activin regulates glucagon-like action in the fat body and glycemic control. Cell Metab. 25, 386-399. 10.1016/j.cmet.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T. C. (2002). Synaptotagmins: why so many? J. Biol. Chem. 277, 7629-7632. 10.1074/jbc.R100052200 [DOI] [PubMed] [Google Scholar]
- Topalidou I., Cattin-Ortolá J., Pappas A. L., Cooper K., Merrihew G. E., MacCoss M. J. and Ailion M. (2016). The EARP complex and its interactor EIPR-1 are required for cargo sorting to dense-core vesicles. PLoS Genet. 12, e1006074 10.1371/journal.pgen.1006074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran D. T., Masedunskas A., Weigert R. and Hagen K. G. and Ten, (2015). Arp2/3-mediated F-actin formation controls regulated exocytosis in vivo. Nat. Commun. 6, 10098 10.1038/ncomms10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Evans J., Andrews H. K., Beckstead R. B., Thummel C. S. and Bashirullah A. (2008). A genetic screen identifies new regulators of steroid-triggered programmed cell death in Drosophila. Genetics 180, 269-281. 10.1534/genetics.108.092478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-W., Beck E. S. and McCabe B. D. (2012). A modular toolset for recombination transgenesis and neurogenetic analysis of Drosophila. PLoS ONE 7, e42102 10.1371/journal.pone.0042102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T. H., Polishchuk R. S., Caplan S., Hirschberg K. and Lippincott-Schwartz J. (2001). Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155, 557-570. 10.1083/jcb.200107045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M. and Barton G. J. (2009). Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189-1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.