SUMMARY
In both academia and the pharmaceutical industry, large-scale assays for drug discovery are expensive and often impractical, particularly for the increasingly important physiologically relevant model systems that require primary cells, organoids, whole organisms, or expensive or rare reagents. We hypothesized that data from a single high-throughput imaging assay can be repurposed to predict the biological activity of compounds in other assays, even those targeting alternate pathways or biological processes. Indeed, quantitative information extracted from a three-channel microscopy-based screen for glucocorticoid receptor translocation was able to predict assay-specific biological activity in two ongoing drug discovery projects. In these projects, repurposing increased hit rates by 60- to 250-fold over that of the initial project assays while increasing the chemical structure diversity of the hits. Our results suggest that data from high-content screens are a rich source of information that can be used to predict and replace customized biological assays.
INTRODUCTION
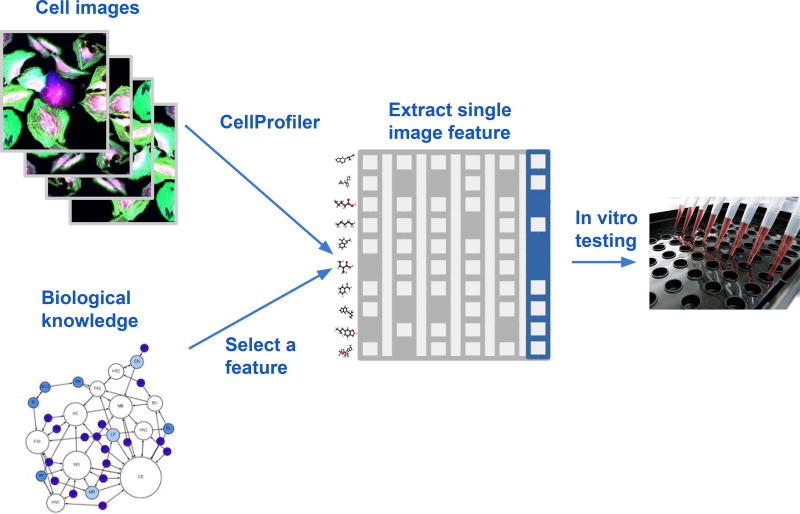
High-throughput imaging (HTI), also known as high-content screening (HCS), captures the morphology of the cell and its organelles by microscopy and has yielded diverse biological discoveries (Pepperkok and Ellenberg, 2006; Starkuviene and Pepperkok, 2007; Walter et al., 2010). HTI is often applied to screen chemical compounds based on morphological changes they induce (Held et al., 2010; Yarrow et al., 2003). Currently, most HTI screens are designed to evaluate one specific biological process and exploit only a handful of morphological features from the image, chosen to best measure that process (Singh et al., 2014) (Figure 1).
Figure 1. A Typical HTI Screen Approach.
Few or single features are extracted from cellular images; the remainder of information (gray) is ignored (Ansbro et al., 2013; Evensen et al., 2010).
However, any cellular system hosts many more biochemical processes and thousands of potential drug targets, all of which are exposed to the screened chemical compounds. Many of these targets and processes impact cell morphology and that morphology can to a large extent be extracted from the images (Carpenter et al., 2006). The resulting set of features, which include not just shape and spatial metrics but also the intensity and patterning of fluorescently labeled markers, can be used to describe chemical compounds and can be considered as an image-based compound fingerprint. Such fingerprints are powerful enough to accomplish a variety of important biological aims including optimizing the diversity of compound libraries, grouping compounds by pharmacological mechanism, and grouping genes based on functional similarity (Caicedo et al., 2016).
Motivation
We therefore hypothesized that image-based fingerprints of compounds derived from a given image-based cellular assay, might be leveraged to predict compound activity in seemingly unrelated assays. Effective predictors of biological activity already exist; virtual screening and Quantitative Structure–Activity Relationship (QSAR) analyses typically rely on features derived from the chemical structure of compounds to predict their activity in assays. Structure-based models are predictively performant (Cumming et al., 2013), but only for those parts of chemical space for which sufficient assay activity data is available. Unfortunately, compounds that are chemically very different from any known active compound are unlikely to be predicted as active. Because cell morphology can reflect compound-induced modulation of diverse targets and biochemical processes regardless of compound structure, we suspected that image-based models would avoid this limitation and may complement chemistry-based models in novel and poorly annotated chemical space.
Decades of high-content screening experience indicate an ability of image-based readouts to generalize over multiple unrelated targets. Yet, most academic and commercial imaging campaigns have followed a narrowly focused classical setup depicted in Figure 1, leaving a large volume of biological information untapped. Therefore, we aimed to repurpose pre-existing imaging screens to generically predict compound activities in assays that may be unrelated to the original screening assay.
RESULTS
Overview of Proposed Repurposing Approach
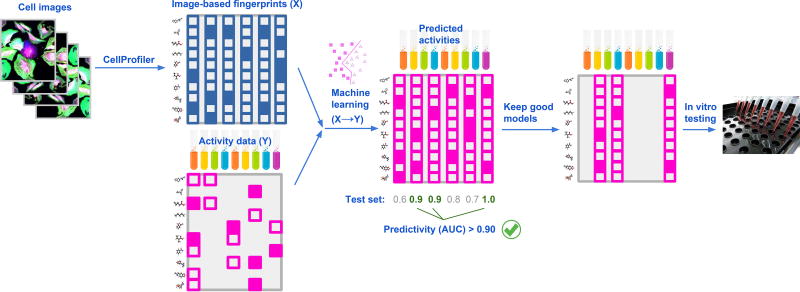
We propose a pipeline (Figure 2) to leverage the rich information in existing image screens for the prediction of activity in a variety of orthogonal assays directed at seemingly unrelated proteins and processes. First, we extract an extensive image-based fingerprint of morphological features for each compound in a single, already completed large-scale imaging screen (X in Figure 2), aiming for maximal and unbiased information capture (Section Extracting Image-based Fingerprints). Second, we introduce existing activity data for orthogonal assays of interest on these compounds (Y in Figure 2). Then, we train supervised machine learning models to predict Y from X and choose models with high predictive performance. Finally, we use these high-quality models to select compounds for in-vitro testing. Next we will describe each of these steps in detail.
Figure 2. Strategy to Repurpose Imaging Screens to Efficiently Predict Biological Activity.
Features extracted from images of cells are used by machine learning methods to model all available activity data from previously performed assays. Assays with good predictivity on the test data are then selected for testing a relatively small number of predicted-active compounds, chosen from a large set of compounds profiled in the imaging assay.
Extracting Image-based Fingerprints
The goal of extracting image-based fingerprints is to capture all available information about the biological state of the cell from the image. In this work, we use previously developed software (CellProfiler) and methods (Gustafsdottir et al., 2013) to produce a feature vector for each cell, capturing general morphology, shape and biologically important parameters (e.g., micronucleus count). For the three-channel glucocorticoid receptor (GR) HTI assay used in the evaluation, this produced an 842-dimensional feature vector per cell. Then for each plate we normalize each feature using the mean and the standard deviation of the corresponding feature from the negative controls (cells without treatment). Finally, for each compound we compute a vector of feature medians across all cells in its image, producing a single image-based fingerprint.
We note that an attractive alternative procedure is to use convolutional neural networks (CNNs) to learn feature representation directly from the raw pixels of cell images. This strategy shows promise but is still exploratory for image-based profiling; together with the high computational cost and hardware requirements, we leave this direction to future research.
Machine Learning for Image-based Fingerprints
We next use machine learning to take image-based fingerprints (X in Figure 2) and the existing bioactivity measurements on the assays of interest (Y in Figure 2) to learn a model to predict bioactivity of new compounds given their image-based fingerprints.
The simplest approach would be to model each column of the activity data separately (single task learning). However, we can take advantage of the existence of multiple related prediction tasks by modeling them jointly (multitask learning). In the case of related tasks multitask learning is known to improve the overall performance significantly (Caruana, 1997).
Both regression and classification methods could be used in the repurposing workflow we propose. Here we describe two which yielded good computational and predictive performance. To document the compatibility of this generic concept with other machine learning methods, we also carried out additional experiments with random forest (Breiman, 2001) and k-nearest neighbor classifier in our validation setup (Methods S3).
Bayesian Matrix Factorization
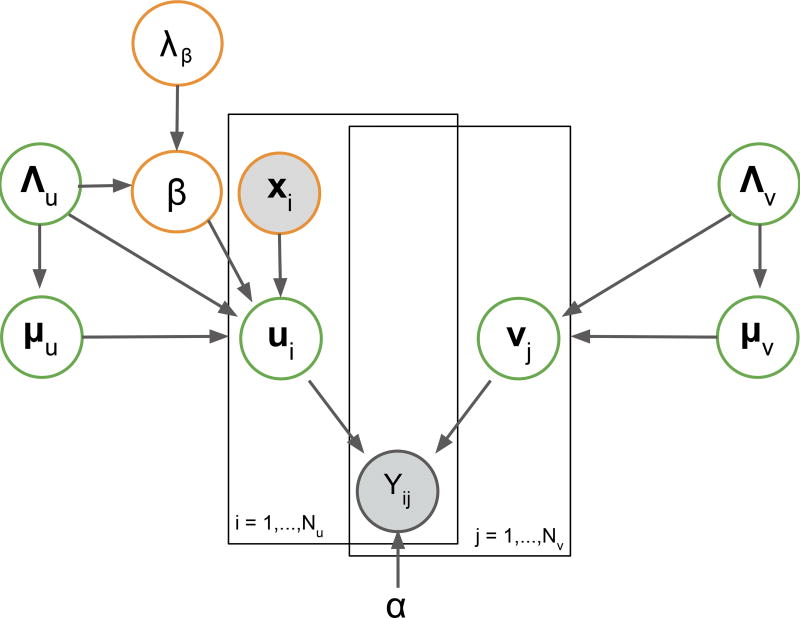
First, we explored Bayesian matrix factorization, a multitask method that does not require hyperparameterization (like regularization) and provides uncertainty estimates for predictions. Specifically, we used the Bayesian matrix factorization method Macau, which can account for side information (in this case image features) as side information. To factorize the N times M activity matrix Y, Macau represents each compound and each assay by D-dimensional latent vectors ui and vj, respectively. The prediction for the element Yij, corresponding to the activity of compound i on assay j, is given by the scalar product uiTvj. The features xi is an F-dimensional vector (F=842) corresponding to the image-based fingerprint (Section Extracting Image-based Fingerprints) and is added to the prior of the latent vectors of compounds ui. Macau maps all tasks to the same D-dimensional latent space, therefore enabling sharing of parts of the model.
This results in a probabilistic model of
where α is the precision of the observations, parameters μu and Λu model the mean and precision of the compound latent vectors, similarly μv and Λv model the latent vectors for assays. The parameter β is a D times F dimensional matrix that maps the image features to the compound latent space. To learn β we apply a Gaussian prior on it:
where ⊗ is the Kronecker product, λβ is a precision parameter and IF is the identity matrix of size F. Figure 3 depicts the plate diagram for the probabilistic model.
Figure 3. Diagram for the Probabilistic Model for the Bayesian Matrix Factorization Approach Macau.
The shaded circles denote observed variables and the transparent circles are inferred from the data.
By deriving conditional distributions for all model variables, we obtain a Gibbs sampler that iterates over all model variables, as in (Simm et al., 2017). For each variable, it samples a value from the conditional distribution by fixing all the others. Finally, to compute the predictions for Yij we use each sample to compute the scalar products uiTvj and then average over the samples. We observed that the performance of the method does not degrade with choosing a high latent dimensionality D. In practice, this implies the choice of a large enough latent space; in our case D=150.
The Macau model described here is for the regression setting, i.e., Yij are real-valued. The model can be easily modified to handle the classification setting by replacing the normal prior on Yij with a probit one. We have made the implementations for both settings available open source.1
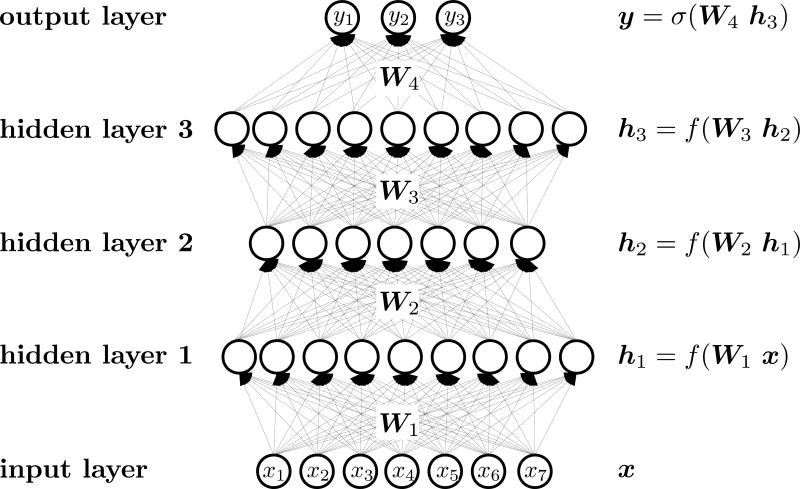
Deep Neural Networks
The matrix factorization model described above is linear and may lack the flexibility to capture all important biological effects. Therefore, we additionally tested a multitask deep learning architecture. We implemented Deep Neural Networks (DNNs), concretely feed-forward artificial neural networks, with many layers comprising a large number of neurons and rectified linear units (Mayr et al., 2016). DNNs (Figure 4) consists of interconnected neurons that are arranged hierarchically in layers. In the first layer of the network (the input layer), the neurons obtain an input vector that is the image-based fingerprint. The intermediate layers (the hidden layers) comprise the hidden neurons that have weighted connections to the neurons of the previous level layer, and can be considered as abstract features, built from features below. The last layer (the output layer) supplies the predictions of the model. Typical DNNs comprise several layers which consists of thousands of neurons.
Figure 4. General Architecture of Deep Neural Networks.
Variable x denotes the image-based fingerprint, y corresponds to biological activity. The tested hyperparameters of DNN are shown in Table S1.
We used rectified linear units (ReLUs) as activation functions in the hidden layers. The output layer has sigmoid activation functions. To avoid overfitting, we employed multiple regularization techniques, concretely dropout (Srivastava et al., 2014) and early stopping. Both the dropout rate and the early-stopping parameter, i.e. the number of epochs after which learning is stopped, were determined on a validation data set.
Deep learning naturally enables multitask learning (Caruana, 1997). In our setting each assay is a task. Commonalities across the assays translate to shared representation in the hidden layers and can yield performance improvements (Mayr et al., 2016). We modeled each assay by a separate output unit.
We used cross-entropy as a loss function for our DNNs:
where Ỹij is the prediction for compound i and assay j and the actual label is Yij, which indicates whether the compound was active (Yij = 1) or inactive (Yij = 0) in the given assay. The binary variable mij indicates whether a measurement is present (mij = 1) or missing (mij = 1). The implementation details, optimization of architecture and hyperparameters are given in Supplemental Information.
Selection of High Quality Models
Next, we select only assays yielding a highly reliable model. To this end we employ cross-validation, i.e., we split the compounds into k folds (here, k = 3). In cross-validation, the activity data for each fold is predicted using a model built on the data from the other folds. The resulting predictions enable the computation of an AUC-ROC score, or some other performance metric of choice. We used the average of the k folds as the evaluation metric for each model, and focused on models with an AUC-ROC > 0.9. If a machine learning method required an optimization of hyperparameters (e.g. choices of model architecture, kernel, dropout), we applied nested cross-validation (Mayr et al., 2016).
The simplest splitting scenario would be the random assignment of compounds to folds. However, in the case of chemistry-based modeling of pharmaceutical data sets, where compounds tend to be concentrated around attractive chemical backbones, this approach results in overoptimistic performance estimates (as close structural analogs get spread over test and validation, and performance metrics are boosted but do not hold up when applied to new chemistry). One popular mitigation approach is the use of temporal or roll-back splitting, where a timestamp is used to separate test and validation folds. In a multitask setting, however, temporal splitting is impractical because the order of measurement of the same compounds in different assays is not guaranteed to be aligned. Instead, we clustered the compounds based on chemical similarity and randomly assigned the clusters into folds (see Figure S1). Here, a Tanimoto similarity cutoff on ECFP6 features was used to ensure close analogs end up in the same test or validation fold. A high choice of cutoff may fall short of addressing the overoptimistic performance estimation, while a low cutoff may restrict the learning potential (as machine learning relies on recognizing similarities). In our experience, a similarity cutoff of 0.7 offers an optimal tradeoff.
Image-based fingerprints are insulated from the underlying chemistry. Thus, performance estimates for the resulting models are not expected to be skewed by the above mentioned pharmaceutical chemistry bias. However, for consistency reasons we still followed the clustered cross-validation approach.
Compound Selection for In-vitro Testing
Finally, we select compounds highly ranked by good quality models. There are two main selection strategies. The first is to select all the highest ranked compounds for in-vitro testing. Although simple the strategy may select sets that are too homogeneous or too chemically similar to the original training set. The second strategy is to apply diversity maximization (for example sphere exclusion clustering) on the highly ranked compounds, and only test a diverse set. This strategy can result in novel hits, but only if the model can generalize across scaffolds. As indicated before, we hypothesized that this is the case for models that use image-based fingerprints.
Experimental Evaluation
In the following we evaluate our HTI assay repurposing approach in a large-scale industrial context. To begin, we chose a high-throughput imaging screen of 524,371 proprietary compounds originally used for the detection of glucocorticoid receptor (GCR) nuclear translocation. In this assay, each compound was applied at a concentration of 10µM to H4 brain neuroglioma cells, incubated for one hour, then exposed to 1µM hydrocortisone for one hour to stimulate translocation of the GCR. Cells were then fixed and imaged in 3-channel fluorescence, with Hoechst to label the nucleus, CellMask Deep Red to delineate cell boundaries, and indirect immunofluorescence to detect GCR. From these images, our pipeline extracted 842-dimensional vectors for each compound representing the feature matrix X (Section Extracting Image-based Fingerprints).
The bioactivity matrix Y documents the available experimental activities of 524,371 imaged compounds in about 1,200 biochemical or cellular assays that can all be interpreted as an activity on a protein target. This also means that a single compound can be measured on multiple targets. The activity is expressed as the pXC50 of the given compound in the given assay. The pXC50 is defined as −log10 of the molarity concentration of the compound yielding a half-maximal effect in the experimentally measured dose-response curve. Typically, a given compound is measured in a handful of assays, such that Y is sparsely populated, i.e. has many missing values. In total, more than 10 million pXC50 values were available for the roughly 1,200 prediction tasks, corresponding to a fill rate of Y of about 1.6%.
We evaluated all protein assays at four different thresholds of pXC50 (here activity is defined as a pXC50 value exceeding the threshold), namely 5.5, 6.5, 7.5 and 8.5. We only used assay-threshold pairs with at least 25 actives and 25 inactives. For 535 assays, at least one pXC50 threshold resulted in a data subset meeting this criterion. In the step of selecting high quality models (Section Selection of High Quality Models) we used AUC-ROC higher than 0.9 as the cutoff. We additionally report results for a cutoff of 0.7.
Results of In-silico Experiments
Of the 535 assays, the described pipeline yielded 31 assays with high quality models using Macau (run for 2000 iterations, discarding the first 400 as burn-in) and 43 using DNN (f or details, including hyperparameter tuning, see Table S1). An AUC-ROC threshold of 0.7 yielded 6–7 times as many assays (Table 1).
Table 1.
The number of protein assays above the AUC-ROC threshold for machine learning methods Macau and deep neural networks (DNN).
| AUC-ROC threshold | Macau | DNN | Common |
|---|---|---|---|
| 0.9 | 31 (5.8%) | 43 (8.0%) | 26 (4.9%) |
| 0.7 | 218 (40.7%) | 245 (45.8%) | 209 (39.1%) |
The percentage is calculated relative to the total number of 535 assays. The Common column depicts the number of assays well predicted by both of the methods. Venn diagrams of the predicted targets are shown in Figure S2 and S3. The tested hyperparameters are described in Table S1 and S2. The mean AUC-ROC values for Macau, DNN, random forest and k-nearest neighbor are given in Table S3.
Both methods can successfully repurpose the original GR HTI assay for predicting activity towards more than 30 unrelated protein targets (AUC-ROC > 0.9), and provide models of sufficient quality to enrich compound sets for (or deplete them of) activity towards a further 200 targets (AUC-ROC > 0.7). Therefore, the image-based fingerprinting of HTI assays prove a rich and hitherto untapped source of information on biological activity, which can be picked up by several machine learning methods. This means that if computational resources are restricted one can use non deep learning methods like Macau or random forest with closely comparable performance (see Table S3 for mean AUC-ROC values for Macau and random forest).
Results for In-vitro Validation
As the Macau results were readily available during the early phase of the research, we proceeded with the in-vitro validation using these models. Among these 31 assays with high-quality predictions from Macau, two were connected to ongoing discovery projects: one oncology project and one central nervous system (CNS) project. For these two projects, we selected compounds for testing.
Results for Oncology Project
For the oncology project, the target was a kinase with no known direct relation to the glucocorticoid receptor. Using our Macau model, we ranked about 60,000 compounds tested in the GR assay but for which no activity measurement was available in the oncology screen. We selected 342 highest ranking compounds for experimental follow-up (Section Compound Selection for In-vitro Testing). We found that 124 of them were submicromolar (XC50 < 1µM) hits (36.3% hit rate), which corresponds to a 50-fold enrichment over the initial high-throughput screen (0.725% hit rate).
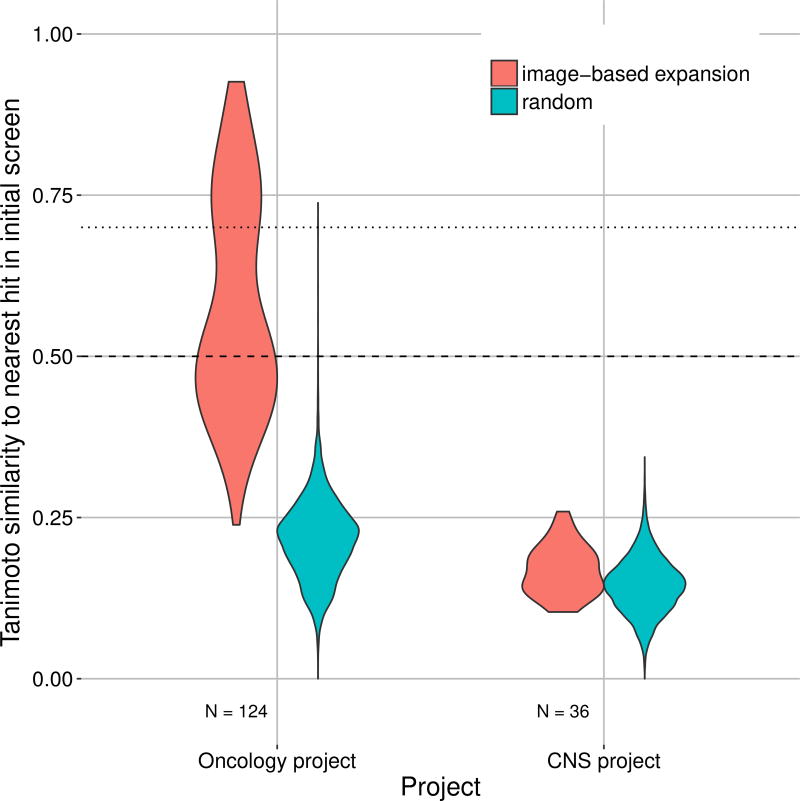
To evaluate the chemical diversity of the hits, we computed the Tanimoto similarity (based on extended-connectivity fingerprints (ECFP), (Rogers and Hahn, 2010)) of each hit to the nearest hit identified by the initial high-throughput screen (red distribution in Figure 5). 70% of the hits are below the 0.7 similarity line, and a significant proportion is even below 0.5. Per definition, a similarity search based on the initial hits would rarely yield analogs below the 0.7 line, and extremely rarely below the 0.5 line. We also found 108 novel Murcko scaffolds among the new hits (Table 2). Together these two facts imply that our repurposing pipeline can result in a hit set with high chemical diversity. For reference, the figure also shows the distribution for randomly selected compounds (blue distribution in Figure 5).
Figure 5. Image-based Profiling Strategy Yields More Chemically Diverse Compounds than Would be Expected for Chemical Extension.
In an oncology project (left) and a CNS project (right), we calculated the ECFP (radius 4) based Tanimoto similarity of each hit to the nearest hit identified by the initial high-throughput screen (red). For reference, the blue distributions show the similarity of randomly selected compounds to the closest hit identified by the initial high-throughput screen. Note that in the CNS project, unlike the oncology project, the selection procedure involved an additional step to reduce representatives from the same chemical-structural class. The horizontal dotted lines depict the 0.5 and 0.7 similarity levels.
Table 2.
Number of Murcko Scaffolds in the Two Follow-ups.
| Project name | #Murcko of initial screen |
#Murcko of new hits (novel / all) |
#New hits |
|---|---|---|---|
| Oncology | 2660 | 108 / 117 | 124 |
| CNS | 57 | 34 / 34 | 36 |
Additionally, we compared the top ranked compounds to those retrieved by chemical fingerprint based approaches. Specifically, we used the exact same activity data with chemical structure based features (ECFP) to train a Macau model and then ranked the untested 60,000 compounds. From the above mentioned top 342 compounds ranked by the image features 113 (33%) compounds were retrieved by ECFP model in its top 342. From the 124 actives 44 (35%) compounds were found in the top 342 of ECFP model. Moreover, to identify all 124 active compounds using the ECFP ranking one would need to test more than 21% of the 60,000 candidate compounds, i.e., at least 13,000 compounds. This shows that the image-fingerprints clearly provided additional source of information that is not encoded in the chemical fingerprints.
Results for CNS Project
For the CNS project, the target was a non-kinase enzyme, again without obvious relation to the glucocorticoid receptor. Using our Macau model, activity was predicted for all 500,000 image-annotated compounds and we selected all compounds with submicromolar prediction, resulting in 1,715 compounds. Next, we kept only compounds without unfavorable properties, like PAINS filter (Baell and Holloway, 2010) and low predicted central nervous system availability (Methods S1). For this project, to maximize compound diversity, we employed the selection strategy of grouping the remaining compounds into clusters based on structure, using sphere exclusion clustering with similarity cutoff 0.7 (Section Compound Selection for In-vitro Testing). We then selected a handful of representatives from each cluster resulting in 141 compounds. We experimentally tested them and found that 36 of them were submicromolar hits (25.5% hit rate), which corresponds to a 280-fold enrichment over the hit rate of the initial high-throughput screen (0.088% rate). These compounds were highly diverse (Tanimoto similarity < 0.3; Figure 5) while maintaining a relatively high hit rate. The 36 hits resulted in 34 novel Murcko scaffolds (Table 2).
DISCUSSION
In this work, we have demonstrated that HTI data enables the identification of diverse hits without the need to test the entire library in the target assay. By accessing rich morphological features of the cell, imaging screens capture diverse cellular processes, resulting in a fingerprint of biological action. Our results indicate that images from HTI screening projects that are conducted in many institutions can be repurposed for dramatically reducing the scale of screens required for other projects, even those that seem unrelated to the primary purpose of the HTI screen.
We emphasize that our approach relies on a supervised machine learning method and hence activity measurements and imaging data must be acquired for a reasonably sized library of compounds to train the model. Subsequently, however, it seems possible to replace many particular assays with the potentially more cost-efficient imaging technology together with machine learning models. Specifically, one would execute one or a few image screens on the library instead of dozens of target-focused assays. This raises an interesting question of the breadth of drug targets that could be accessed by imaging screens if the screen were optimized for that purpose, or if a combination of screens was used that explored multiple cell lines or sources, culturing conditions, staining of organelles and/or incubation times.
We leave for future work the head-to-head comparison of chemistry-based and image-based fingerprints, but can speculate based on our results. In the case of a well-covered chemical space we would not expect image-based fingerprints to outperform a well-designed chemical fingerprint like ECFP. For example, if the compound in question has several close enough neighbors we expect chemical fingerprints to prove predictively performant. In contrast, we expect the performance of image-based fingerprints that do not depend on chemical closeness to be superior for scaffold hopping, i.e., identifying active compounds with novel backbones, given it does not depend on the chemical closeness. Evidence for this idea includes the high chemical diversity of active compounds and the ability to pick up actives that are not detect even by chemistry-based machine learning (Section Results for Oncology Project). Moreover, image-based fingerprinting is a feasible approach to predict the activity of not just small molecule compounds, but any agent, such as antibodies, RNA interference agents or other biologics.
We also anticipate that improvements in the computational pipeline may increase the power of the method. For example, convolutional neural networks could predict activity from raw images directly rather than from features extracted from each cell using classical image processing. This would allow the model to learn the best image features for the specific task at hand and may improve results. Another future direction is to maintain the native single-cell resolution of image-based profiles instead of aggregating values. Finally, our current results are based on a single HTI screen and we envision that data fusion across a collection of multiple HTI screens could even be more powerful for assay activity prediction. We leave as a follow up work to investigate how adding new HTI assays improves the predictive performance.
Our results also encourage the creation of a sufficiently large public datasets of compounds annotated with chemical structures, activity measurements in validated assays and images. While a few efforts have publicly documented up to about 30,000 compounds with cellular images (Wawer et al., 2014), only a tenth of the compounds have been annotated with some assay activities, yielding a very sparse annotation matrix.
Supplementary Material
SIGNIFICANCE.
High-throughput imaging is an affordable screening technology most often used to read out a handful of morphological features that document a single biological process of interest. Leveraging access to a large private set of activity and image-annotated compounds, we here establish proof-of-concept that images from one given cellular assay support activity prediction across a spectrum of seemingly unrelated biological assays. Hence, images can inform on biological activity far beyond the intended focus of the original screen. Once a chemical library is documented with image-based fingerprints, a medium scale screening in an expensive or tedious assay may suffice to train an image-based model that can predict the outcome for the rest of the library and enable cost-effective targeted experimental validation. Effective predictive approaches that rely on the chemical structure of compounds are well established in the context of the gradual virtualization of screening and drug discovery. Our study suggests image-based approaches can complement these structure-based ones, particularly in those cases where the latter suffer from chemical biases in training data. Moreover, they can extend predictive modeling options to agents with (bio)chemistry that elude standard structure-based approaches, like antibodies, RNA interference agents and other biologics. Importantly, given that the field of structure-based prediction already exploits decades of optimization and research, the pace of predictive performance gain has slowed down. In contrast, advancements like convolutional neural networks have recently boosted the performance of generic image-based machine learning. The proof of concept described in this paper justifies further research in optimizing the specific application of image-based machine learning in drug discovery. Future lines of research may aim to maximize the generic informativity by screen design or by data fusion over pre-existing screens that cover a broader range of biological contexts, and to improve feature extraction and additional learning from microscopy images.
Acknowledgments
This work was supported by research grants IWT135122 ChemBioBridge, IWT130405 ExaScience Life Pharma and IWT150865 Exaptation from the Flanders Innovation and Entrepreneurship agency. The NVIDIA Corporation generously donated a GPU. J.S., A.A., and Y.M. were additionally supported by Research Council KU Leuven: CoE PFV/10/016 SymBioSys, PhD grants and imec strategic funding 2017.
P.B., H.C., Y.T.C., E.G., A.V., J.V., I.V. and J.K.W. are or were at the time of writing employees of Janssen Pharmaceutica N.V. A.A., G.K., J.S., M.S., S.S. and R.W. are funded for industrial collaboration with Janssen Pharmaceutica N.V.
Footnotes
The C++/Python package is available at https://github.com/jaak-s/macau
AUTHOR CONTRIBUTIONS
J.S., G.K., A.A., S.H., Y.M., and H.C. conceived this study and designed the experiments. J.S., G.K., A.A., M.S., J.K.W, E.G., V.C., Y.T.C., J.V., P.B., I.V., A.V., S.S., A.E.C., R.W., and H.C. conducted the experiments and interpreted data. S.H., Y.M., and H.C. supervised this project. J.S., G.K., A.A., A.E.C., and H.C. wrote the manuscript with input from all authors.
DECLARATION OF INTERESTS
The authors declare no competing interest.
References
- Ansbro MR, Shukla S, Ambudkar SV, Yuspa SH, Li L. Screening compounds with a novel high-throughput ABCB1-mediated efflux assay identifies drugs with known therapeutic targets at risk for multidrug resistance interference. PLoS One. 2013;8:e60334. doi: 10.1371/journal.pone.0060334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of medicinal chemistry. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Baumann D, Baumann K. Reliable estimation of prediction errors for QSAR models under model uncertainty using double cross-validation. Journal of cheminformatics. 2014;6:47. doi: 10.1186/s13321-014-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Machine learning. 2001;45:5–32. [Google Scholar]
- Caicedo JC, Singh S, Carpenter AE. Applications in image-based profiling of perturbations. Current opinion in biotechnology. 2016;39:134–142. doi: 10.1016/j.copbio.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana R. Multitask Learning. Machine Learning. 1997;28:41–75. [Google Scholar]
- Cumming JG, Davis AM, Muresan S, Haeberlein M, Chen H. Chemical predictive modelling to improve compound quality. Nature reviews Drug discovery. 2013;12:948. doi: 10.1038/nrd4128. [DOI] [PubMed] [Google Scholar]
- Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. Journal of medicinal chemistry. 2000;43:3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- Evensen L, Link W, B Lorens J. Imaged-based high-throughput screening for anti-angiogenic drug discovery. Current pharmaceutical design. 2010;16:3958–3963. doi: 10.2174/138161210794455030. [DOI] [PubMed] [Google Scholar]
- Garg P, Verma J. In silico prediction of blood brain barrier permeability: an artificial neural network model. Journal of chemical information and modeling. 2006;46:289–297. doi: 10.1021/ci050303i. [DOI] [PubMed] [Google Scholar]
- Gustafsdottir SM, Ljosa V, Sokolnicki KL, Wilson JA, Walpita D, Kemp MM, Seiler KP, Carrel HA, Golub TR, Schreiber SL. Multiplex cytological profiling assay to measure diverse cellular states. PloS one. 2013;8:e80999. doi: 10.1371/journal.pone.0080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held M, Schmitz MH, Fischer B, Walter T, Neumann B, Olma MH, Peter M, Ellenberg J, Gerlich DW. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nature methods. 2010;7:747–754. doi: 10.1038/nmeth.1486. [DOI] [PubMed] [Google Scholar]
- Hochreiter S, Obermayer K. 15 Gene Selection for Microarray Data. Kernel methods in computational biology. 2004:319. [Google Scholar]
- Louppe G. Understanding random forests: From theory to practice. arXiv preprint arXiv:14077502 2014 [Google Scholar]
- Mayr A, Klambauer G, Unterthiner T, Hochreiter S. DeepTox: Toxicity Prediction using Deep Learning. Frontiers in Environmental Science. 2016;3 [Google Scholar]
- Oshiro TM, Perez PS, Baranauskas JA. How many trees in a random forest? Paper presented at: MLDM (Springer) 2012 [Google Scholar]
- Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nature reviews Molecular cell biology. 2006;7:690. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- Polishchuk PG, Muratov EN, Artemenko AG, Kolumbin OG, Muratov NN, Kuz’min VE. Application of random forest approach to QSAR prediction of aquatic toxicity. Journal of chemical information and modeling. 2009;49:2481–2488. doi: 10.1021/ci900203n. [DOI] [PubMed] [Google Scholar]
- Rogers D, Hahn M. Extended-connectivity fingerprints. Journal of chemical information and modeling. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Simm J, Arany A, Zakeri P, Haber T, Wegner JK, Chupakhin V, Ceulemans H, Moreau Y. Macau: Scalable Bayesian Factorization with High-dimensional side information using MCMC; Proceeding of 2017 IEEE International Workshop on Machine Learning for Signal Processing.2017. [Google Scholar]
- Singh S, Carpenter AE, Genovesio A. Increasing the content of high-content screening: an overview. Journal of biomolecular screening. 2014;19:640–650. doi: 10.1177/1087057114528537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava N, Hinton GE, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. Journal of machine learning research. 2014;15:1929–1958. [Google Scholar]
- Starkuviene V, Pepperkok R. The potential of high-content high-throughput microscopy in drug discovery. British journal of pharmacology. 2007;152:62–71. doi: 10.1038/sj.bjp.0707346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter T, Shattuck DW, Baldock R, Bastin ME, Carpenter AE, Duce S, Ellenberg J, Fraser A, Hamilton N, Pieper S. Visualization of image data from cells to organisms. Nature methods. 2010;7:S26–S41. doi: 10.1038/nmeth.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xing J, Xu Y, Zhou N, Peng J, Xiong Z, Liu X, Luo X, Luo C, Chen K. In silico ADME/T modelling for rational drug design. Quarterly reviews of biophysics. 2015;48:488–515. doi: 10.1017/S0033583515000190. [DOI] [PubMed] [Google Scholar]
- Wawer MJ, Li K, Gustafsdottir SM, Ljosa V, Bodycombe NE, Marton MA, Sokolnicki KL, Bray M-A, Kemp MM, Winchester E. Toward performance-diverse small-molecule libraries for cell-based phenotypic screening using multiplexed high-dimensional profiling. Proceedings of the National Academy of Sciences. 2014;111:10911–10916. doi: 10.1073/pnas.1410933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MN, Ziegler A. ranger: A fast implementation of random forests for high dimensional data in C++ and R. arXiv preprint arXiv:150804409 2015 [Google Scholar]
- Yarrow J, Feng Y, Perlman Z, Kirchhausen T, Mitchison T. Phenotypic screening of small molecule libraries by high throughput cell imaging. Combinatorial chemistry & high throughput screening. 2003;6:279–286. doi: 10.2174/138620703106298527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.