ABSTRACT
The cell biology field has outstanding working knowledge of the fundamentals of membrane-trafficking pathways, which are of critical importance in health and disease. Current challenges include understanding how trafficking pathways are fine-tuned for specialized tissue functions in vivo and during development. In parallel, the ENCODE project and numerous genetic studies have revealed that alternative splicing regulates gene expression in tissues and throughout development at a post-transcriptional level. This Review summarizes recent discoveries demonstrating that alternative splicing affects tissue specialization and membrane-trafficking proteins during development, and examines how this regulation is altered in human disease. We first discuss how alternative splicing of clathrin, SNAREs and BAR-domain proteins influences endocytosis, secretion and membrane dynamics, respectively. We then focus on the role of RNA-binding proteins in the regulation of splicing of membrane-trafficking proteins in health and disease. Overall, our aim is to comprehensively summarize how trafficking is molecularly influenced by alternative splicing and identify future directions centered on its physiological relevance.
KEY WORDS: RNA-binding proteins, Alternative splicing, Membrane dynamics, Trafficking
Summary: This Review summarizes how trafficking is molecularly influenced by alternative splicing and how this interplay impacts physiology and pathological conditions.
Introduction
Membrane trafficking controls multiple cellular functions. Trafficking to and from the plasma membrane modulates cell communication during organ development and function. Trafficking from the cell exterior comprises internalization of ion channels, receptors and ligands to control homeostasis and signaling. Trafficking from the inside controls the transport of newly synthetized proteins from the endoplasmic reticulum to their final destinations. Numerous human diseases are caused by mutations in membrane-trafficking genes (Dowling et al., 2008; Sigismund et al., 2012), highlighting the physiological importance of these proteins.
Membrane-trafficking genes are developmentally and tissue-specifically regulated by alternative splicing (Brinegar et al., 2017; Dillman et al., 2013; Giudice et al., 2014; Hannigan et al., 2017; Irimia et al., 2014), a post-transcriptional mechanism used by single genes to produce multiple transcripts and, thus, several protein isoforms with different features. Brain, heart and skeletal muscle, together with the testes, are the organs where most of the tissue-specific and conserved alternative splicing takes place (Merkin et al., 2012). Tissue-specific exons encode disordered segments within proteins that function in microtubule-based transport, endocytosis and membrane deformation (Buljan et al., 2012; Ellis et al., 2012) (Box 1).
Box 1. Alternative splicing of trafficking proteins is tissue specific and affects short disordered motifs.
Brain, heart and skeletal muscle, as well as the testes, are the organs where most of the tissue-specific and conserved alternative splicing takes place (Merkin et al., 2012). Tissue-specific alternative exons encode regions of proteins that tend to be centrally located within protein-protein interaction networks (Buljan et al., 2012; Ellis et al., 2012) and comprise short intrinsically disordered motifs that confer functional diversity onto splice variants (Weatheritt and Gibson, 2012; Weatheritt et al., 2012). In general, the pairs of variants tend to behave like distinct proteins in terms of their interaction with other proteins, and the interaction partners that are specific to each splice isoform tend to be highly tissue specific (Yang et al., 2016). Tissue-specific alternative exons are present in genes encoding proteins that control microtubule-based transport, endocytosis, membrane deformation and endosome formation (Buljan et al., 2012; Ellis et al., 2012). Table S2 summarizes several of these tissue-specific alternative exons in membrane-trafficking genes that are predicted to impact protein-protein interactions and below we highlight two examples that have been experimentally demonstrated.
(1) Growth factor receptor bound protein-2 (GRB2) is involved in internalization of receptor tyrosine kinases. GRB2 homodimerizes through an interaction between its SH2 and SH3 domains (Maignan et al., 1995), which is important for signaling (McDonald et al., 2008). Exon 4 of GRB2 is alternatively spliced and encodes a region that overlaps the SH2 domain. When exon 4 is skipped, the self-interaction of GRB2 is lost, suggesting that alternative splicing controls GRB2 homodimerization and thus signaling activity (Ellis et al., 2012).
(2) Kinesin-1 is a molecular motor protein associated with microtubules; it is formed by two heavy chains that control motor activity and two light chains (KLC1 and KLC2) involved in cargo binding. KLC1 contains five alternative exons (13-17). Inclusion of exon 15 reduces the interaction of KLC1 with Marlin1, which controls transport of the GABA-B receptor towards dendrites (Vidal et al., 2007). Therefore, regulation of exon 15 splicing might control distribution of the GABA-B receptor, and thus neurotransmission (Ellis et al., 2012).
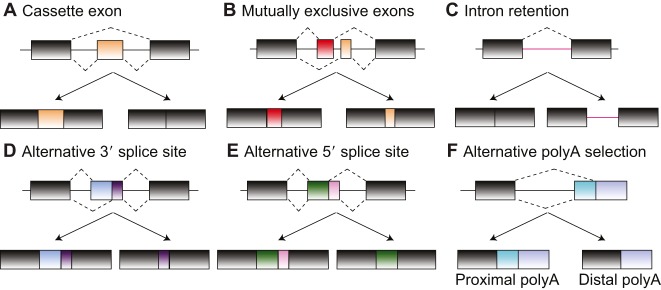
In humans, 90-95% of genes undergo alternative splicing, expanding protein function beyond genetic diversity (Pan et al., 2008; Wang et al., 2008). Splicing of intronic regions is regulated by the strength of the splice sites; strong splice sites lead to constitutive splicing, whereas weak splice sites are used in a context-dependent manner (alternative splicing). Usage of weak splice sites is regulated by cis-regulatory sequences, trans-acting factors such as RNA-binding proteins (RBPs) and epigenetics (Kornblihtt et al., 2013). Depending on splice site locations, different types of alternative splicing events are produced, which comprise insertion of alternative cassette exons or mutually exclusive exons, selection between alternative 5′- or 3′-splice sites, poly-adenylation sites and intron retention (Fig. 1). Alternative splicing can dramatically impact protein function or affect the expression, localization, and/or stability of mRNAs (Irimia and Blencowe, 2012). Coordination of alternative splicing contributes to cell differentiation, lineage determination, tissue identity acquisition and, ultimately, organ development (Baralle and Giudice, 2017; Wang et al., 2008). The physiological relevance of splicing is evident from the vast number of mutations in cis-regulatory elements, RBPs or spliceosome components, which cause a broad spectrum of human diseases (Scotti and Swanson, 2016).
Fig. 1.
Types of alternative splicing events. The types of splicing events are defined by the location of the alternative splice sites. (A) Cassette exons can be either included or spliced out. (B) Mutually exclusive exons are consecutive exons that are included in a mutually exclusive manner, i.e. when one of them is included the other one is skipped and vice versa. (C) An intron or a portion of an intronic region can be included in the mRNA. (D,E) When more than one splice site exists at the end or beginning of an exon, alternative 5′ or 3′ splice sites can be selected. (F) The polyA tail at the 3′ end of an RNA can be added in some genes at different poly-adenylation sites (polyA), leading to the production of more than one transcript. Constitutive exons are shown in gray boxes, introns as lines and alternative regions as colored boxes.
Here, we review the molecular connection between alternative splicing and membrane trafficking from both physiological and disease perspectives. First, we discuss how alternative splicing impacts membrane-trafficking proteins involved in clathrin-mediated endocytosis (CME), secretory pathways and membrane dynamics. Then, we discuss the role of RBPs in controlling alternative splicing of trafficking proteins and how this regulation contributes to health and disease. The final section identifies the major questions still outstanding in the field.
Alternative splicing regulation and CME
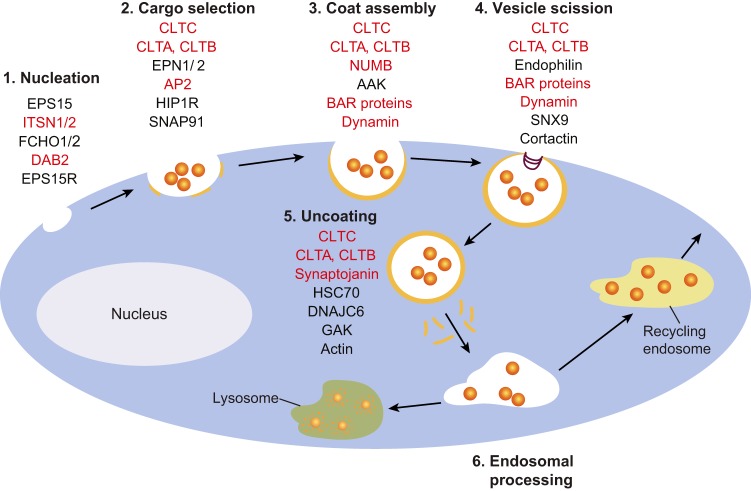
CME is one of the most common mechanisms that cells employ to absorb nutrients, hormones or proteins from the exterior and involves clathrin-coated vesicles. In addition, CME regulates the protein content of the plasma membrane, monitors external cues from the surrounding environment, modulates signaling pathways and directs protein recycling and degradation (McMahon and Boucrot, 2011). Loss of function of central components of the CME machinery, such as clathrin, AP2, epsin or dynamin, is embryonically lethal and alterations in other CME proteins are often present in cancer, neurological disorders, genetic syndromes and muscle pathologies. CME occurs in multiple steps (Fig. 2): (1) the formation of clathrin-coated vesicles starts by membrane invagination (nucleation); (2) proteins are recruited to the nucleation site by the AP2 complex, which together with other cargo-specific adaptors mediates cargo selection; (3) clathrin polymerization stabilizes the curvature of the forming vesicle; (4) dynamin, endophilin and BAR-domain proteins drive vesicle scission from the membrane; (5) vesicles are uncoated to fuse with target endosomes; (6) cargo is transported through early endosomes into either recycling vesicles, or late endosomes and lysosomes for degradation (McMahon and Boucrot, 2011). Several of the CME proteins undergo alternative splicing (highlighted in red in Fig. 2) and some are discussed below.
Fig. 2.
Regulatory mechanisms for alternative splicing impact numerous components of the CME. CME occurs in multiple steps: (1) clathrin-coated vesicles are formed; (2) proteins are recruited to the nucleation site by the AP2 complex, which together with other cargo-specific adaptors mediates cargo selection; (3) the clathrin triskelion facilitates vesicle assembly and clathrin polymerization stabilizes the curvature of the forming vesicle; (4) dynamin, endophilin and BAR-domain proteins drive vesicle scission from the membrane; (5) vesicles are uncoated to fuse with target endosomes; (6) cargo is transported through early-endosomes into either recycling vesicles, or late endosomes and lysosomes for degradation. Alternative splicing regulates several of the proteins that control different CME steps, and the known splicing-regulated proteins are shown in red. AAK, 5′-AMP-activated protein kinase catalytic subunit α-1; AP2, adaptor-related protein complex-2; CLTA, clathrin light chain-a; CLTB, clathrin light chain-b; CLTC, clathrin heavy chain; DNAJC6, DNAJ heat shock protein family (Hsp40) member C6; DAB2, disabled-2; EPN1/2, epsin-1 and epsin-2; EPS15, epidermal growth factor receptor pathway substrate-15; EPS15R, epidermal growth factor receptor pathway substrate 15 recombinant; FCHO1/2, FCH-domain only 1 and FCH-domain only 2; GAK, Cyclin G-associated kinase; HIP1R, Huntington interacting protein 1 related; HSC70, heatshock protein family A (Hsp70) member 8; ITSN1/2, intersectin- 1 and intersectin-2; NUMB, endocytic adapter protein; SNAP91, synaptosome-associated protein-91; SNX9, sorting nexin 9.
Clathrin chains
Non-assembled clathrin comprises a three-legged structure called a triskelion (Unanue et al., 1981), which is formed by three heavy chain polypeptides bound to light chain subunits arranged along the triskelion leg (Brodsky, 2012). Humans have two genes encoding clathrin heavy chain, CLTC (also known as CHC or CHC17) and CLTCL1 (also known as CHC22) (see Table S1, for a list of official and alternative gene and protein names) (Kedra et al., 1996; Vassilopoulos et al., 2009). CLTC predominates, except in skeletal muscle where the two proteins are equally expressed.
By self-assembly, the triskelia form a polyhedral lattice that coats the transport vesicles (Brodsky et al., 2001; Ungewickell and Hinrichsen, 2007). While CLTC constitutes the backbone of the vesicle lattice, the light chains regulate clathrin recruitment in the cell (Majeed et al., 2014). In humans and mice, the CLTC gene contains 33 exons, of which exon 31 is postnatally regulated by alternative splicing in cardiomyocytes and skeletal muscles, with it being skipped in neonates and included in adults (Brinegar et al., 2017; Giudice et al., 2014, 2016). CLTC regulates the formation and maintenance of myofiber architecture (Vassilopoulos et al., 2014); thus, the tissue-specificity of CLTC splicing suggests that it might be important for muscle structure. Alternative splicing is not limited to CLTC, but also regulates the clathrin light chain genes CLTA and CLTB (also known as LCA/CLCa and LCB/CLCb, respectively). In comparison with the ubiquitous isoforms, brain splice variants of CLTA contain 18 or 30 additional residues, and a further different but homologous 18 residues were found in CLTB (Jackson et al., 1987; Kirchhausen et al., 1987). Alternative splicing regulates CLTA exon 5, which encodes the 18 residues and CLTA exon 6, which encodes the 12 residues, as well as CLTB exon 5, which encodes 18 residues. In mouse hearts, Clta exon 6 is skipped in neonates and included in adults, whereas Clta exon 5 and Cltb exon 6 are skipped at all developmental stages (Giudice et al., 2014). Although CLTA and CLTB are regulated by alternative splicing in a tissue- and developmental-stage-specific manner, we still lack a complete description of the tissue expression patterns of transcript and protein isoforms and their functions.
Dynamin and DAB2
In addition to the clathrin triskelion, other CME proteins are also alternatively spliced, including the adaptor protein disabled-2 (DAB2), which controls endocytosis of the low-density lipoprotein receptor family (Keyel et al., 2006; Maurer and Cooper, 2006). DAB2 has two tissue-specific splice isoforms known as P96 and P67 (Xu et al., 1995). P96 contains two clathrin-binding sites and one AP2-binding site, which are absent in P67, explaining why P96 binds to clathrin and AP2 and localizes at clathrin-coated pits, whereas P67 does not (Buljan et al., 2012; Morris and Cooper, 2001). DAB2 depletion results in alterations in the surface levels of integrins that give rise to migration and polarization defects, which can be rescued by re-expression of P96, but not P67 (Teckchandani et al., 2009).
In mammals, there are three dynamin genes (DNM1, DNM2 and DNM3), which encode members of a family of GTPases involved in membrane fission and the release of clathrin-coated vesicles from the plasma membrane. The three genes are highly regulated by alternative splicing, giving rise to more than 25 splice isoforms. The neuron-specific DNM1 is essential for synaptic vesicle endocytosis and is alternatively spliced at two regions: the middle domain and the C-terminus. Splicing results in either DNM1ax or DNM1bx isoforms, where ‘a’ and ‘b’ are the middle domain variants and ‘x’ is one of the three alternative terminal exons. In mice, a spontaneous missense mutation (fitful) (p.A408T) that only affects the DNM1ax isoforms (the ‘a’ exon is spliced out in the DNM1bx variants) alters endocytosis, DNM1 self-assembly, which is required for dynamin function and, ultimately, synaptic transmission (Boumil et al., 2010).
DNM2 is ubiquitously expressed and has several splice variants. In particular, two alternative regions generate four splice variants, DNM2aa, DNM2ab, DNM2ba and DNM2bb (Cao et al., 1998; Cook et al., 1994; Sontag et al., 1994). All isoforms localize to clathrin-coated pits at the plasma membrane but only DNM2ba and DNM2bb localize to the Golgi. All isoforms rescue the endocytosis defects in Dyn2-depleted cells, but DNM2ba and DNM2bb were more effective than the DNM2aa and DNM2bb variants with regard to rescuing the export of the neutrophin receptor p75 from the trans-Golgi network (Liu et al., 2008).
In summary, the regulation of CME factors by alternative splicing has been demonstrated to be tissue- and developmental-stage-specific. However, further studies are required to better describe the molecular details of splicing events and their physiological implications.
Alternative splicing of SNARE proteins
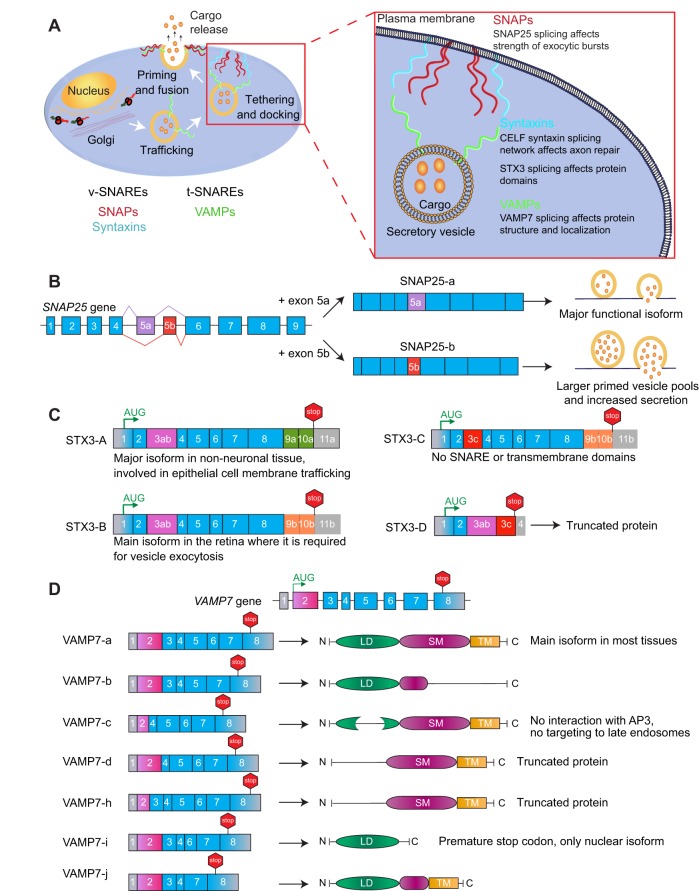
The SNARE machinery controls the interaction between vesicular v-SNAREs (VAMPs) and target t-SNAREs (such as SNAPs and syntaxins) during vesicle fusion (Fig. 3A). Several components of the SNARE machinery undergo alternative splicing, as discussed below.
Fig. 3.
Alternative splicing of SNARE-related proteins. (A) Steps of vesicle-mediated exocytosis that require the assembly of the SNARE complex. The SNARE machinery controls the interaction between vesicular v-SNAREs and target t-SNAREs during vesicle fusion. (B-D) Multiple components of the SNARE machinery undergo alternative splicing, including synaptosome associated protein-25 (SNAP25) (B), syntaxin-3 (STX3) (C) and vesicle associated membrane protein-7 (VAMP7) (D). CELF, CUGBP Elav-like family member-1; LD, Longin domain; SM, SNARE motif; TM, transmembrane domain.
SNAPs
Synaptosome-associated protein-25 (SNAP25) bridges synaptic vesicles to the plasma membrane during exocytosis. In higher vertebrates, two mutually exclusive exons, 5a and 5b, give rise to the SNAP25-a and SNAP25-b isoforms (Bark and Wilson, 1994) (Fig. 3B). These variants differ by nine centrally located residues, two of which alter the relative positioning of clustered cysteines whose palmitoylation controls membrane anchoring (Vogel and Roche, 1999). Snap25 null mice exhibit defects in primed vesicle pools and fast calcium-triggered release in chromaffin cells. Here, re-expression of SNAP25-b results in larger primed vesicle pools compared with those seen upon re-expression of SNAP25-a, suggesting that alternative splicing regulates the ability of SNAP25 to stabilize primed vesicles (Sørensen et al., 2003). A developmental switch from the fetal SNAP25-a isoform to adult SNAP25-b occurs in brain (Bark et al., 1995), and its impairment induces lethality in mice three to five weeks after birth (Bark et al., 2004). The exclusive expression of the SNAP25-a isoform through replacement of exon 5b with a copy of exon 5a leads to developmental defects, spontaneous seizures, impaired short-term synaptic plasticity, morphological alterations in adult hippocampus, changes in neuropeptide expression and impairment of spatial learning (Johansson et al., 2008). Furthermore, when provided with a high-fat diet, these mice develop a metabolic syndrome accompanied by weight gain, thus linking the neuronal defect in the exocytosis machinery with dyslipidemia and disrupted glucose homeostasis (Valladolid-Acebes et al., 2015). A lack of the SNAP25-b isoform in endocrine β-cells increases insulin secretion and alters calcium dynamics (Daraio et al., 2017). Recently, hippocampal lysates have revealed that SNAP25-a is less efficient than SNAP25-b in forming complexes with Munc18-1 and the Gβ1 and Gβ2 subunits of heterotrimeric G-proteins. As these interactions play important roles in presynaptic inhibition, the results suggest a less inhibitory role of SNAP25-a (Daraio et al., 2018).
SNAP23 is a SNAP25 homologue that is almost ubiquitously expressed and has been implicated in insulin sensitivity (Boström et al., 2007), lipid-raft actions (Puri and Roche, 2006; Yoon et al., 2016) and endothelial exocytosis of the von Willebrand factor (Zhu et al., 2015). The human SNAP23 gene contains eight exons and different splice isoforms have been reported; for instance, SNAP23-a differs from SNAP23-b in 53 residues encoded by exon 6 (Mollinedo and Lazo, 1997). This region contains sites for post-translational fatty acid acylation, suggesting differences in membrane interaction between the isoforms. In vitro, both isoforms bind to syntaxin-6 (STX6), but SNAP23-b has an apparently higher affinity (Lazo et al., 2001). In addition, the SNAP23-c variant lacks exons 5 to 7, giving rise to a truncated 50-residue protein. Another isoform, SNAP23-d, lacks exons 6 and 7, and, here, a frame-shift changes the C-terminus of the protein. The SNAP23-e isoform also lacks exons 6 and 7, but it uses an alternative 5′-splice site within exon 8 and shares the same last eight residues as SNAP23-a and SNAP23-b (Shukla et al., 2001). All isoforms, except SNAP23-c, contain the cysteine-rich domain that is palmitoylated. Transfected SNAP23-a and SNAP23-b mainly localize to the plasma membrane, while the other isoforms also exhibit intracellular localizations (Shukla et al., 2001).
Syntaxins
In the brain, alternative splicing of syntaxins is regulated by the CUGBP Elav-like family (CELF) of RBPs. In Caenorhabditis elegans, the CELF homolog UNC-75 promotes the expression of the neuronal syntaxin isoform UNC-64A, which includes exon 8a, and represses the non-neuronal variant UNC-64B, which includes exon 8b. These isoforms differ in their C-terminal hydrophobic membrane anchors (Ogawa et al., 1998; Saifee et al., 1998). The mutants lacking unc-75 only express UNC-64B and show axon regeneration and locomotion defects similar to those observed in unc-64 loss-of-function mutants. While overexpression of either UNC-64A or UNC-64B isoforms rescues axon regeneration defects, only UNC-64A rescues locomotion defects (Chen et al., 2016; Norris et al., 2014). CELF proteins also control axon regeneration in rodents. The Celf2-knockout mouse exhibits axon regeneration defects and mis-splicing of several syntaxins (Stx1a, Stx2, Stx16 and Stx18) and syntaxin-binding proteins (Stxbp1, Stxbp2 and Stxbp5), suggesting that CELF-regulated splicing of syntaxins is an evolutionarily conserved mechanism involved in axon regeneration (Chen et al., 2016; Norris et al., 2014).
STX3 is highly expressed in spleen, lung, kidney, retina and brain, and functions in vesicle trafficking. The mouse Stx3 gene contains several mutually exclusive exons: 3ab and 3c, 9a and 9b, 10a and 10b, and 11a and 11b, generating four splice variants (shown in Fig. 3C) that differ in their domain organization and biochemical properties (Ibaraki et al., 1995). STX3-A and STX3-B share the N-terminus but differ in the second half of the SNARE domain and the C-terminal transmembrane domain. In STX3-D, the inclusion of both exons 3ab and 3c introduces a premature stop-codon and thus produces a truncated protein (Curtis et al., 2008). STX3-A is the only isoform detected in the kidney with proposed functions in epithelial cell membrane trafficking, whereas STX3-B is the exclusive variant found in retinal ribbon synapses (Curtis et al., 2008) where it is required for synaptic vesicle exocytosis (Curtis et al., 2010). Recently, a translatome study in mouse retinal cells revealed that axons and soma use different last exons in the Stx3 gene, leading to differences in the C-terminus of the encoded STX3 proteins and the 3′UTR. Notably, the axon-specific exon is sufficient to promote axonal mRNA translation, providing evidence for a potential mechanism underlying the selective and dynamic mRNA translation in axons (Shigeoka et al., 2016).
VAMPs
Human VAMP7 (previously known as SYBL1) encodes the v-SNARE protein VAMP7, which controls intracellular trafficking (Galli et al., 1998). In the full-length VAMP7-a isoform, exons 2 to 4 encode the N-terminal Longin domain, which negatively regulates membrane fusion and neurite outgrowth (Martinez-Arca et al., 2000, 2001), whereas exons 5 to 8 encode the transmembrane region and the SNARE motif (Fig. 3D). Skipping of exon 3 and the use of an alternative 5′-splice site in exon 2 lead to the truncated proteins VAMP7-d and VAMP7-h, respectively (Vacca et al., 2011). VAMP7-c uses the same 5′-splice site in exon 2 as VAMP7-h, but lacks exon 3. Thus, in comparison with VAMP7-a, VAMP7-c lacks 40 residues within the Longin domain, losing the interaction with the adaptor protein AP3 and its targeting to late endosomes (Martinez-Arca et al., 2003). Splicing of exons 5 and 6 generates three additional isoforms with intact Longin domains, but altered SNARE domains. VAMP7-b lacks exon 6, and thus has a different C-terminus than VAMP7-a. In VAMP7-i, skipping of exon 5 introduces a premature stop-codon, and thus only the Longin domain is expressed. In VAMP7-j, skipping of both exons maintains the original reading frame; therefore, in addition to the Longin domain, VAMP7-j contains a short hinge region, and the original transmembrane and intra-vesicular tail regions (Vacca et al., 2011) (Fig. 3D). The different VAMP7 isoforms exhibit differential subcellular and tissue localizations. VAMP7-b is widely distributed, whereas VAMP7-i is the only nuclear isoform. Furthermore, VAMP7-a is the main isoform in most tissues, whereas other variants show some tissue specificity (Vacca et al., 2011).
Based on all the studies discussed above, it is clear that alternative splicing regulates different members of the SNARE machinery and so contributes to the control of exocytosis.
Alternative splicing impacts membrane dynamics
Cellular membranes are flexible and extensively remodeled during numerous processes, including cell movement, cell division, endocytosis, muscle contraction and repair, transverse tubule (T-tubule) and sarcomere formation, mitochondria fusion and fission, as well as axon and dendrite growth. During membrane remodeling, reversible curvature changes and areas of high tension exist for limited periods of time and are controlled by trafficking proteins, such as dynamin, amphiphysin, endophilin and epsin (McMahon and Gallop, 2005). BAR-domain proteins are cytoplasmic molecules with membrane-bending properties (Mim and Unger, 2012). Below, we describe two BAR-domain proteins that are regulated by alternative splicing: CDC42-interacting protein-4 (TRIP10, also known as CIP4) and bridging integrator protein-1 (BIN1, also known as amphiphysin-2).
TRIP10
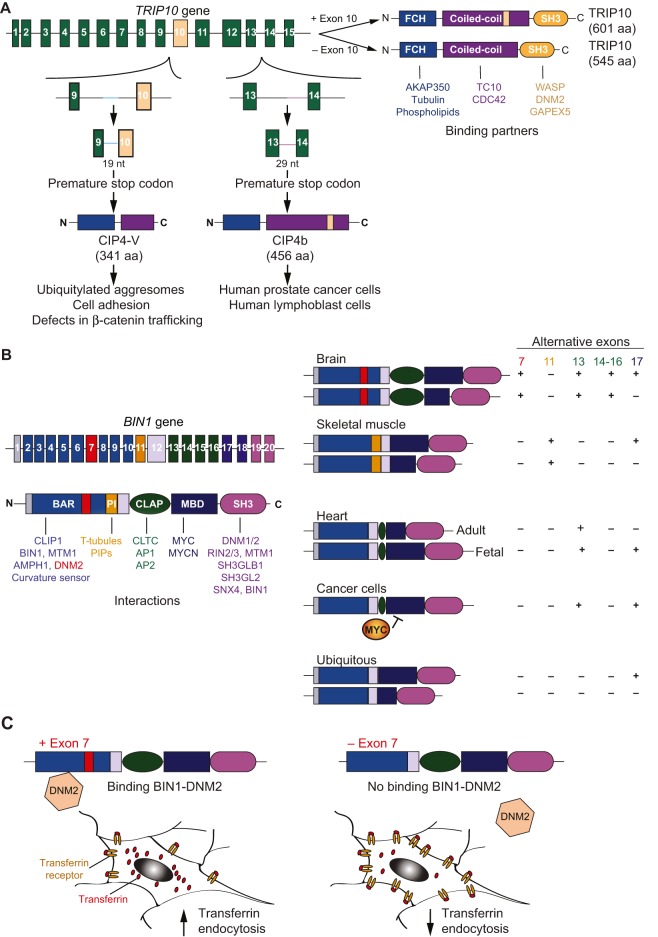
TRIP10 controls centrosome and Golgi polarization in migratory cells (Tonucci et al., 2015), E-cadherin trafficking during epithelial morphogenesis (Zobel et al., 2015), cell growth and invasion in cancer metastasis (Chander et al., 2012; Rolland et al., 2014; Truesdell et al., 2014), hypertrophy in neonatal cardiomyocytes (Rusconi et al., 2013), and GLUT4 trafficking in adipocytes (Chang et al., 2002). The N-terminus of TRIP10 contains the FCH domain and two coiled-coil domains, and mediates the interaction with AKAP350, tubulin and phospholipids. Its internal GTP-binding homology region mediates binding to the GTPases TC10 (Chang et al., 2002) and CDC42 (Aspenström, 1997) (Fig. 4A, top). The Src-homology-3 (SH3) domain controls the interaction of TRIP10 with the Wiskott-Aldrich syndrome protein (WASP) and DNM2, thus connecting actin polymerization with membrane deformation to promote GLUT4 trafficking (Hartig et al., 2009) and initiation and scission of endocytic vesicles (Feng et al., 2010). Exon 10 of the TRIP10 gene is alternatively spliced generating a 545-residue isoform (CIP4a) when it is skipped and a 601-residue variant (known as CIP4h or CIP4/2) when it is included (Fig. 4A). The isoform lacking exon 10 is the most ubiquitously expressed. TRIP10 variant containing exon 10 was identified in a two-hybrid screening as a TC10-interacting protein (Chang et al., 2002; Wang et al., 2002) and found to be expressed in adult skeletal muscles and hearts (Feng et al., 2010; Giudice et al., 2014, 2016). CIP4b (also known as Felic) is a TRIP10 isoform that is exclusively expressed in human prostate cancer and lymphoblast cells and is generated by a 29-nucleotide insertion of intron 13 that destroys the SH3 domain (Wang et al., 2002). Furthermore, aberrant splicing in renal carcinoma generates the CIP4-V variant by a 19-nucleotide retention of intron 9, which introduces a premature stop codon (Tsuji et al., 2006) (Fig. 4A). CIP4-V lacks the CDC42-binding region and the SH3 domain, and its overexpression induces the formation of ubiquitylated aggresomes and defects in β-catenin trafficking that ultimately reduce cell adhesion and contribute to cancer progression and metastasis (Tsuji et al., 2006).
Fig. 4.
Alternative splicing of the BAR-domain proteins TRIP10 and BIN1. (A) The TRIP10 (CDC42-interacting protein-4) gene is regulated by alternative splicing. Alternative splicing of exon 10 gives rise to two protein isoforms, one containing 601 amino acids (aa) and the other 545 aa. Retention of portions of intron 9 or 13 generates CIP4-V and CIP4b (Felic) variants, respectively, which are present in the context of cancer. (B) The bridging integrator protein-1 BIN1 is also highly regulated by alternative splicing in a tissue- and developmental-stage-specific manner. BIN1 domain structure is shown on the left with the diverse splice isoforms in different tissues illustrated on the right. (C) Mis-splicing of exon 7 of BIN1 affects its interaction with dynamin-2 (DNM2) and thus endocytosis, as illustrated here for the model cargo transferrin. AKAP350, A-kinase anchoring protein-9; AMPH1, amphiphysin; AP1/2, adaptor complex-1/2; CDC42, cell division cycle 42; CLAP, clathrin- and AP2-binding domain; CLIP1, CAP-Gly domain containing linker protein-1; CLTC, clathrin heavy chain; DM, myotonic dystrophy; DNM1, dynamin-1; GAPEX5, GTPase activating protein and VPS9 domains 1; MBD, MYC-binding domain; MTM1, myotubularin-1; MYC, MYC proto-oncogene; bHLH transcription factor; nt, nucleotides; PI, phosphoinositide binding domain; PIPs, phosphoinositides; RIN2, Ras and Rab interactor 2; RIN3, Ras and Rab interactor 2; SH3, Src-homology-3 domain; SH3GL2, SH3-domain containing GRB2-like or endophilin A1; SH3GLB1, SH3-domain containing GRB2 like or endophilin B1; SNX4, sorting nexin-4; TC10-Ras homolog family member-Q; T-tubules, transverse tubules; WASP, Wiskott-Aldrich syndrome protein family.
BIN1 in brain and cancer
BIN1 is a ubiquitous BAR-domain protein that is highly expressed in striated muscles and brain (Butler et al., 1997; Sakamuro et al., 1996). BIN1 regulates a number of cellular processes, including endocytosis, cytoskeleton organization, DNA repair, the cell cycle, tumor suppression, membrane invagination and T-tubule organization (Prokic et al., 2014). BIN1 has multiple domains (Fig. 4B): (1) the BAR domain (exons 1-10), which binds to membrane lipids and senses membrane curvature; (2) a polybasic segment encoded by the muscle-specific exon 11 that mediates its affinity to phosphoinositides and T-tubules (Kojima et al., 2004; Lee et al., 2002); (3) a brain-specific clathrin- and AP2-binding domain (CLAP) (exons 13-16) (Ramjaun and McPherson, 1998); (4) a MYC-binding domain (MBD) (exons 17-18) that confers tumor-suppressor functions; and (5) the SH3-domain (exons 19-20), which mediates binding to proline-rich motifs. BIN1 contains seven alternatively spliced exons and mis-splicing occurs in disease contexts. In the brain, exon 7 and exons 13-17 are included (Prokic et al., 2014); here, inclusion of exon 7 promotes transferrin endocytosis and interaction between BIN1 with DNM2 (Ellis et al., 2012) (Fig. 4C). In melanoma and breast cancer, the aberrant inclusion of exon 13 disrupts the tumor-suppressor functions of BIN1 (Ge et al., 1999), and exogenous expression of a BIN1 variant lacking exon 13 restores its binding with MYC and thus its tumor-suppressor functions (Anczuków et al., 2012).
BIN1 in striated muscles
Vesicle-mediated transport, membrane remodeling and cytoskeletal genes are globally regulated by alternative splicing in mouse striated muscles during the first four postnatal weeks (Brinegar et al., 2017; Giudice et al., 2014). This is the period of maturation of the sarcoplasmic reticulum and T-tubules that are necessary for excitation-contraction coupling and adult contractility. In striated muscle cells, T-tubules facilitate the access of environmental signals and the propagation of membrane depolarization deep into cells during excitation-contraction coupling. Growing evidence demonstrates that trafficking proteins have structural roles in striated muscles. For example, CLTC regulates the formation and maintenance of the costameres (Vassilopoulos et al., 2014), which are the lateral attachment sites between the sarcolemma and the sarcomere. Another example from a recent study in Drosophila melanogaster revealed that autophagy is required for T-tubule remodeling through a mechanism that implicates the GTPase RAB2 (Fujita et al., 2017). Overall, these and other studies have revealed unconventional but crucial roles of trafficking proteins in muscle cell architecture.
BIN1 induces membrane tubulation in muscle cells, and misregulation of its alternative splicing contributes to T-tubule alterations seen in muscular dystrophies and cardiac diseases (Böhm et al., 2013; Fugier et al., 2011; Hong et al., 2014). In patients with myotonic dystrophy (DM), exon 11 is skipped, contributing to muscle weakness and T-tubule defects (Fugier et al., 2011). Reintroduction of the BIN1 isoform that contains exon 11 restores T-tubule organization in myofibers isolated from DM patients, whereas, by contrast, the variant lacking exon 11 is unable to recover the phenotype (Fugier et al., 2011). Further evidence for the importance of exon 11 comes from the human BIN1 mutation (IVS10-1G>A) that affects the acceptor splice site in a consanguineous family with rapidly progressive and fatal centronuclear myopathy (Böhm et al., 2013). Interestingly, the same splice site is mutated (IVS10-2A>G) in a spontaneous dog model of the disease. Humans and dogs carrying these mutations lack exon 11 and exhibit a similar histopathology with ultrastructural and membrane-triad defects (Böhm et al., 2013). In the heart, exons 7 and 11 are skipped, and exons 13 and 17 are developmentally regulated, contributing to T-tubule organization (Hong et al., 2014). In cardiomyocytes, T-tubules contain dense protective inner membrane folds, which are formed by the BIN1 isoform containing exons 13 and 17 (Hong et al., 2014). Accordingly, in Bin1-depleted mice, T-tubule folding is decreased, leading to free diffusion of local calcium and potassium ions, which prolongs the action potential and increases susceptibility to ventricular arrhythmias. These T-tubule defects can be rescued by expression of the BIN1 isoform that contains exons 13 and 17, which facilitates the folding of the T-tubule membrane (Hong et al., 2014) and so creates a local region of ion accumulation that restricts ion flux.
Therefore, the fact that T-tubule maturation and alternative splicing of membrane-trafficking genes follow similar developmental time-courses in striated muscles, suggests that splicing contributes to T-tubule biogenesis. In support of this notion, developmental splicing networks coordinately revert to neonatal patterns in disease contexts, such as heart failure, cardiomyopathies, congenital muscular dystrophy and DM, as discussed below.
Role of RBPs in alternative splicing of membrane-trafficking proteins in health and disease
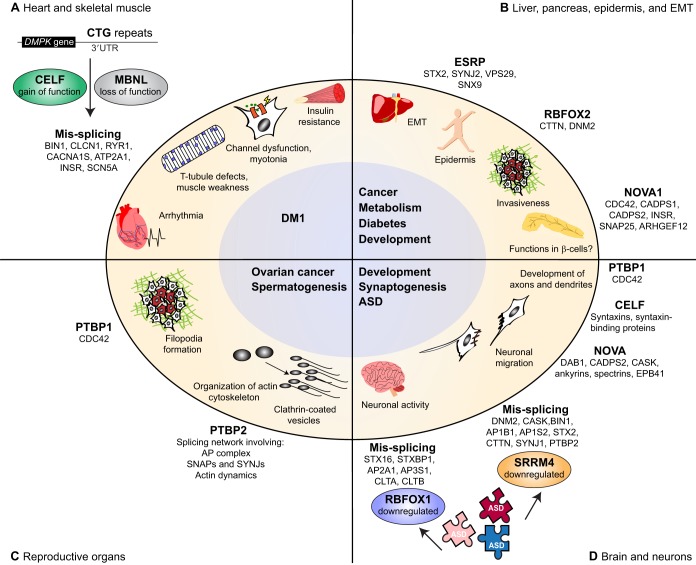
In this section, we discuss how alternative splicing of trafficking proteins is regulated by RBPs and misregulated in neurological and muscle disease (Fig. 5).
Fig. 5.
RBPs regulate alternative splicing of membrane-trafficking proteins in health and disease. (A) In myotonic dystrophy type-1 (DM1), CTG repeat expansion in the 3′UTR region of the DMPK gene leads to the activation of CUGBP Elav-like family member-1 (CELF1) (gain of function) and the sequestration of muscleblind-like protein (MBNL), which causes its loss of function. Alterations in the functions of CELF and MBNL lead to mis-splicing of their targets (listed below). (B) Epithelial ESRP1 and ESRP2 along with the RNA-binding fox-1 homolog-2 protein (RBFOX2) and the neuro-oncological ventral antigen-1 (NOVA1) regulate splicing during epithelial-mesenchymal transition (EMT) in liver development and in metastatic cancers. (C) Polypyrimidine-tract binding proteins (PTBPs) regulate alternative splicing in spermatogenesis and ovarian cancer. PTBP2 is required for mouse spermatogenesis and proper splicing regulation of a network of genes encoding subunits of the AP1 and AP2 complexes, members of the SNARE complex and synaptojanins, and proteins involved in actin dynamics. In ovarian cancer cells, PTBP1 depletion inhibits the formation of filopodia and affects CDC42 (cell division cycle 42) splicing. (D) In the brain, NOVA proteins regulate neuron-specific alternative splicing events that are essential for motor neuron survival, synapse, and axon guidance. Brains of people with autism spectrum disorder (ASD) show downregulation of RBFOX1 and the serine/arginine repetitive matrix-4 (SRRM4), and thus dysregulation of the dependent alternative exons in the factors listed. AP2A1, adaptor-related protein complex-2 α1 subunit; AP3S1, adaptor-related protein complex-3 sigma-1 subunit; ARHGEF12, Rho-GEF12; ATP2A1, ATPase sarcoplasmic/endoplasmic reticulum calcium transporting-1; BIN1, bridging integrator protein-1; CACNA1S, calcium voltage-gated channel subunit alpha-1S; CADPS, calcium-dependent secretion activators; CASK, calcium-/calmodulin-dependent serine protein kinase; CLTA, clathrin light chain-a; CLTB, clathrin light chain-b;CTTN, cortactin; DAB1, reelin adaptor protein; DNM2, dynamin-2; EPB4.1, erythrocyte membrane protein band 4.1; INSR, insulin receptor; RYR1, ryanodine receptor-1; SCN5A, sodium voltage-gated channel alpha subunit-5; SNX9, sorting nexin-9; STX16, syntaxin-16; STXBP1, syntaxin binding protein-1; SYNJ1, synaptojanin-1; SYNJ2, synaptojanin-2; VPS29, retromer complex component.
CELF and MBNL splicing networks in DM1
The muscleblind-like (MBNL) and CELF family of RBPs antagonistically regulate alternative splicing in striated muscles (Wang et al., 2015) and are associated with DM, which is an inherited neuromuscular disease and the most common adult muscular dystrophy. DM patients suffer from skeletal myopathy, cardiac arrhythmia, cataracts, hypogonadism, hyper-somnolence and insulin resistance, among other symptoms (Harper, 2001). DM1 is caused by expansion of CTG repeats in the 3′UTR region of the DMPK gene that leads to activation of CELF1 (gain of function) and sequestration of MBNL, causing its loss of function. Alterations in the functions of CELF and MBNL result in mis-splicing of the targets of these two RBPs, which include several trafficking genes (Fig. 5A) (Dixon et al., 2015; Freyermuth et al., 2016; Fugier et al., 2011). CELF and MBNL misregulation results in the global re-expression of the fetal splicing isoforms of several proteins that we discuss below. This reprogramming in adult tissues takes place mainly in the heart and skeletal muscle. The fetal isoforms are functional in embryonic and neonatal muscles; however, adult muscle cells are large and require a precise internal architecture for proper excitation-contraction coupling. In adults, skipping of exon 7a of the muscle chloride channel CLCN1 generates the functional full-length protein but loss of function of MBNL in DM1 leads to the inclusion of exon 7a, which generates a frameshift and thus non-functional CLCN1 (Charlet-Berguerand et al., 2002; Mankodi et al., 2002). Chloride channels are dispensable in neonatal small myofibers with undeveloped T-tubules, but their absence in large adult myofibers disrupts the potassium counterbalance within T-tubules, resulting in depolarization and myotonia, as seen in DM1 patients (Charlet-Berguerand et al., 2002; Mankodi et al., 2002). Furthermore, the alterations in MBNL and CELF function result in the mis-splicing of the ryanodine receptor-1 (RYR1) (exon 70), the ATPase sarcoplasmic/endoplasmic reticulum calcium transporting-1 (ATP2A1) (exon 22), the calcium voltage-gated channel subunit-alpha1S (CACNA1S) (exon 29) and BIN1 (exon 11), which is thought to partially contribute to muscle weakness associated with DM1 (Fugier et al., 2011; Kimura et al., 2005). In particular, aberrant skipping of exon 29 of the channel CACN1S correlates with the severity of muscle weakness in DM patients (Tang et al., 2012). Finally, insulin insensitivity is associated with mis-splicing of exon 11 of the insulin receptor (INSR), which is due to the dysfunction of MBNL and CELF proteins (Dansithong et al., 2005; Echeverria and Cooper, 2014; Savkur et al., 2001, 2004).
Heart samples from DM1 adult patients exhibit global splicing changes in genes encoding proteins that control ion handling, cell architecture and vesicle-mediated transport (Freyermuth et al., 2016). One of these genes is the sodium channel protein type-5 subunit-alpha (SCN5A), which is critical for cardiomyocyte excitability and impulse propagation through the conduction system. The SCN5A isoform containing exon 6a is predominant in human fetal hearts, exons 6a and 6b are used to similar extents in infants, and the isoform containing exon 6b is preferred in adulthood. The two isoforms differ in seven residues in one of the voltage-sensing domains, and thus in their electrophysiological properties. DM1 patients express the fetal isoform of SCN5A and exhibit severe cardiac conduction defects and arrhythmias that are reproduced when this isoform is re-expressed in adult murine hearts (Freyermuth et al., 2016; Onkal et al., 2008).
ESRPs and RBFOX proteins
The epithelial splicing regulatory proteins ESRP1 and ESRP2, along with the RNA-binding fox-1 homolog-2 protein (RBFOX2) and the neuro-oncological ventral antigen-1 (NOVA1) regulate splicing during epithelial-mesenchymal transition (EMT) in development and in metastatic cancer (Lu et al., 2015). ESRP2 is a driver of liver development, where it regulates splicing of the retromer complex component VPS29, synaptojanin-2 (SYNJ2) and STX2, among other trafficking genes (Bhate et al., 2015). In the embryonic epidermis in humans, mice and fish, ESRP1 and ESRP2 regulate an evolutionarily conserved splicing-trafficking network that includes the sortin nexin-9 (SNX9), STX2 and VPS29 (Burguera et al., 2017) (Fig. 5B). RBFOX2 is induced in EMT and regulates splicing of the endocytic proteins cortactin and DNM2. Depletion of RBFOX2 after EMT significantly reduces cell invasive potential, suggesting that RBFOX2-regulated splicing controls tissue invasiveness (Braeutigam et al., 2014).
NOVA proteins
In the pancreas, NOVA1 has been implicated in regulating splicing of a number of proteins, including the INSR, exocytosis proteins such as SNAP25, the calcium-dependent secretion activator (CADPS), as well as Rho-GTPases, such as cell division cycle-42 (CDC42) and the RhoGEF protein ARHGEF12 (Villate et al., 2014) (Fig. 5B). However, the functional implications of this splicing network that is regulated by NOVA1 in the β-cells were not investigated.
In the brain, NOVA1 regulates neuron-specific alternative splicing and is essential for motor neuron survival after birth (Jensen et al., 2000). Furthermore, the Nova2-knockout mouse exhibits major splicing defects in genes that encode proteins with well-defined functions at the synapse, including neurotransmitter receptors, cation channels, adhesion and scaffold proteins, or in axon guidance (Ule et al., 2005). NOVA proteins regulate splicing of all members of the spectrin–ankyrin–Protein-4.1–CASK scaffold complex, which acts as a linker between membrane proteins and the actin-tropomyosin cytoskeleton, and contributes to organization of the GABAergic synapse (Jensen et al., 2000; Ule et al., 2003; Ule et al., 2005). The functional implications of NOVA-regulated splicing have been convincingly demonstrated for the reelin adaptor protein DAB1, where NOVA2 controls neuronal migration by regulating the splicing of exons 7b and 7c of DAB1 (Yano et al., 2010).
PTBP coordinates splicing trafficking networks in spermatogenesis and ovarian cancer
Polypyrimidine-tract binding proteins (PTBPs) regulate alternative splicing in the brain (Vuong et al., 2016), during spermatogenesis (Hannigan et al., 2017; Zagore et al., 2015), in myogenesis (Hall et al., 2013) and in ovarian cancer (He et al., 2015). PTBP2 is required for mouse spermatogenesis and proper splicing regulation of a network of genes that encode proteins involved in nearly all steps of vesicle-mediated trafficking. This includes subunits of the AP1 and AP2 complexes and DENND1A, which is a component of clathrin-coated vesicles that binds to the AP2 complex, proteins involved in tethering vesicles to microtubules and target membranes, and factors involved in actin dynamics (Hannigan et al., 2017; Zagore et al., 2015) (Fig. 5C). PTBP2 depletion in germ cells results in a disorganization of the F-actin cytoskeleton in Sertoli cells, indicating that regulation of alternative splicing is necessary for cellular crosstalk during germ cell development (Hannigan et al., 2017).
In ovarian cancer cells, PTBP1 depletion has been shown to inhibit the formation of filopodia and to alter CDC42 splicing (Fig. 5C). The Cdc42 gene encodes two protein isoforms, CDC42-v1 and CDC42-v2, which differ in their terminal exons. While CDC42-v1 includes exon 6a, CDC42-v2 includes exon 6b. Because PTBP1 represses exon 6b, it reduces expression of the CDC42-v2 variant, which has been shown to have inhibitory effects on ovarian cancer cell growth and invasiveness, thus functioning as a tumor suppressor (He et al., 2015). More recently, it has been shown that perturbing the ratio between the CDC42-v1 and CDC42-v2 variants in neurons results in alterations in axons and dendrites because CDC42-v1 is required for the development of dendritic spines, whereas CDC42-v2 functions in axogenesis (Yap et al., 2016) (Fig. 5D).
RBFOX1 and SRRM4 in autism spectrum diseases
Autism spectrum disorder (ASD) is a highly heritable neurodevelopmental condition which is characterized by genetic heterogeneity. High-throughput studies have revealed common patterns of misregulated gene expression and alternative splicing in ASD (Gupta et al., 2014; Irimia et al., 2014; Quesnel-Vallières et al., 2016; Voineagu et al., 2011). Brains from ASD patients exhibit a downregulation of the RBP RBFOX1 (also known as A2BP1 and FOX1) and, accordingly, a dysregulation of the RBFOX1-dependent alternative exons in genes that encode proteins involved in cytoskeleton organization and vesicle-mediated transport such as CLTA, CLTB, STX16, STXBP1, AP2A1, AP3S1, among several others (Voineagu et al., 2011) (Fig. 5D).
Another RBP that is downregulated in ASD is the serine/arginine repetitive matrix-4 (SRRM4, also known as nSR100). SRRM4 controls the inclusion of a large set of brain-specific alternative exons that are significantly enriched in genes associated with membrane dynamics, endocytosis and cytoskeleton remodeling (Calarco et al., 2009). SRRM4 controls the neural specificity of alternative splicing by activating the expression of the neuronal isoform of PTBP2 (nPTBP), which includes its exon 10. Therefore, nPTB and SRRM4 act in concert to control neuronal alternative splicing (Calarco et al., 2009). Numerous neural microexons (3-15 nucleotides) are frequently misregulated in ASD brains, and this has been shown to be associated with a reduced expression of SRRM4 (Irimia et al., 2014). Gene ontology analysis revealed that the microexons that are misregulated in ASD are significantly enriched for those present in vesicle-mediated transport genes, among a few other categories (Irimia et al., 2014). Furthermore, mutant mice lacking one functional copy of the Srrm4 gene exhibit several autistic-like features and recapitulate the misregulated splicing patterns that are observed in brains of people with ASD (Irimia et al., 2014; Quesnel-Vallières et al., 2016) (Fig. 5D).
Taken together, it has become clear that the regulation of trafficking proteins by alternative splicing is not limited to one specific RBP and in most cases, RNA processing results from the action of multiple RBPs that function in concert.
Perspectives
The physiological relevance of alternative splicing was first investigated for individual genes; however, genome-wide studies have exponentially increased the number of splicing isoforms, and highlighted networks with unknown and unexplored functions (Baralle and Giudice, 2017). High-resolution imaging and state-of-the-art genetic tools are now available to study trafficking within entire organisms and recent progress in the splicing and trafficking fields pinpoints the questions that remain unanswered at the intersection between these two scientific fields. First, the physiological roles of numerous splicing events in trafficking proteins are still unknown within in vivo contexts. The second challenge arises from the fact that alternative splicing is regulated or misregulated in a coordinated manner in development and disease. Therefore, we need to investigate how splicing trafficking networks modulate intracellular functions, thereby contributing to tissue identity, and ultimately organ maturation and function. Finally, we believe that expanding our knowledge of the contribution of mis-splicing of membrane-trafficking genes in disease will open new possibilities for therapeutic approaches.
Supplementary Material
Acknowledgements
We thank Drs Thomas Cooper (Baylor College of Medicine) and Frances Brodsky (University College London) for their feedback on the initial draft.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The authors are funded by National Institutes of Health/NIGMS grant (R25-GM089569) to E.G.C., Junior Faculty Development Award, Pilot & Feasibility Research Grant (Nutrition and Obesity Research Center, P30DK056350), and start-up funds from The University of North Carolina at Chapel Hill (to J.G.) and in part by the March of Dimes Foundation (5-FY18-36) to J.G. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.216465.supplemental
References
- Anczuków O., Rosenberg A. Z., Akerman M., Das S., Zhan L., Karni R., Muthuswamy S. K. and Krainer A. R. (2012). The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 19, 220-228. 10.1038/nsmb.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenström P. (1997). A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr. Biol. 7, 479-487. 10.1016/S0960-9822(06)00219-3 [DOI] [PubMed] [Google Scholar]
- Baralle F. E. and Giudice J. (2017). Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437-451. 10.1038/nrm.2017.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bark I. C. and Wilson M. C. (1994). Human cDNA clones encoding two different isoforms of the nerve terminal protein SNAP-25. Gene 139, 291-292. 10.1016/0378-1119(94)90773-0 [DOI] [PubMed] [Google Scholar]
- Bark I. C., Hahn K. M., Ryabinin A. E. and Wilson M. C. (1995). Differential expression of SNAP-25 protein isoforms during divergent vesicle fusion events of neural development. Proc. Natl. Acad. Sci. USA 92, 1510-1514. 10.1073/pnas.92.5.1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bark C., Bellinger F. P., Kaushal A., Mathews J. R., Partridge L. D. and Wilson M. C. (2004). Developmentally regulated switch in alternatively spliced SNAP-25 isoforms alters facilitation of synaptic transmission. J. Neurosci. 24, 8796-8805. 10.1523/JNEUROSCI.1940-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhate A., Parker D. J., Bebee T. W., Ahn J., Arif W., Rashan E. H., Chorghade S., Chau A., Lee J.-H., Anakk S. et al. (2015). ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Nat. Commun. 6, 8768 10.1038/ncomms9768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm J., Vasli N., Maurer M., Cowling B., Shelton G. D., Kress W., Toussaint A., Prokic I., Schara U., Anderson T. J. et al. (2013). Altered splicing of the BIN1 muscle-specific exon in humans and dogs with highly progressive centronuclear myopathy. PLoS Genet. 9, e1003430 10.1371/journal.pgen.1003430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boström P., Andersson L., Rutberg M., Perman J., Lidberg U., Johansson B. R., Fernandez-Rodriguez J., Ericson J., Nilsson T., Borén J. et al. (2007). SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat. Cell Biol. 9, 1286-1293. 10.1038/ncb1648 [DOI] [PubMed] [Google Scholar]
- Boumil R. M., Letts V. A., Roberts M. C., Lenz C., Mahaffey C. L., Zhang Z., Moser T. and Frankel W. N. (2010). A missense mutation in a highly conserved alternate exon of dynamin-1 causes epilepsy in fitful mice. PLoS Genet. 6, e1001046 10.1371/journal.pgen.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeutigam C., Rago L., Rolke A., Waldmeier L., Christofori G. and Winter J. (2014). The RNA-binding protein Rbfox2: An essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene 33, 1082-1092. 10.1038/onc.2013.50 [DOI] [PubMed] [Google Scholar]
- Brinegar A. E., Xia Z., Loehr J. A., Li W., Rodney G. G. and Cooper T. A. (2017). Extensive alternative splicing transitions during postnatal skeletal muscle development are required for Calcium handling functions. eLife 6, e27192 10.7554/eLife.27192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky F. M. (2012). Diversity of clathrin function: new tricks for an old protein. Annu. Rev. Cell Dev. Biol. 28, 309-336. 10.1146/annurev-cellbio-101011-155716 [DOI] [PubMed] [Google Scholar]
- Brodsky F. M., Chen C.-Y., Knuehl C., Towler M. C. and Wakeham D. E. (2001). Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 17, 517-568. 10.1146/annurev.cellbio.17.1.517 [DOI] [PubMed] [Google Scholar]
- Buljan M., Chalancon G., Eustermann S., Wagner G. P., Fuxreiter M., Bateman A. and Babu M. M. (2012). Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell 46, 871-883. 10.1016/j.molcel.2012.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguera D., Marquez Y., Racioppi C., Permanyer J., Torres-Méndez A., Esposito R., Albuixech-Crespo B., Fanlo L., D'Agostino Y., Gohr A. et al. (2017). Evolutionary recruitment of flexible Esrp-dependent splicing programs into diverse embryonic morphogenetic processes. Nat. Commun. 8, 1799 10.1038/s41467-017-01961-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M. H., David C., Ochoa G.-C., Freyberg Z., Daniell L., Grabs D., Cremona O. and De Camilli P. (1997). Amphiphysin II (SH3p9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle. J. Cell Biol. 137, 1355-1367. 10.1083/jcb.137.6.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco J. A., Superina S., O'Hanlon D., Gabut M., Raj B., Pan Q., Skalska U., Clarke L., Gelinas D., van der Kooy D. et al. (2009). Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 138, 898-910. 10.1016/j.cell.2009.06.012 [DOI] [PubMed] [Google Scholar]
- Cao H., Garcia F. and Mcniven M. A. (1998). Differential distribution of dynamin isoforms in mammalian cells. Mol. Biol. Cell 9, 2595-2609. 10.1091/mbc.9.9.2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chander H., Truesdell P., Meens J. and Craig A. W. B. (2012). Transducer of Cdc42-dependent actin assembly promotes breast cancer invasion and metastasis. Oncogene 32, 3080-3090. 10.1038/onc.2012.317 [DOI] [PubMed] [Google Scholar]
- Chang L., Adams R. D. and Saltiel A. R. (2002). The TC10-interacting protein CIP4/2 is required for insulin-stimulated Glut4 translocation in 3T3L1 adipocytes. Proc. Natl. Acad. Sci. USA 99, 12835-12840. 10.1073/pnas.202495599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet-Berguerand N., Savkur R. S., Singh G., Philips A. V., Grice E. A. and Cooper T. A. (2002). Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell 10, 45-53. 10.1016/S1097-2765(02)00572-5 [DOI] [PubMed] [Google Scholar]
- Chen L., Liu Z., Zhou B., Wei C., Zhou Y., Rosenfeld M. G., Fu X.-D., Chisholm A. D. and Jin Y. (2016). CELF RNA binding proteins promote axon regeneration in C. elegans and mammals through alternative splicing of syntaxins. eLife 5, e16072 10.7554/eLife.16072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook T. A., Urrutia R. and McNiven M. A. (1994). Identification of dynamin 2, an isoform ubiquitously expressed in rat tissues. Proc. Natl. Acad. Sci. USA 91, 644-648. 10.1073/pnas.91.2.644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis L. B., Doneske B., Liu X., Thaller C., McNew J. A. and Janz R. (2008). Syntaxin 3b is a t-SNARE specific for ribbon synapses of the retina. J. Comp. Neurol. 510, 550-559. 10.1002/cne.21806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis L., Datta P., Liu X., Bogdanova N., Heidelberger R. and Janz R. (2010). Syntaxin 3B is essential for the exocytosis of synaptic vesicles in ribbon synapses of the retina. Neuroscience 166, 832-841. 10.1016/j.neuroscience.2009.12.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansithong W., Paul S., Comai L. and Reddy S. (2005). MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 280, 5773-5780. 10.1074/jbc.M410781200 [DOI] [PubMed] [Google Scholar]
- Daraio T., Bombek L. K., Gosak M., Valladolid-Acebes I., Klemen M. S., Refai E., Berggren P.-O., Brismar K., Rupnik M. S. and Bark C. (2017). SNAP-25b-deficiency increases insulin secretion and changes spatiotemporal profile of Ca2+oscillations in β cell networks. Sci. Rep. 7, 7744 10.1038/s41598-017-08082-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daraio T., Valladolid-Acebes I., Brismar K. and Bark C. (2018). SNAP-25a and SNAP-25b differently mediate interactions with Munc18-1 and Gβγ subunits. Neurosci. Lett. 674, 75-80. 10.1016/j.neulet.2018.03.024 [DOI] [PubMed] [Google Scholar]
- Dillman A. A., Hauser D. N., Gibbs J. R., Nalls M. A., McCoy M. K., Rudenko I. N., Galter D. and Cookson M. R. (2013). mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat. Neurosci. 16, 499-506. 10.1038/nn.3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. M., Choi J., El-Ghazali A., Park S. Y., Roos K. P., Jordan M. C., Fishbein M. C., Comai L. and Reddy S. (2015). Loss of muscleblind-like 1 results in cardiac pathology and persistence of embryonic splice isoforms. Sci. Rep. 5, 9042 10.1038/srep09042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling J. J., Gibbs E. M. and Feldman E. L. (2008). Membrane traffic and muscle: lessons from human disease. Traffic 9, 1035-1043. 10.1111/j.1600-0854.2008.00716.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria G. V. and Cooper T. A. (2014). Muscleblind-like 1 activates insulin receptor exon 11 inclusion by enhancing U2AF65 binding and splicing of the upstream intron. Nucleic Acids Res. 42, 1893-1903. 10.1093/nar/gkt1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. D., Barrios-Rodiles M., Çolak R., Irimia M., Kim T. H., Calarco J. A., Wang X., Pan Q., O'Hanlon D., Kim P. M. et al. (2012). Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol. Cell 46, 884-892. 10.1016/j.molcel.2012.05.037 [DOI] [PubMed] [Google Scholar]
- Feng Y., Hartig S. M., Bechill J. E., Blanchard E. G., Caudell E. and Corey S. J. (2010). The Cdc42-interacting protein-4 (CIP4) gene knock-out mouse reveals delayed and decreased endocytosis. J. Biol. Chem. 285, 4348-4354. 10.1074/jbc.M109.041038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyermuth F., Rau F., Kokunai Y., Linke T., Sellier C., Nakamori M., Kino Y., Arandel L., Jollet A., Thibault C. et al. (2016). Splicing misregulation of SCN5A contributes to cardiac-conduction delay and heart arrhythmia in myotonic dystrophy. Nat. Commun. 7, 11067 10.1038/ncomms11067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugier C., Klein A. F., Hammer C., Vassilopoulos S., Ivarsson Y., Toussaint A., Tosch V., Vignaud A., Ferry A., Messaddeq N. et al. (2011). Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 17, 720-725. 10.1038/nm.2374 [DOI] [PubMed] [Google Scholar]
- Fujita N., Huang W., Lin T.-H., Groulx J.-F., Jean S., Nguyen J., Kuchitsu Y., Koyama-Honda I., Mizushima N., Fukuda M. et al. (2017). Genetic screen in drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. eLife 6, e23367 10.7554/eLife.23367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli T., Zahraoui A., Vaidyanathan V. V., Raposo G., Tian J. M., Karin M., Niemann H. and Louvard D. (1998). A novel tetanus neurotoxin-insensitive vesicle-associated membrane protein in SNARE complexes of the apical plasma membrane of epithelial cells. Mol. Biol. Cell 9, 1437-1448. 10.1091/mbc.9.6.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K., DuHadaway J., Du W., Herlyn M., Rodeck U. and Prendergast G. C. (1999). Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc. Natl. Acad. Sci. USA 96, 9689-9694. 10.1073/pnas.96.17.9689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice J., Xia Z., Wang E. T., Scavuzzo M. A., Ward A. J., Kalsotra A., Wang W., Wehrens X. H. T., Burge C. B., Li W. et al. (2014). Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nat. Commun. 5, 3603 10.1038/ncomms4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice J., Loehr J. A., Rodney G. G. and Cooper T. A. (2016). Alternative splicing of four trafficking genes regulates myofiber structure and skeletal muscle physiology. Cell Rep. 17, 1923-1933. 10.1016/j.celrep.2016.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Ellis S. E., Ashar F. N., Moes A., Bader J. S., Zhan J., West A. B. and Arking D. E. (2014). Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat. Commun. 5, 5748 10.1038/ncomms6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. P., Nagel R. J., Fagg W. S., Shiue L., Cline M. S., Perriman R. J., Donohue J. P. and Ares M. (2013). Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA 19, 627-638. 10.1261/rna.038422.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan M. M., Zagore L. L. and Licatalosi D. D. (2017). Ptbp2 controls an alternative splicing network required for cell communication during spermatogenesis. Cell Rep. 19, 2598-2612. 10.1016/j.celrep.2017.05.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper P. (2001). Myotonic Dystrophy. London: W.B. Saund. [Google Scholar]
- Hartig S. M., Ishikura S., Hicklen R. S., Feng Y., Blanchard E. G., Voelker K. A., Pichot C. S., Grange R. W., Raphael R. M., Klip A. et al. (2009). The F-BAR protein CIP4 promotes GLUT4 endocytosis through bidirectional interactions with N-WASp and Dynamin-2. J. Cell Sci. 122, 2283-2291. 10.1242/jcs.041343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Yuan C. and Yang J. (2015). Regulation and functional significance of CDC42 alternative splicing in ovarian cancer. Oncotarget 6, 29651-29663. 10.18632/oncotarget.4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong T. T., Yang H., Zhang S.-S., Cho H. C., Kalashnikova M., Sun B., Zhang H., Bhargava A., Grabe M., Olgin J. et al. (2014). Cardiac BIN1 folds T-tubule membrane, controlling ion flux and limiting arrhythmia. Nat. Med. 20, 624-632. 10.1038/nm.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibaraki K., Horikawa H. P. M., Morita T., Mori H., Sakimura K., Mishina M., Saisu H. and Abe T. (1995). Identification of four different forms of syntaxin 3. Biochem. Biophys. Res. Commun. 211, 997-1005. 10.1006/bbrc.1995.1910 [DOI] [PubMed] [Google Scholar]
- Irimia M. and Blencowe B. J. (2012). Alternative splicing: decoding an expansive regulatory layer. Curr. Opin. Cell Biol. 24, 323-332. 10.1016/j.ceb.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Irimia M., Weatheritt R. J., Ellis J. D., Parikshak N. N., Gonatopoulos-Pournatzis T., Babor M., Quesnel-Vallières M., Tapial J., Raj B., O'Hanlon D. et al. (2014). A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159, 1511-1523. 10.1016/j.cell.2014.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P., Seow H.-F., Holmes N., Drickamer K. and Parham P. (1987). Clathrin light chains contain brain-specific insertion sequences and a region of homology with intermediate filaments. Nature 326, 154-159. 10.1038/326154a0 [DOI] [PubMed] [Google Scholar]
- Jensen K. B., Dredge B. K., Stefani G., Zhong R., Buckanovich R. J., Okano H. J., Yang Y. Y. L. and Darnell R. B. (2000). Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25, 359-371. 10.1016/S0896-6273(00)80900-9 [DOI] [PubMed] [Google Scholar]
- Johansson J. U., Ericsson J., Janson J., Beraki S., Stanić D., Mandic S. A., Wikström M. A., Hökfelt T., Ögren S. O., Rozell B. et al. (2008). An ancient duplication of exon 5 in the Snap25 gene is required for complex neuronal development/function. PLoS Genet. 4, e1000278 10.1371/journal.pgen.1000278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedra D., Peyrard M., Fransson I., Collins J. E., Dunham I., Roe B. A. and Dumanski J. P. (1996). Characterization of a second human clathrin heavy chain polypeptide gene (CLH-22) from chromosome 22q11. Hum. Mol. Genet. 5, 625-631. 10.1093/hmg/5.5.625 [DOI] [PubMed] [Google Scholar]
- Keyel P. A., Mishra S. K., Roth R., Heuser J. E., Watkins S. C. and Traub L. M. (2006). A single common portal for clathrin-mediated endocytosis of distinct cargo governed by cargo-selective adaptors. Mol. Biol. Cell 17, 4300-4317. 10.1091/mbc.E06-05-0421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Nakamori M., Lueck J. D., Pouliquin P., Aoike F., Fujimura H., Dirksen R. T., Takahashi M. P., Dulhunty A. F. and Sakoda S. (2005). Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 14, 2189-2200. 10.1093/hmg/ddi223 [DOI] [PubMed] [Google Scholar]
- Kirchhausen T., Scarmato P., Harrison S., Monroe J., Chow E., Mattaliano R., Ramachandran K., Smart J., Ahn A. and Brosius J. (1987). Clathrin light chains LCA and LCB are similar, polymorphic, and share repeated heptad motifs. Science 236, 320-324. 10.1126/science.3563513 [DOI] [PubMed] [Google Scholar]
- Kojima C., Hashimoto A., Yabuta I., Hirose M., Hashimoto S., Kanaho Y., Sumimoto H., Ikegami T. and Sabe H. (2004). Regulation of Bin1 SH3 domain binding by phosphoinositides. EMBO J. 23, 4413-4422. 10.1038/sj.emboj.7600442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A. R., Schor I. E., Alló M., Dujardin G., Petrillo E. and Muñoz M. J. (2013). Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 14, 153-165. 10.1038/nrm3525 [DOI] [PubMed] [Google Scholar]
- Lazo P., Nadal M., Ferrer M., Area E., Hernández-Torres J., Nabokina S., Mollinedo F. and Estivill X. (2001). Genomic organization, chromosomal localization, alternative splicing, and isoforms of the human synaptosome-associated protein-23 gene implicated in vesicle-membrane fusion processes. Hum. Genet. 108, 211-215. 10.1007/s004390100480 [DOI] [PubMed] [Google Scholar]
- Lee E., Marcucci M., Daniell L., Pypaert M., Weisz O. A., Ochoa G.-C., Farsad K., Wenk M. R. and de Camilli P. (2002). Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science 297, 1193-1196. 10.1126/science.1071362 [DOI] [PubMed] [Google Scholar]
- Liu Y.-W., Surka M. C., Schroeter T., Lukiyanchuk V. and Schmid S. L. (2008). Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knock-out cells. Mol. Biol. Cell 19, 5347-5359. 10.1091/mbc.E08-08-0890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Huang Q., Park J. W., Shen S., Lin L., Tokheim C. J., Henry M. D. and Xing Y. (2015). Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. Mol. Cancer Res. 13, 305-318. 10.1158/1541-7786.MCR-14-0366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maignan S., Guilloteau J., Fromage N., Arnoux B., Becquart J. and Ducruix A. (1995). Crystal structure of the mammalian Grb2 adaptor. Science 268, 291-293. 10.1126/science.7716522 [DOI] [PubMed] [Google Scholar]
- Majeed S. R., Vasudevan L., Chen C.-Y., Luo Y., Torres J. A., Evans T. M., Sharkey A., Foraker A. B., Wong N. M. L., Esk C. et al. (2014). Clathrin light chains are required for the gyrating-clathrin recycling pathway and thereby promote cell migration. Nat. Commun. 5, 3891 10.1038/ncomms4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankodi A., Takahashi M. P., Jiang H., Beck C. L., Bowers W. J., Moxley R. T., Cannon S. C. and Thornton C. A. (2002). Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell 10, 35-44. 10.1016/S1097-2765(02)00563-4 [DOI] [PubMed] [Google Scholar]
- Martinez-Arca S., Alberts P., Zahraoui A., Louvard D. and Galli T. (2000). Role of tetanus neurotoxin insensitive vesicle-associated membrane protein (TI-VAMP) in vesicular transport mediating neurite outgrowth. J. Cell Biol. 149, 889-900. 10.1083/jcb.149.4.889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arca S., Coco S., Mainguy G., Schenk U., Alberts P., Bouillé P., Mezzina M., Prochiantz A., Matteoli M., Louvard D. et al. (2001). A common exocytotic mechanism mediates axonal and dendritic outgrowth. J. Neurosci. 21, 3830-3838. 10.1523/JNEUROSCI.21-11-03830.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arca S., Rudge R., Vacca M., Raposo G., Camonis J., Proux-Gillardeaux V., Daviet L., Formstecher E., Hamburger A., Filippini F. et al. (2003). A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc. Natl. Acad. Sci. USA 100, 9011-9016. 10.1073/pnas.1431910100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer M. E. and Cooper J. A. (2006). The adaptor protein Dab2 sorts LDL receptors into coated pits independently of AP-2 and ARH. J. Cell Sci. 119, 4235-4246. 10.1242/jcs.03217 [DOI] [PubMed] [Google Scholar]
- McDonald C. B., Seldeen K. L., Deegan B. J., Lewis M. S. and Farooq A. (2008). Grb2 adaptor undergoes conformational change upon dimerization. Arch. Biochem. Biophys. 475, 25-35. 10.1016/j.abb.2008.04.008 [DOI] [PubMed] [Google Scholar]
- McMahon H. T. and Boucrot E. (2011). Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517-533. 10.1038/nrm3151 [DOI] [PubMed] [Google Scholar]
- McMahon H. T. and Gallop J. L. (2005). Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590-596. 10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- Merkin J., Russell C., Chen P. and Burge C. B. (2012). Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338, 1593-1599. 10.1126/science.1228186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C. and Unger V. M. (2012). Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci. 37, 526-533. 10.1016/j.tibs.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinedo F. and Lazo P. A. (1997). Identification of two isoforms of the vesicle-membrane fusion protein SNAP-23 in human neutrophils and HL-60 cells. Biochem. Biophys. Res. Commun. 231, 808-812. 10.1006/bbrc.1997.6196 [DOI] [PubMed] [Google Scholar]
- Morris S. M. and Cooper J. A. (2001). Disabled-2 colocalizes with the LDLR in clathrin-coated pits and interacts with AP-2. Traffic 2, 111-123. 10.1034/j.1600-0854.2001.020206.x [DOI] [PubMed] [Google Scholar]
- Norris A. D., Gao S., Norris M. L., Ray D., Ramani A. K., Fraser A. G., Morris Q., Hughes T. R., Zhen M. and Calarco J. A. (2014). A Pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Mol. Cell 54, 946-959. 10.1016/j.molcel.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H., Harada S.-I., Sassa T., Yamamoto H. and Hosono R. (1998). Functional properties of the unc-64 gene encoding a Caenorhabditis elegans syntaxin. J. Biol. Chem. 273, 2192-2198. 10.1074/jbc.273.4.2192 [DOI] [PubMed] [Google Scholar]
- Onkal R., Mattis J. H., Fraser S. P., Diss J. K. J., Shao D., Okuse K. and Djamgoz M. B. A. (2008). Alternative splicing of Nav1.5: an electrophysiological comparison of “neonatal” and “adult” isoforms and critical involvement of a lysine residue. J. Cell. Physiol. 216, 716-726. 10.1002/jcp.21451 [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L. J., Frey B. J. and Blencowe B. J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413-1415. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Prokic I., Cowling B. S. and Laporte J. (2014). Amphiphysin 2 (BIN1) in physiology and diseases. J. Mol. Med. 92, 453-463. 10.1007/s00109-014-1138-1 [DOI] [PubMed] [Google Scholar]
- Puri N. and Roche P. A. (2006). Ternary SNARE complexes are enriched in lipid rafts during mast cell exocytosis. Traffic 7, 1482-1494. 10.1111/j.1600-0854.2006.00490.x [DOI] [PubMed] [Google Scholar]
- Quesnel-Vallières M., Dargaei Z., Irimia M., Gonatopoulos-Pournatzis T., Ip J. Y., Wu M., Sterne-Weiler T., Nakagawa S., Woodin M. A., Blencowe B. J. et al. (2016). Misregulation of an activity-dependent splicing network as a common mechanism underlying autism spectrum disorders. Mol. Cell 64, 1023-1034. 10.1016/j.molcel.2016.11.033 [DOI] [PubMed] [Google Scholar]
- Ramjaun A. R. and McPherson P. S. (1998). Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites. J. Neurochem. 70, 2369-2376. 10.1046/j.1471-4159.1998.70062369.x [DOI] [PubMed] [Google Scholar]
- Rolland Y., Marighetti P., Malinverno C., Confalonieri S., Luise C., Ducano N., Palamidessi A., Bisi S., Kajiho H., Troglio F. et al. (2014). The CDC42-interacting protein 4 controls epithelial cell cohesion and tumor dissemination. Dev. Cell 30, 553-568. 10.1016/j.devcel.2014.08.006 [DOI] [PubMed] [Google Scholar]
- Rusconi F., Thakur H., Li J. and Kapiloff M. S. (2013). CIP4 is required for the hypertrophic growth of neonatal cardiac myocytes. J. Biomed. Sci. 20, 56 10.1186/1423-0127-20-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifee O., Wei L. and Nonet M. L. (1998). The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Mol. Biol. Cell 9, 1235-1252. 10.1091/mbc.9.6.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamuro D., Elliott K. J., Wechsler-Reya R. and Prendergast G. C. (1996). BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat. Genet. 14, 69-77. 10.1038/ng0996-69 [DOI] [PubMed] [Google Scholar]
- Savkur R. S., Philips A. V. and Cooper T. A. (2001). Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 29, 40-47. 10.1038/ng704 [DOI] [PubMed] [Google Scholar]
- Savkur R. S., Philips A. V., Cooper T. A., Dalton J. C., Moseley M. L., Ranum L. P. W. and Day J. W. (2004). Insulin receptor splicing alteration in myotonic dystrophy type 2. Am. J. Hum. Genet. 74, 1309-1313. 10.1086/421528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti M. M. and Swanson M. S. (2016). RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19-32. 10.1038/nrg.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T., Jung H., Jung J., Turner-Bridger B., Ohk J., Lin J. Q., Amieux P. S. and Holt C. E. (2016). Dynamic axonal translation in developing and mature visual circuits. Cell 166, 181-192. 10.1016/j.cell.2016.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Corydon T. J., Nielsen S., Hoffmann H. J. and Dahl R. (2001). Identification of three new splice variants of the SNARE protein SNAP-23. Biochem. Biophys. Res. Commun. 285, 320-327. 10.1006/bbrc.2001.5144 [DOI] [PubMed] [Google Scholar]
- Sigismund S., Confalonieri S., Ciliberto A., Polo S., Scita G. and Di Fiore P. P. (2012). Endocytosis and signaling: cell logistics shape the eukaryotic cell plan. Physiol. Rev. 92, 273-366. 10.1152/physrev.00005.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag J. M., Fykse E. M., Ushkaryov Y., Liu J. P., Robinson P. J. and Südhof T. C. (1994). Differential expression and regulation of multiple dynamins. J. Biol. Chem. 269, 4547-4554. [PubMed] [Google Scholar]
- Sørensen J. B., Nagy G., Varoqueaux F., Nehring R. B., Brose N., Wilson M. C. and Neher E. (2003). Differential control of the releasable vesicle pools by SNAP-25 splice variants and SNAP-23. Cell 114, 75-86. 10.1016/S0092-8674(03)00477-X [DOI] [PubMed] [Google Scholar]
- Tang Z. Z., Yarotskyy V., Wei L., Sobczak K., Nakamori M., Eichinger K., Moxley R. T., Dirksen R. T. and Thornton C. A. (2012). Muscle weakness in myotonic dystrophy associated with misregulated splicing and altered gating of Cav1.1 calcium channel. Hum. Mol. Genet. 21, 1312-1324. 10.1093/hmg/ddr568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teckchandani A., Toida N., Goodchild J., Henderson C., Watts J., Wollscheid B. and Cooper J. A. (2009). Quantitative proteomics identifies a Dab2/integrin module regulating cell migration. J. Cell Biol. 186, 99-111. 10.1083/jcb.200812160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonucci F. M., Hidalgo F., Ferretti A., Almada E., Favre C., Goldenring J. R., Kaverina I., Kierbel A. and Larocca M. C. (2015). Centrosomal AKAP350 and CIP4 act in concert to define the polarized localization of the centrosome and Golgi in migratory cells. J. Cell Sci. 128, 3277-3289. 10.1242/jcs.170878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truesdell P., Ahn J., Chander H., Meens J., Watt K., Yang X. and Craig A. W. B. (2014). CIP4 promotes lung adenocarcinoma metastasis and is associated with poor prognosis. Oncogene 34, 1-9. 10.1038/onc.2014.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji E., Tsuji Y., Fujiwara T., Ogata S., Tsukamoto K. and Saku K. (2006). Splicing variant of Cdc42 interacting protein-4 disrupts beta-catenin-mediated cell-cell adhesion: Expression and function in renal cell carcinoma. Biochem. Biophys. Res. Commun. 339, 1083-1088. 10.1016/j.bbrc.2005.11.117 [DOI] [PubMed] [Google Scholar]
- Ule J., Jensen K. B., Ruggiu M., Mele A., Ule A. and Darnell R. B. (2003). CLIP identifies nova-regulated RNA networks in the brain. Science 302, 1212-1215. 10.1126/science.1090095 [DOI] [PubMed] [Google Scholar]
- Ule J., Ule A., Spencer J., Williams A., Hu J.-S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M. et al. (2005). Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 37, 844-852. 10.1038/ng1610 [DOI] [PubMed] [Google Scholar]
- Unanue E. R., Ungewickell E. and Branton D. (1981). The binding of clathrin triskelions to membranes from coated vesicles. Cell 26, 439-446. 10.1016/0092-8674(81)90213-0 [DOI] [PubMed] [Google Scholar]
- Ungewickell E. J. and Hinrichsen L. (2007). Endocytosis: clathrin-mediated membrane budding. Curr. Opin. Cell Biol. 19, 417-425. 10.1016/j.ceb.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Vacca M., Albania L., Della Ragione F., Carpi A., Rossi V., Strazzullo M., De Franceschi N., Rossetto O., Filippini F. and D'Esposito M. (2011). Alternative splicing of the human gene SYBL1 modulates protein domain architecture of Longin VAMP7/TI-VAMP, showing both non-SNARE and synaptobrevin-like isoforms. BMC Mol. Biol. 12, 26 10.1186/1471-2199-12-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladolid-Acebes I., Daraio T., Brismar K., Harkany T., Ögren S. O., Hökfelt T. G. M. and Bark C. (2015). Replacing SNAP-25b with SNAP-25a expression results in metabolic disease. Proc. Natl. Acad. Sci. USA 112, E4326-E4335. 10.1073/pnas.1511951112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilopoulos S., Esk C., Hoshino S., Funke B. H., Chen C.-Y., Plocik A. M., Wright W. E., Kucherlapati R. and Brodsky F. M. (2009). A Role for the CHC22 clathrin heavy-chain isoform in human glucose metabolism. Science 324, 1192-1196. 10.1126/science.1171529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilopoulos S., Gentil C., Lainé J., Buclez P.-O., Franck A., Ferry A., Précigout G., Roth R., Heuser J. E., Brodsky F. M. et al. (2014). Actin scaffolding by clathrin heavy chain is required for skeletal muscle sarcomere organization. J. Cell Biol. 205, 377-393. 10.1083/jcb.201309096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R. L., Ramírez O. A., Sandoval L., Koenig-Robert R., Härtel S. and Couve A. (2007). Marlin-1 and conventional kinesin link GABAB receptors to the cytoskeleton and regulate receptor transport. Mol. Cell. Neurosci. 35, 501-512. 10.1016/j.mcn.2007.04.008 [DOI] [PubMed] [Google Scholar]
- Villate O., Turatsinze J.-V., Mascali L. G., Grieco F. A., Nogueira T. C., Cunha D. A., Nardelli T. R., Sammeth M., Salunkhe V. A., Esguerra J. L. S. et al. (2014). Nova1 is a master regulator of alternative splicing in pancreatic beta cells. Nucleic Acids Res. 42, 11818-11830. 10.1093/nar/gku861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K. and Roche P. A. (1999). SNAP-23 and SNAP-25 are palmitoylated in vivo. Biochem. Biophys. Res. Commun. 258, 407-410. 10.1006/bbrc.1999.0652 [DOI] [PubMed] [Google Scholar]
- Voineagu I., Wang X., Johnston P., Lowe J. K., Tian Y., Horvath S., Mill J., Cantor R. M., Blencowe B. J. and Geschwind D. H. (2011). Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380-384. 10.1038/nature10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C. K., Black D. L. and Zheng S. (2016). The neurogenetics of alternative splicing. Nat. Rev. Neurosci. 17, 265-281. 10.1038/nrn.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Rudert W. A., Grishin A., Dombrosky-Ferlan P., Sullivan K., Deng X., Whitcomb D. and Corey S. (2002). Identification and genetic analysis of human and mouse activated Cdc42 interacting protein-4 isoforms. Biochem. Biophys. Res. Commun. 293, 1426-1430. 10.1016/S0006-291X(02)00398-4 [DOI] [PubMed] [Google Scholar]
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P. and Burge C. B. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470-476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. T., Ward A. J., Cherone J. M., Giudice J., Wang T. T., Treacy D. J., Lambert N. J., Freese P., Saxena T., Cooper T. A. et al. (2015). Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 25, 858-871. 10.1101/gr.184390.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatheritt R. J. and Gibson T. J. (2012). Linear motifs: lost in (pre)translation. Trends Biochem. Sci. 37, 333-341. 10.1016/j.tibs.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Weatheritt R. J., Davey N. E. and Gibson T. J. (2012). Linear motifs confer functional diversity onto splice variants. Nucleic Acids Res. 40, 7123-7131. 10.1093/nar/gks442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.-X., Yang W., Jackowski S. and Rock C. O. (1995). Cloning of a novel phosphoprotein regulated by colony-stimulating factor 1 shares a domain with the Drosophila disabled gene product. J. Biol. Chem. 270, 14184-14191. 10.1074/jbc.270.23.14184 [DOI] [PubMed] [Google Scholar]
- Yang X., Coulombe-Huntington J., Kang S., Sheynkman G. M., Hao T., Richardson A., Sun S., Yang F., Shen Y. A., Murray R. R. et al. (2016). Widespread expansion of protein interaction capabilities by alternative splicing. Cell 164, 805-817. 10.1016/j.cell.2016.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M., Hayakawa-Yano Y., Mele A. and Darnell R. B. (2010). Nova2 regulates neuronal migration through an RNA switch in Disabled-1 signaling. Neuron 66, 848-858. 10.1016/j.neuron.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K., Xiao Y., Friedman B. A., Je H. S. and Makeyev E. V. (2016). Polarizing the neuron through sustained co-expression of alternatively spliced isoforms. Cell Rep. 15, 1316-1328. 10.1016/j.celrep.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M. S., Won K.-J., Kim D.-Y., Hwang D. I., Yoon S. W., Jung S. H., Lee K. P., Jung D., Choi W. S., Kim B. et al. (2016). Diminished lipid raft SNAP23 increases blood pressure by inhibiting the membrane fluidity of vascular smooth-muscle cells. J. Vasc. Res. 52, 321-333. 10.1159/000443888 [DOI] [PubMed] [Google Scholar]
- Zagore L. L., Grabinski S. E., Sweet T. J., Hannigan M. M., Sramkoski R. M., Li Q. and Licatalosi D. D. (2015). RNA binding protein Ptbp2 is essential for male germ cell development. Mol. Cell. Biol. 35, 4030-4042. 10.1128/MCB.00676-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Yamakuchi M. and Lowenstein C. J. (2015). SNAP23 regulates endothelial exocytosis of von Willebrand Factor. PLoS ONE 10, 14-22. 10.1371/journal.pone.0118737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel T., Brinkmann K., Koch N., Schneider K., Seemann E., Fleige A., Qualmann B., Kessels M. M. and Bogdan S. (2015). Cooperative functions of the two F-BAR proteins Cip4 and Nostrin in the regulation of E-cadherin in epithelial morphogenesis. J. Cell Sci. 128, 1453-1453 10.1242/jcs.170944 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.