ABSTRACT
Lysosomes receive and degrade cargo from endocytosis, phagocytosis and autophagy. They also play an important role in sensing and instructing cells on their metabolic state. The lipid kinase PIKfyve generates phosphatidylinositol-3,5-bisphosphate to modulate lysosome function. PIKfyve inhibition leads to impaired degradative capacity, ion dysregulation, abated autophagic flux and a massive enlargement of lysosomes. Collectively, this leads to various physiological defects, including embryonic lethality, neurodegeneration and overt inflammation. The reasons for such drastic lysosome enlargement remain unclear. Here, we examined whether biosynthesis and/or fusion-fission dynamics contribute to swelling. First, we show that PIKfyve inhibition activates TFEB, TFE3 and MITF, enhancing lysosome gene expression. However, this did not augment lysosomal protein levels during acute PIKfyve inhibition, and deletion of TFEB and/or related proteins did not impair lysosome swelling. Instead, PIKfyve inhibition led to fewer but enlarged lysosomes, suggesting that an imbalance favouring lysosome fusion over fission causes lysosome enlargement. Indeed, conditions that abated fusion curtailed lysosome swelling in PIKfyve-inhibited cells.
KEY WORDS: Transcription factors, Phosphoinositides, Lysosomes, Organelles, Fusion, Fission
Summary: PIKfyve inhibition causes lysosomes to coalesce, resulting in fewer, enlarged lysosomes; TFEB-mediated lysosome biosynthesis does not contribute to swelling.
INTRODUCTION
Lysosomes are organelles responsible for the degradation of a large variety of molecules that originate from within and from outside cells. Lysosomes accomplish this by fusing with cargo-containing organelles such as autophagosomes, Golgi-derived vesicles, endosomes and phagosomes (Luzio et al., 2007, 2009; Schwake et al., 2013). To enable their degradative capacity, lysosomes are equipped with a plethora of hydrolytic enzymes that digest proteins, lipids, nucleic acids and carbohydrates. In addition, the vacuolar H+-pump ATPase generates a highly acidic lumen optimal for hydrolytic activity (Mindell, 2012; Schwake et al., 2013; Xiong and Zhu, 2016; Luzio et al., 2007). Nonetheless, the function of lysosomes is not limited to molecular degradation. Among other roles, lysosomes are an important storage organelle, they mediate antigen presentation, they undergo exocytosis to repair membrane damage, and are a genuine signalling platform that senses and informs the cell of its nutrient and stress conditions (Medina et al., 2015; Settembre and Ballabio, 2014; Sancak et al., 2010; Delamarre et al., 2005; Xiong and Zhu, 2016; Reddy et al., 2001; Settembre et al., 2012; Roczniak-Ferguson et al., 2012).
Strikingly, the lysosome is now considered to be a highly dynamic and heterogeneous organelle. Not only can individual cells host tens to hundreds of lysosomes, lysosomes themselves are highly heterogeneous in their pH (Johnson et al., 2016), subcellular position (Li et al., 2016a; Pu et al., 2015; Bagshaw et al., 2006) and morphology (Swanson et al., 1987; Saric et al., 2016; Vyas et al., 2007). Lastly, lysosomes can adapt to their environment. For example, in macrophages and dendritic cells, lysosomes are transformed from individual puncta into a highly tubular network (Saric et al., 2016; Vyas et al., 2007; Swanson et al., 1987). In addition, nutrient depletion, protein aggregation and phagocytosis activate a family of transcription factors that can enhance expression of lysosome genes and adjust lysosome activity (Gray et al., 2016; Pastore et al., 2016; Sardiello et al., 2009; Tsunemi et al., 2012; Medina et al., 2011; Settembre et al., 2012; Roczniak-Ferguson et al., 2012). This is best understood for the related transcription factors, TFEB and TFE3, which are recruited to the surface of lysosomes and subject to phosphorylation by various kinases, including mTORC1 to modulate their nucleocytoplasmic transport (Li et al., 2016b; Martina et al., 2012; Peña-Llopis et al., 2011; Martina et al., 2014, 2016; Roczniak-Ferguson et al., 2012).
Given the above, lysosomes are highly regulated organelles. A master modulator of lysosomes is the lipid kinase PIKfyve, which converts phosphatidylinositol-3-phosphate [PtdIns(3)P] into phosphatidylinositol-3,5-bisphosphate [PtdIns(3,5)P2] (Sbrissa et al., 1999). Inactivation of PIKfyve or of its regulators, Vac14/ArPIKfyve and Fig4/Sac3, causes a variety of physiological problems, including embryonic lethality, neurodegeneration and immune malfunction (Cai et al., 2013; Chow et al., 2007; Jin et al., 2008; Ho et al., 2012; Mccartney et al., 2014; Sbrissa et al., 2007; Ferguson et al., 2010; Min et al., 2014; Lenk et al., 2016; Ikonomov et al., 2011). At the cellular level, these defects relate to impaired autophagic flux, altered delivery to lysosomes, dysregulation of lysosomal Ca2+ transport and massive enlargement of lysosomes (Ferguson et al., 2009; Dong et al., 2010; de Lartigue et al., 2009; Ferguson et al., 2010; Kim et al., 2014, 2016; Ho et al., 2012; Mccartney et al., 2014). Remarkably, it remains unclear how lysosomes enlarge in cells with defective PIKfyve activity. It has been suggested that PIKfyve, or loss of the yeast orthologue Fab1 impairs recycling from endosomes, lysosomes or from the yeast vacuole, thus causing an influx of membrane into lysosomes and their enlargement (Rutherford et al., 2006; Bryant et al., 1998; Dove et al., 2004). This recycling process is assumed to occur through vesicular-tubular transport intermediates like those induced by clathrin, adaptor proteins and retromer (Currinn et al., 2016; de Lartigue et al., 2009; Ho et al., 2012; Phelan et al., 2006), but remains speculative. This type of membrane fission is distinct from the ‘kiss-and-run’ process, where lysosomes undertake transient homotypic and heterotypic fusion events that are followed by fission to avoid coalescence of lysosomes (Bright et al., 2005). Membrane fission can also occur as organelle splitting, rather than budding, as is the case of mitochondrial fission (Chan, 2006).
Recently, impairment of PIKfyve was shown to cause nuclear accumulation of TFEB, suggesting that increased lysosomal biogenesis may contribute to lysosome swelling (Gayle et al., 2017; Wang et al., 2015). Here, we examined the role of TFEB and the related proteins TFE3 and MITF in lysosome enlargement and conclude that biosynthesis does not contribute to lysosome enlargement, at least acutely. Instead, we observed that PIKfyve inhibition causes a significant reduction in lysosome number concurrent with lysosome enlargement. We propose that lysosome swelling proceeds through lysosome coalescence, probably as a result of reduced ‘run’ or fission events during lysosome kiss-and-run and/or full fusion and fission cycles.
RESULTS
PIKfyve inhibition activates the transcription factor TFEB independently of mTOR
PIKfyve inhibition causes lysosome enlargement (Ikonomov et al., 2001; Mccartney et al., 2014) but the mechanism by which this happens remains unclear. There is a growing body of literature that links TFEB and related transcription factors to enhanced lysosome and protein expression (Settembre and Ballabio, 2014; Raben and Puertollano, 2016). Thus, we set out to test if TFEB activation might contribute to lysosome swelling. Indeed, PIKfyve inhibition using apilimod, an exquisitely specific inhibitor of PIKfyve (Cai et al., 2013; Gayle et al., 2017), in RAW cells triggered a rapid and robust nuclear translocation of GFP-tagged TFEB and endogenous TFEB (Fig. 1A,B). To complement our pharmacological treatment, we also demonstrated that siRNA-mediated silencing of PIKfyve significantly boosted the number of RAW cells with nuclear TFEB-GFP relative to control cells (Fig. 1C). In addition, HeLa cells treated with apilimod also exhibited nuclear TFEB relative to resting cells, showing that activation of TFEB in the absence of PIKfyve occurs across distinct cell types (Fig. 1D). Lastly, we showed that GFP fusions of TFE3 and MITF, which are related to TFEB (Ploper and De Robertis, 2015; Raben and Puertollano, 2016), also translocated to the nucleus upon PIKfyve abrogation in RAW cells (Fig. S1A,B). Overall, our data are consistent with recent reports that PIKfyve activity robustly controls nuclear localization of TFEB and related factors (Gayle et al., 2017; Wang et al., 2015).
Fig. 1.
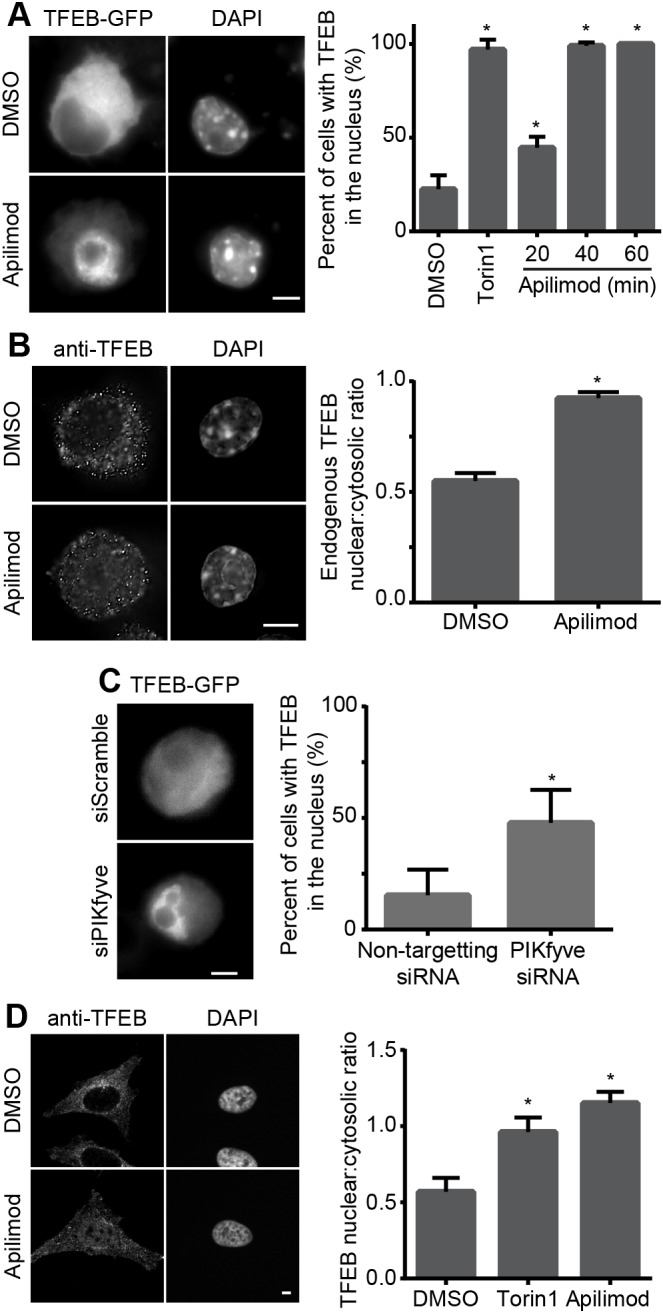
PIKfyve inhibition causes nuclear translocation of TFEB. (A,B) RAW cells expressing TFEB-GFP (A) or stained for endogenous TFEB (B) treated with vehicle or 20 nM apilimod for 1 h. (C) RAW cells silenced for PIKfyve or mock-silenced and expressing TFEB-GFP. (D) HeLa cells stained for endogenous TFEB treated with vehicle or 20 nM apilimod for 1 h. DAPI was used to stain the nuclei. Nuclear translocation of TFEB was quantified either as percentage of cells containing nuclear GFP-tagged TFEB (A,C) or as the nuclear-to-cytosol fluorescence intensity ratio for endogenous TFEB (B,D). Shown is the mean and s.e.m. from at least three independent experiments and based on 50-300 cells per condition per experiment. *P<0.05 compared with respective control conditions using one-way ANOVA and Tukey's post hoc test or with an unpaired Student's t-test, as appropriate. Scale bars: 5 µm.
TFEB is controlled by several factors including mTOR, which phosphorylates and maintains TFEB in the cytosol (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012). In addition, PIKfyve has been suggested to control mTORC1 in adipocytes (Bridges et al., 2012), although others have noted that mTOR activity is sustained in PIKfyve-inhibited cells (Wang et al., 2015; Krishna et al., 2016). To better assess the possibility that PIKfyve controls TFEB by coordinating with mTOR in our cells, we examined the phosphorylation state of S6K, a major mTORC1 target, in cells treated with apilimod. Strikingly, we could not observe a significant difference in the phosphorylation levels of S6K between control and apilimod-treated cells (Fig. 2A). This suggests that mTORC1 retains its activity in PIKfyve-inhibited RAW macrophages. We also wondered whether mTOR could control TFEB by regulating PIKfyve activity instead. To test this, we exposed cells to torin1, an inhibitor of mTOR. First, we showed that apilimod reduced PtdIns(3,5)P2 by >70% with a concomitant increase in its precursor phosphatidylinositol-3-phosphate (Fig. 2B). In comparison, we did not observe a decrease in the levels of either phosphatidylinositol-3-phosphate or PtdIns(3,5)P2 in torin1-treated cells relative to control cells; in fact, there was a statistically significant increase in PtdIns(3,5)P2 levels in torin1-treated cells (Fig. 2B).
Fig. 2.
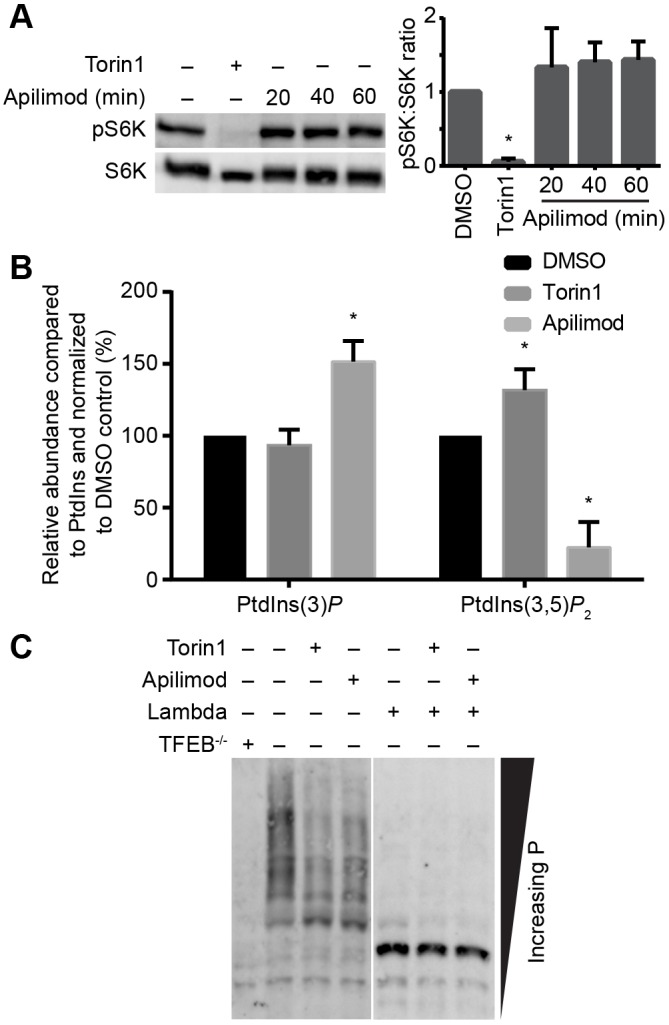
mTOR and PIKfyve function independently. (A) Western blot detection of phosphoThr389-p70S6K and total p70S6K in PIKfyve-inhibited cells treated as indicated. The ratio of phospho-S6K to total S6K was quantified from three independent blots. Shown is the mean ratio and s.d. normalized to DMSO-treated cells. (B) Quantification of PtdIns(3)P and PtdIns(3,5)P2 levels using 3H-myo-inositol incorporation and HPLC-coupled flow scintillation. Results are shown as the mean and s.d. from three independent experiments, normalized to their PtdInsP levels and to the respective DMSO-treated controls. *P<0.05 compared with respective control conditions using multiple Student's t-test with the Bonferroni correction. (C) Western blot showing TFEB expression in whole-cell lysates resolved on a Phos-tag gel. Shown in a representative blot from three independent experiments showing differential migration of TFEB in lysates from control relative to torin1- and apilimod-treated cells. Anti-TFEB antibody specificity was confirmed by the loss of most bands in Tfeb−/− RAW whole-cell lysates. In addition, most bands represent phosphorylated isoforms of TFEB since pre-treatment of lysates with λ protein phosphatase led to a single fast-running band.
Since TFEB is regulated by phosphorylation at various sites (Dephoure et al., 2008; Mayya et al., 2009), we decided to assess whether PIKfyve activity might govern TFEB through phosphorylation. To achieve this, we examined changes in TFEB phosphorylation using Phos-tag gels, which can resolve multiple phospho-isoforms of a target protein. Notably, apilimod treatment caused a change in the phosphorylation of TFEB when resolved on a Phos-tag gel to a pattern comparable to that in torin1-treated cells (Fig. 2C). Importantly, lysates from Tfeb−/− RAW cells lacked most bands resolved by Phos-tag gels and detected with anti-TFEB antibodies, identifying the signal as specific to TFEB (Fig. 2C). In addition, pre-incubation of lysates with λ-phosphatase collapsed the signal into a single fast-running TFEB band (Fig. 2C), suggesting that we specifically detected a variety of phospho-TFEB isoforms. Overall, these data suggest that PIKfyve regulates kinases or phosphatases that impinge on TFEB, but these are seemingly in parallel and/or independent of mTOR.
PIKfyve inhibition boosts lysosome gene expression but not lysosomal protein levels
Our observations suggest that PIKfyve may stimulate lysosomal gene and protein expression by activating TFEB and related factors. To test this, we measured mRNA levels of genes encoding selected lysosomal proteins, including LAMP1, MCOLN1, cathepsin D and the V-ATPase H and D subunits using qRT-PCR. As expected, we observed augmented mRNA levels for these genes after incubation with apilimod for 3 h (Fig. 3A). We next examined if the enhanced mRNA levels corresponded to an increase in protein levels. Surprisingly, the levels of the V-ATPase H subunit, LAMP1 and cathepsin D in apilimod-treated RAW macrophages were similar to control cells after 3 or 6 h of PIKfyve inhibition, although the small increase in LAMP1 protein levels after treatment for 6 h was statistically higher than that in resting cells (Fig. 3B,C). The decoupling between enhanced mRNA levels and unchanged protein levels may be due to the role of PIKfyve in targeting lysosomal proteins to lysosomes (Ikonomov et al., 2009a; Rutherford et al., 2006; Min et al., 2014). Given this, we suspected that TFEB activation may not contribute to lysosome swelling in cells acutely suppressed for PIKfyve.
Fig. 3.
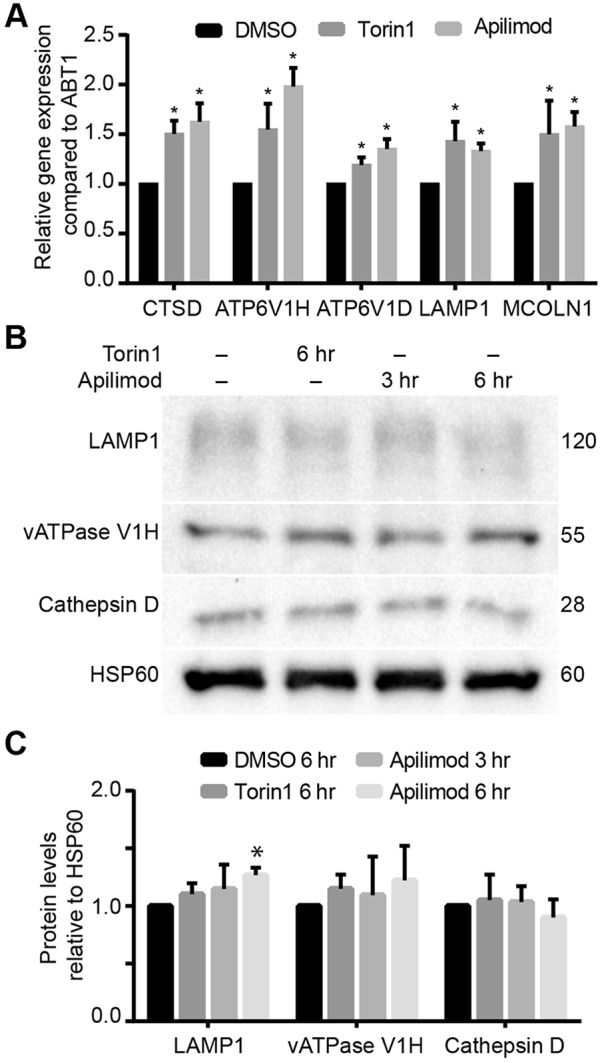
Acute PIKfyve inhibition enhances lysosomal gene transcription but not protein levels. (A) Expression of selected lysosomal genes quantified by qRT-PCR and normalized against Abt1. Shown is the mean±s.e.m. from seven independent experiments. (B) Western blot of selected lysosomal proteins in RAW cells treated as indicated. Numbers on the right represent relative molecular mass (kDa). (C) Quantification of indicated proteins demonstrated in B and shown as the mean±s.e.m. intensity from four independent blots. *P<0.05 compared with respective control conditions using multiple Student's t-test with the Bonferroni correction.
PIKfyve inhibition induces lysosome swelling in cells deleted for TFEB and related proteins
Next, we tested whether expression of TFEB and/or TFE3 is dispensable for lysosome swelling during PIKfyve inhibition. To do this, RAW macrophages deleted for TFEB, TFE3 or both (Pastore et al., 2016) were treated with vehicle or apilimod for 1 h and manually assessed for the average diameter and number of swollen vacuoles using differential interference contrast (DIC) optics. As suspected, RAW cells carrying deletions for TFEB and/or TFE3 were not impaired in apilimod-induced vacuolation, with vacuole number and size comparable to values in wild-type RAW cells (Fig. S2A).
We then followed manual assessment with a more robust volumetric analysis to quantify the size and number of endo/lysosomes by decorating their luminal volume with fluid-phase pinocytic tracers fluorescent-tagged dextrans and Lucifer Yellow. Notably, our labelling method likely decorates a collection of late endosomes, lysosomes and endolysosomes, a term that describes hybrid endosome-lysosome organelles (Huotari and Helenius, 2011). Thus, we use the term endo/lysosome to reflect the heterogeneous mixture of endosomes and lysosomes likely to be labelled here and to differentiate from endolysosomes. First, we showed that fluorescent-dextran colocalized to LAMP1-positive structures similarly well between all RAW strains, confirming that deletion of TFEB and/or TFE3 did not disrupt trafficking of the label to endo/lysosomes (Fig. S2B). Second, we defined parameters such as thresholding and minimum particle size to estimate endo/lysosome number and volume (see Materials and Methods and Fig. S3 for optimization testing). Using this approach, we conclude that the average number and volume of individual endo/lysosomes were similar between apilimod-treated wild-type and Tfeb−/−, Tfe3−/− and Tfeb−/− Tfe3−/− RAW macrophages (Fig. 4A), consistent with the manual analysis (Fig. S2A). Nevertheless, Tfeb−/− Tfe3−/− RAW macrophages retain expression of the related MITF protein, which becomes nuclear upon addition of apilimod (Fig. S1); this raised the possibility that MITF may be sufficiently redundant with TFEB and TFE3 to promote lysosome expansion during PIKfyve inhibition. To test this, we opted to compare HeLa cells to counterpart HeLa cells deleted for all three proteins, previously described by Nezich et al. (2015). Despite deletion of genes encoding TFEB, TFE3 and MITF, endo/lysosome enlargement caused by apilimod was indistinguishable between wild-type and mutant HeLa strains (Fig. S4). Finally, we then asked whether protein biosynthesis was necessary for lysosomes to enlarge during PIKfyve repression. Strikingly, cycloheximide, which arrests protein biosynthesis, did not interfere with apilimod-induced lysosome swelling (Fig. 4B). As a control for the effectiveness of cycloheximide inhibition of protein synthesis, we assayed for translation activity with puromycylation and steady-state levels of p53, which is a high turnover protein. As shown in Fig. 4C, resting cells exhibited a high rate of puromycylation, which is a marker for ribosome-mediated protein synthesis, whereas cycloheximide ablated this. In addition, cycloheximide rapidly depleted p53 levels, demonstrating that protein synthesis was arrested (Fig. 4D). Overall, our data suggest that protein biosynthesis and lysosome biogenesis controlled by TFEB and related transcription factors do not contribute substantially to endo/lysosome swelling during acute PIKfyve inhibition.
Fig. 4.
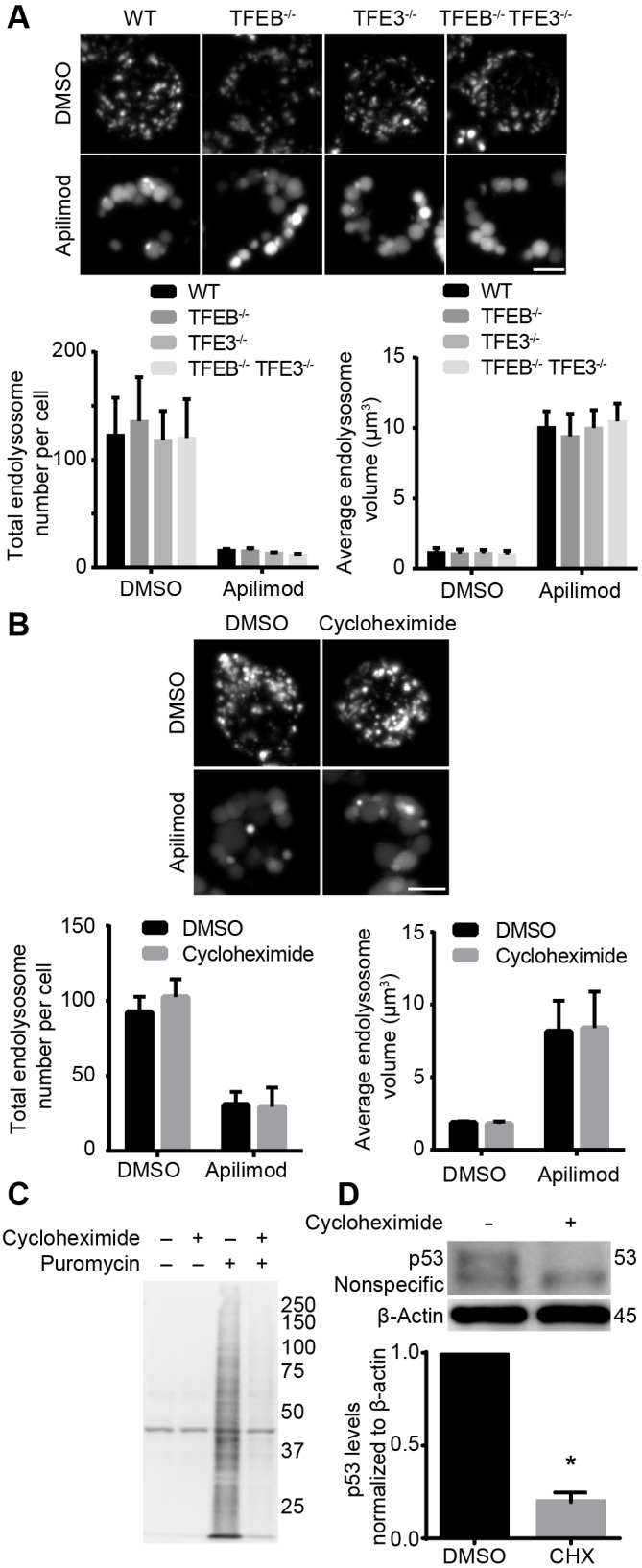
Volumetric analysis of endo/lysosomes in PIKfyve-curtailed wild-type, Tfeb−/−, Tfe3−/− and Tfeb−/−Tfe3−/− RAW cells. (A) Cells were pre-labelled with Lucifer Yellow and then treated with apilimod to induce lysosome vacuolation. Lysosome number and lysosome volume were quantified from 100 cells from three independent experiments. (B) Lysosome morphology in wild-type RAW cells treated as indicated. Lysosome volume and number were quantified. In all cases, data shown are the mean±s.e.m. from three independent experiments, with at least 100 cells per condition per experiment. (C) Puromycylation of proteins in the absence and presence of cycloheximide. Puromycin is incorporated into elongating proteins by the ribosome, which can be detected by anti-puromycin antibodies during western blotting. Cycloheximide treatment eradicated puromycylation of proteins by blocking protein synthesis. The first two lanes resolved lysates unexposed to puromycin and thus accounts for non-specific bands detected by anti-puromycin antibodies. Representative blot shown from three independent experiments. (D) Cycloheximide reduces p53 abundance relative to β-actin. Representative blot shown from three independent experiments. Numbers on the right represent relative molecular mass (in kDa). Data are mean±s.e.m. abundance by measuring p53 signal as a ratio against β-actin. *P<0.05 compared with respective control conditions using Student's t-test. Scale bars: 5 µm.
PIKfyve inhibition concurrently increases endo/lysosome volume while decreasing endo/lysosome number
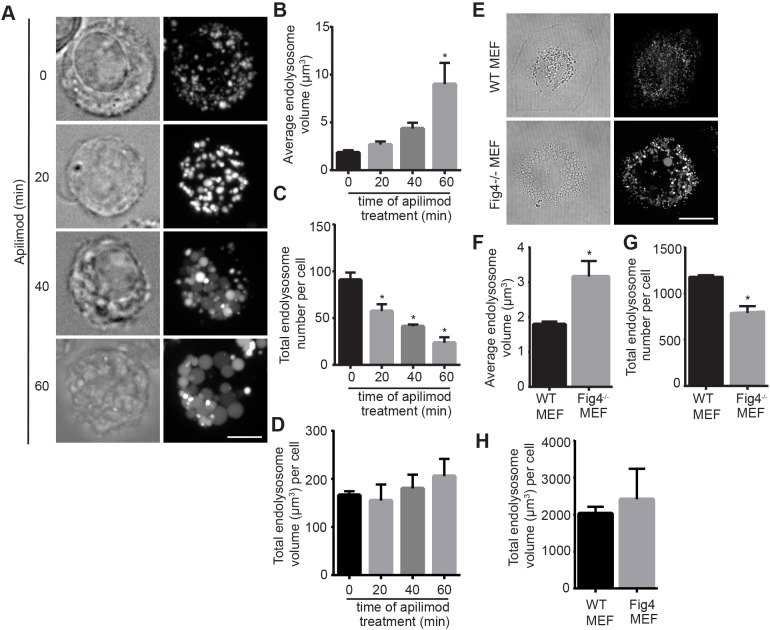
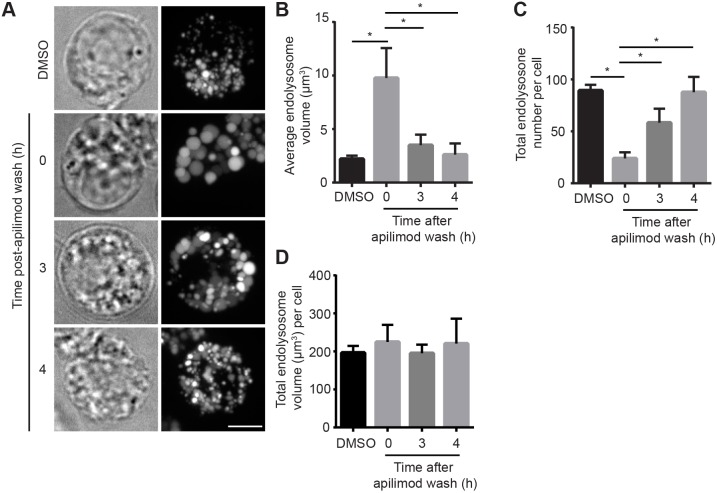
To better understand the process of endo/lysosome enlargement during PIKfyve inhibition, we further employed quantitative volumetric image analysis. Using this approach, we observed that the average volume of individual endo/lysosomes in RAW cells treated with apilimod increased with incubation period relative to vehicle-only cells (Fig. 5A,B). Remarkably, the average number of endo/lysosomes per cell in apilimod-treated cells decreased considerably with incubation period and relative to vehicle-exposed cells (Fig. 5A,C). Despite the decrease in lysosome number, the total lysosome volume per cell remained unperturbed (Fig. 5A,D). As suggested above, we also observed similar trends with HeLa cells exposed to apilimod and Fig4−/− mouse embryonic fibroblasts; there was an increase in the average volume of individual endo/lysosomes, a concurrent decline in their number, whereas total lysosome volume per cell was not significantly altered (Fig. S4, Fig. 5D-H). Finally, and as implied above, there was no significant difference in the average size, number and total volume of endo/lysosomes between wild-type RAW and HeLa cells and counterpart strains deleted for TFEB, TFE3 and/or MITF before and after PIKfyve suppression (Fig. 4; Fig. S4). Interestingly, replacing apilimod-containing medium with fresh medium reversed the effects on endo/lysosome volume and number per cell (Fig. 6). Overall, our results collectively suggest that acute PIKfyve inhibition not only enlarges endo/lysosomes, but also decreases their numbers without disturbing the total endo/lysosome volume.
Fig. 5.
Volumetric analysis of endo/lysosomes during acute and chronic PIKfyve suppression. (A) RAW cells were pre-labelled with Lucifer Yellow and exposed to vehicle only or to 20 nM apilimod for the indicated times. Fluorescence micrographs are z-projections of 45-55 z-planes acquired by spinning disc confocal microscopy. Scale bar: 5 µm. (B-D) Quantification of individual endo/lysosome volume (B), endo/lysosome number per RAW macrophage (C) and total endo/lysosome volume (D). (E) Wild-type and Fig4−/− mouse embryonic fibroblasts labelled with Lucifer Yellow. Scale bar: 30 µm. (F-H) Analysis of individual endo/lysosome volume (F), endo/lysosome number per cell (G) and total endo/lysosome volume per cell (H) in wild-type and Fig4−/− mouse embryonic fibroblasts. In all cases, data shown are the mean±s.e.m. from three independent experiments, with at least 15-20 cells per condition per experiment. *P<0.05 compared with respective control conditions using one-way ANOVA and Tukey's post hoc test for B-D, and unpaired Student's t-test for F-H.
Fig. 6.
Volumetric analysis of endo/lysosome number and volume during PIKfyve reactivation. (A) Cells were pre-labelled with Lucifer Yellow as before and then exposed to vehicle only or to 20 nM apilimod for 1 h, followed by removal and chase in fresh medium for the indicated times. Fluorescence micrographs are z-projections of 45-55 z-planes acquired by spinning disc confocal microscopy. Scale bar: 5 µm. (B-D) Volumetric quantification of individual endo/lysosome volume (B), endo/lysosome number per cell (C) and total endo/lysosome volume per cells (D). Data shown are the mean±s.e.m. from three independent experiments, with at least 15-20 cells per condition per experiment. *P<0.05 between data indicated by lines using one-way ANOVA and Tukey's post hoc test.
Imbalanced fusion-fission cycles may underpin endo/lysosome swelling in PIKfyve-inhibited cells
It is unlikely that endo/lysosome number is reduced during PIKfyve inhibition via lysosome lysis because the total lysosome volume per cell was unchanged relative to control cells. Instead, the reduction in endo/lysosome number coupled to their enlargement suggests that there is a shift towards endo/lysosome homotypic fusion relative to fission in PIKfyve-abrogated cells. Hence, we predicted that conditions that abate homotypic lysosome fusion would lessen enlargement and maintain higher lysosome numbers in PIKfyve-inhibited cells.
To test this hypothesis, we disrupted the microtubule system and motor activity using nocadozole and ciliobrevin, respectively, to impair lysosome-lysosome fusion (Rosa-Ferreira and Munro, 2011; Deng and Storrie, 1988; Jordens et al., 2001). Indeed, cells treated with these compounds resisted apilimod-induced swelling and retained higher endo/lysosome number relative to results in the apilimod-only condition (Fig. 7A-C for nocadozole, Fig. S5A-C for ciliobrevin). Conversely, endo/lysosome shrinkage was accelerated upon washing apilimod in microtubule-disrupted cells (Fig. 7D-F). To complement the pharmacological approach, we transfected cells with the p50 dynamitin subunit or dominant negative kinesin-1 to respectively impair dynein and kinesin. We observed reduced swelling and increased endo/lysosome numbers during apilimod exposure relative to untransfected cells (Fig. S5D-I). Overall these data suggest that endo/lysosomes swell in PIKfyve-inhibited cells through endo/lysosome coalescence.
Fig. 7.
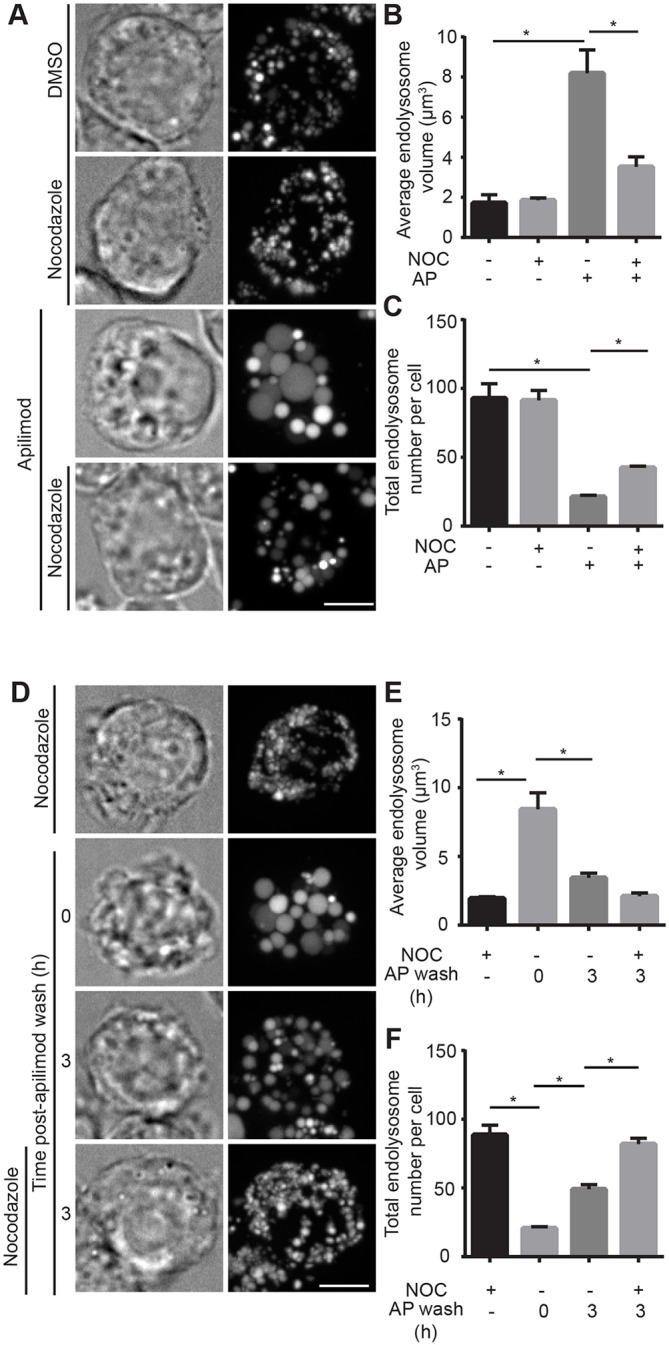
Volumetric analysis of endo/lysosome number and volume in cells disrupted for microtubule function. (A) Cells were pre-labelled with Alexa Fluor 488-conjugated dextran, followed by vehicle alone, 10 µM nocadozole for 60 min, or co-treated with apilimod and vehicle or nocadozole for 1 h. Fluorescence micrographs are z-projections of 45-55 z-planes acquired by spinning disc confocal microscopy. (B,C) Quantification of individual endo/lysosome volume (B) and endo/lysosome number per RAW macrophage (C). (D) Cells were pre-labelled with Alexa Fluor 488-conjugated dextran as before, followed by 20 nM apilimod for 1 h and then replaced with fresh medium or medium containing nocadozole for the indicated time period. (E,F) Volumetric quantification of individual endo/lysosome volume (E) and endo/lysosome number per cell (D). In all cases, data shown are the mean±s.e.m. from three independent experiments, with at least 15-20 cells per condition per experiment. *P<0.05 statistical difference between conditions indicated by lines using one-way ANOVA and Tukey's post hoc test. Scale bars: 5 µm.
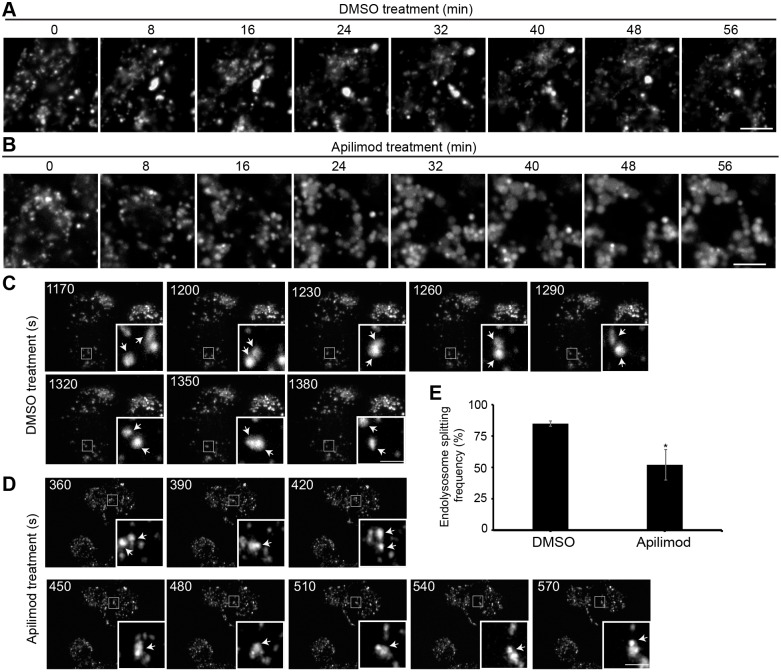
Lysosomes undergo constant rounds of ‘kiss-and-run’ fusion and fission (Duclos et al., 2003; Bright et al., 2005; Pryor et al., 2000). We propose a model in which PIKfyve, probably through synthesis of PtdIns(3,5)P2, controls endo/lysosome fission. Consequently, in PIKfyve-inhibited cells, lysosomes fail to perform fission and/or ‘kiss-and-marry’, resulting in endo/lysosome coalescence, enlargement and a drop in total lysosome number. To support this model, we tried to visualize endo/lysosome swelling during apilimod exposure in RAW cells by using live-cell spinning disc confocal microscopy to obtain z-stacks at high temporal resolution. However, endo/lysosomes under observation failed to swell when acquiring z-stacks even as infrequently as every 2 min (Fig. S6, Movies 1 and 2). In addition, lysosomes also appeared to become less dynamic. Indeed, when we moved to a different field of view within the same coverslip after the period of observation, endo/lysosomes were engorged (Fig. S6). This suggests that photo-toxicity impairs apilimod-dependent endo/lysosome swelling. We could observe dynamic endo/lysosome movement and enlargement when imaging single planes at 1 frame/2 min (Fig. 8A,B, Movies 3 and 4). Unfortunately, this made tracking of individual endo/lysosomes difficult due to their highly dynamic nature.
Fig. 8.
Live-cell imaging of apilimod-induced lysosome swelling. (A,B) Cells were pre-labelled with Alexa Fluor 546-conjugated dextran, followed by (A) vehicle alone or (B) 20 nM apilimod at time 0 min. Single-plane images were obtained by spinning disc microscopy at 1 frame/2 min. Time in min refers to time after adding vehicle or apilimod. See Movies 3 and 4 for full capture. Scale bars: 5 µm. (C,D) Swep-tfield imaging of RAW cells exposed to (C) vehicle or (D) 20 nM apilimod. See Movies 5 and 6 for full capture. Shown are z-collapsed frames at 30 s intervals. The inset is a magnified portion of the field-of-view tracking a few endo/lysosomes (arrows) undergoing contact and split events, which may represent fusion or fission. Scale bars: 3 µm. (E) Rate of ‘splitting’ events over 30 min in vehicle- or apilimod-treated cells showing a reduction in splitting in PIKfyve-inhibited cells, which may reflect reduced fission. *P<0.05 by Student's t-test.
To bypass the photo-toxicity issue, we switched to swept-field confocal microscopy, which employs dramatically less light energy. Using this technology, we could reconstruct cells at high temporal resolution and using z-stacks. Indeed, we could track the dynamics of endo/lysosomes in control cells and those undergoing vacuolation in PIKfyve-inhibited cells over 2 h (Fig. 8C,D, Movies 5 and 6). When imaging untreated cells, endo/lysosomes proved to be highly dynamic structures undertaking significant number of collisions, splitting and ‘tubulation’ events (Fig. 8C, Movie 5). After adding apilimod, endo/lysosomes underwent enlargement and became fewer in number through coalescence (Fig. 8D, Movie 6). However, it is possible to see that even enlarged endo/lysosome vacuoles in PIKfyve-suppressed cells retain their dynamic nature. To try to quantify fission and ‘run’ events, we used particle tracking and splitting functions in Imaris as a proxy. Although this analysis cannot distinguish between fission and separation of overlapping but independent particles, we reasoned that this could estimate differences in fission rates if the defect was sufficiently large. Indeed, while ∼85% of endo/lysosomes ‘split’ in control cells over a period of 30 min, this was reduced to ∼50% in apilimod-treated cells (Fig. 8E). Altogether, we propose that PIKfyve loss of function causes lysosome enlargement by reducing the rate of lysosome fission/‘run’ events relative to lysosome fusion/‘kiss’ events, leading to lysosome coalescence.
DISCUSSION
Disruption of PIKfyve activity causes several major physiological defects, including neurological decay, lethal embryonic development and inflammatory disease, among others (Ikonomov et al., 2011; Min et al., 2014; Chow et al., 2007; McCartney et al., 2014). It is unclear how these defects arise, but they are likely to relate to disruption of endo/lysosome homeostasis, including impaired hydrolytic function, reduced rates of autophagic flux and endo/lysosome swelling (Martin et al., 2013; de Lartigue et al., 2009; Min et al., 2014; Kim et al., 2016). Endo/lysosome vacuolation remains the most dramatic and visible defect in PIKfyve-abrogated cells. Yet, the mechanism by which endo/lysosomes become engorged remains poorly defined. The prevailing model is that recycling from endo/lysosomes is impaired in PIKfyve/Fab1-deficient cells, thus causing endo/lysosome and vacuole swelling (McCartney et al., 2014; Ho et al., 2012). Arguably, this is assumed to proceed through tubulo-vesicular transport intermediates formed during membrane fission catalysed by coat proteins like the retromer (Bryant et al., 1998; de Lartigue et al., 2009; Rutherford et al., 2006; Zhang et al., 2007). Here, we explored two other possibilities that could contribute to endo/lysosome enlargement: enhanced endo/lysosome biosynthesis and/or an imbalance in endo/lysosome fusion-fission dynamics. Collectively, our work intimates that enhanced lysosome biosynthesis does not drive endo/lysosome swelling. Instead, our work suggests that endo/lysosomes coalescence during PIKfyve inhibition, resulting in concurrent enlargement of endo/lysosomes and abatement in their numbers. We provide evidence that coalescence is due to a shift away from fission during endo/lysosome fusion-fission cycling (Fig. S7). Our observations are consistent with recent work by Bissig et al. (2017), who suggest a role for PIKfyve in reforming terminal lysosomes from endolysosomes, a term they use to define a subset of lysosomes that are acidic and proteolytic.
PIKfyve and TFEB function
Inhibition of PIKfyve caused rapid and robust nuclear translocation of TFEB, TFE3 and MITF. Indeed, others have recently observed that TFEB shuttles into the nucleus in PIKfyve-inhibited cells (Gayle et al., 2017; Wang et al., 2015). Consistent with nuclear translocation, we also observed increased mRNA levels of selected lysosomal genes including LAMP1, V-ATPase subunits and cathepsins. This increase is in line with a recent study that looked at transcriptome-wide changes in mRNA levels in B-cell non-Hodgkins lymphoma cells treated with apilimod (Gayle et al., 2017). This suggests that PIKfyve regulates a negative-feedback loop to control lysosome biogenesis through suppression of TFEB and related factors. However, acute inhibition of PIKfyve led either to no change, or in the case of LAMP1, to a small increase in protein levels after 6 h of PIKfyve inhibition, despite the augmented mRNA levels. The decoupling between enhanced transcription and constant protein levels may result from pleiotropic effects of PIKfyve inhibition. For example, PIKfyve is known to govern trafficking of newly synthesized lysosomal genes by controlling recycling of mannose-6-phosphate receptors (Gayle et al., 2017; Ikonomov et al., 2009a; Rutherford et al., 2006; Zhang et al., 2007). By contrast, cells chronically impaired for PIKfyve have augmented LAMP1 levels (Ferguson et al., 2009, 2010). However, this LAMP1 accumulation is thought to occur through impaired autophagosome turnover rather than an increase in biosynthesis of LAMP1 (Ferguson et al., 2009; Kim et al., 2016). This may be consistent with the small increase in LAMP1 after prolonged inhibition of PIKfyve (Fig. 3C). Overall, given that endo/lysosome enlargement was unaffected in cells deleted for TFEB, TFE3 and/or MITF or treated with cycloheximide, we argue that biosynthesis does not significantly contribute to lysosome enlargement in cells acutely inhibited for PIKfyve, although we do not exclude a role in other PIKfyve-mediated functions.
PIKfyve and TFEB regulation
TFEB and TFE3 are maintained in the cytosol via phosphorylation on specific residues that form docking sites for the 14-3-3 scaffolding protein (Roczniak-Ferguson et al., 2012; Martina et al., 2012). We now know that TFEB and related factors are subjected to multiple kinases including mTORC1, PKC, PKD, GSK3β and phosphatases such as the Ca2+-dependent calcineurin (Li et al., 2016b; Martina et al., 2014; Roczniak-Ferguson et al., 2012; Settembre et al., 2012; Najibi et al., 2016; Ploper et al., 2015; Marchand et al., 2015; Wang et al., 2015). However, mTORC1 regulation of TFEB remains the best-understood regulatory circuit. Given that PIKfyve is needed for mTORC1 activity in adipocytes (Bridges et al., 2012), we examined whether there could be a PIKfyve-mTORC1-TFEB axis. However, S6K remained phosphorylated in apilimod-treated cells, suggesting that mTORC1 was active. These observations are consistent with the work of Wang et al. (2015) and Krishna et al. (2016) who also showed that mTORC1 retains its activity in PIKfyve-abrogated cells. We considered the possibility that mTORC1 and starvation control PIKfyve activity, which could then modulate TFEB, perhaps by PtdIns(3,5)P2-dependent localization of TFEB to lysosomes, as has been shown for Tup1, a transcription factor in yeast that controls galactose metabolism (Han and Emr, 2011). However, we did not observe a significant decrease in PtdIns(3,5)P2 levels in mTOR-inhibited cells, although we cannot exclude disruption of a specific pool of PtdIns(3,5)P2 or of PtdIns(5)P, a lipid that is directly or indirectly synthesized by PIKfyve (Zolov et al., 2012; Sbrissa et al., 2012). Overall, our data suggest that mTORC1 and PIKfyve regulate TFEB in parallel. We speculate that PIKfyve may regulate other modulators of TFEB such as PKD, PKC and GSK3β.
Enlargement of lysosomes in PIKfyve-suppressed cells
Loss of PIKfyve causes massive lysosome swelling. This has been partially explained through a defect in membrane recycling from endosomes and/or lysosomes (Ho et al., 2012; McCartney et al., 2014). This model arose through experiments done in yeast expressing an engineered ALP protein carrying a Golgi retrieval sequence (RS-ALP). RS-ALP traffics to the vacuole, where it is subjected to proteolytic maturation. Mature RS-ALP appears in the Golgi membrane fraction in wild-type yeast cells, but not in cells with defective PtdIns(3,5)P2 activity, suggesting a defect in retrieval from the vacuole (Bryant et al., 1998; Dove et al., 2004). In addition, PIKfyve appears to regulate retrieval of mannose-6-phosphate receptors from endo/lysosomes in mammalian cells, which suggests a link between PIKfyve and coat proteins such as retromer (Ikonomov et al., 2009b; Rutherford et al., 2006; Zhang et al., 2007). Hence, endo/lysosome enlargement may partly occur through an imbalance in membrane influx, caused by unabated anterograde membrane flow and hindered recycling due to defects in forming tubulo-vesicular intermediates (Fig. S7). However, our observations also suggest that the number of endo/lysosomes in PIKfyve-suppressed cells is significantly reduced. The diminished endo/lysosome number is unlikely to be due to lysosome lysis since the total endo/lysosome volume was unchanged in PIKfyve-arrested cells. Instead, we propose that PIKfyve suppression causes an imbalance in lysosome fusion-fission cycling, leading to lysosome coalescence and enlargement (Fig. S7).
Lysosomes constantly interact with each other through full fusion and fission, or through ‘kiss-and-run’ events. During the ‘kiss’ event, at least two lysosomes dock and form a transient pore to help exchange and homogenize their luminal content (Bright et al., 2005; Luzio et al., 2009; Duclos et al., 2000). However, complete fusion and coalescence is pre-empted by the ‘run’ event, when the two lysosomes separate, thus preventing coalescence. Our work suggests that PIKfyve, probably through PtdIns(3,5)P2, plays a role in mediating lysosome fission to balance fusion. This role may occur during complete fusion-fission cycles and/or by catalysing the ‘run’ event during ‘kiss-and-run’ (Fig. S7). In either case, failure to perform fission would result in overt coalescence, resulting in lysosome enlargement and reduced numbers. This model is consistent with recent work by Bissig et al. (2017), who suggest a role for PIKfyve in reforming terminal lysosomes from endolysosomes. We do not understand how PIKfyve modulates lysosome fission dynamics. However, we speculate that mechanisms that offset lysosome coalescence are distinct from those that drive vesiculation and recycling (Krishna et al., 2016; Bryant et al., 1998; Rutherford et al., 2006). One interesting possibility may relate to contact sites between the endo/lysosome and the endoplasmic reticulum since these have been shown to demarcate and assemble molecular machines that catalyse endolysosome fission (Friedman et al., 2013; Rowland et al., 2014). Thus, it is appealing to speculate that PIKfyve and PtdIns(3,5)P2 may mediate this process or be regulated by these contact sites to drive endo/lysosome fission.
Overall, we examined two possible mechanisms that might have contributed to endo/lysosome enlargement in PIKfyve-inhibited cells. While TFEB and related factors are controlled by PIKfyve, which probably establishes a negative-feedback loop for lysosome biogenesis, we did not obtain evidence to suggest that biosynthesis contributes to lysosome swelling during acute PIKfyve inhibition. Instead, we observed that loss of PIKfyve activity caused a concomitant reduction in the number and increase in the volume of endo/lysosomes, suggesting that endo/lysosomes undergo coalescence through fusion and impaired fission. Future work should delineate the contribution of lysosome coalescence and vesicular recycling to lysosome enlargement.
MATERIALS AND METHODS
Cell culture and transfection
RAW 264.7 macrophage strains and HeLa cell strains were cultured in Dulbecco's Modified Eagle Medium (DMEM; Wisent, St Bruno, QC) supplemented with 5% heat-inactivated fetal bovine serum (FBS; Wisent). Mouse embryonic fibroblast (MEF) cells were cultured in Roswell Park Memorial Institute medium (RPMI-1640; Gibco, Burlington, ON) with 15% FBS. All cells were authenticated, checked for contamination and cultured at 37°C with 5% CO2. Transient transfections were performed with FuGene HD (Promega, Madison, WI) following the manufacturer's instructions, using a 3:1 DNA to transfection reagent ratio. The transfection mixture was replaced with fresh medium after 4-6 h and cells were used 24 h post-transfection. Silencing RNA oligonucleotides were electroporated into RAW cells with the Neon Electroporation system (Life Technologies, Burlington, ON) following the manufacturer's instructions. Briefly, two rounds of knockdown were performed using ON-TARGETplus PIKfyve SMARTpool siRNA (L-040127-00-0005) or ON-TARGETplus non-targeting control pool (GE Healthcare, Mississauga, ON) over 48 h.
Pharmacological manipulation of cells
Cells were treated with apilimod (Toronto Research Chemicals, Toronto, ON) at indicated concentrations and times. Torin-1 (Tocris Bioscience, Minneapolis, MN) or rapamycin (BioShop, Burlington, ON) were used as positive controls for TFEB nuclear localization and mTORC1 inhibition. Cells were also incubated with 100 µM ciliobrevin D (EMD Millipore, Toronto, ON) or 10 µM nocodazole (Sigma-Aldrich, Oakville, ON).
Western blotting
Whole-cell lysates were generated with 2× Laemmli sample buffer supplemented with protease inhibitor cocktail (Sigma-Aldrich) and PhosSTOP phosphatase inhibitor (Roche, Laval, QC) where necessary. Lysates were passed through a 27-gauge needle six times and heated. Puromycylation assays were carried out with the addition of 10 µg/ml puromycin for 15 min before cell lysis. When protein dephosphorylation was required, cells were lysed with 1% Triton-X100 in PBS supplemented with protease inhibitor cocktail, centrifuged at 16,000 g for 10 min, and the supernatant was subjected to lambda protein phosphatase (NEB, Whitby, ON) as per the manufacturer's instruction. Briefly, 20 µg protein was incubated with 50 mM HEPES, 100 mM NaCl, 2 mM DTT, 0.01% Brij 25, 1 mM MnCl2 and 200 units of phosphatase at 30°C for 60 min. The reaction was stopped with 5× Laemmli sample buffer and heated.
Most samples were subjected to SDS-PAGE through a 10% acrylamide resolving gel. To assay for TFEB mobility shift, we used an 8% acrylamide resolving gel supplemented with 12.5 μM Phos-tag (Wako Chemicals USA, Richmond, VA) and 25 μM MnCl2. Phos-tag gels were washed three times with 100 mM EDTA for 10 min each, followed by a wash with Transfer buffer (25 mM Tris-HCl, 192 mM glycine, 20% methanol). Proteins were transferred to PVDF, blocked and incubated with primary and HRP-linked secondary antibodies using Tris-buffered saline plus 0.1% Tween-20 with either 5% skimmed milk or 3% BSA. Proteins were visualized using Clarity enhanced chemiluminescence (Bio-Rad Laboratories, Mississauga, ON) with a ChemiDoc XRS+ or ChemiDoc Touch imaging system (Bio-Rad). Quantification of proteins were determined using Image Lab software (Bio-Rad) by normalizing against a loading control and then normalizing against the vehicle-treated control. Rabbit monoclonal antibodies used were against cathepsin D (1:1000, GTX62603, GeneTex Inc., Irvine, CA), HSP60 (1:1000, 12165S, Cell Signaling Technologies, Danvers, MA), and p70 S6 kinase (1:1000, 2708S, Cell Signaling). Rabbit polyclonal antibodies were used against ATP6V1H (1:1000, GTX110778, GeneTex), TFEB (1:2000, A303-673A, Bethyl Laboratories, Montgomery, TX), phosphoThr389-p70 S6 kinase (1:1000, 9205S, Cell Signaling) and β-actin (1:1000, 4967S, Cell Signaling). Rat monoclonal antibodies were used against LAMP1 (1:200, 1D4B, Developmental Studies Hybridoma Bank, Iowa City, IO). Mouse monoclonal antibodies were used against puromycin (1:1000, 540411, Millipore, Burlington, MA) and p53 (1:1000, 2524S, Cell Signaling). Secondary antibodies were raised in donkey (Bethyl) and conjugated to HRP.
Quantitative RT-PCR
Total RNA was extracted from RAW cells using the GeneJET RNA purification kit (Thermo Fisher Scientific, Mississauga, ON). Equal quantities of mRNA were reverse transcribed with SuperScript VILO cDNA synthesis kit (Invitrogen) following the manufacturer's guidelines. The subsequent cDNA was diluted 1:100 and amplified for quantitative PCR using the TaqMan Fast Advanced Master Mix (Applied Biosystems, Foster City, CA) in the presence of TaqMan assays with a Step One Plus Real-Time PCR thermal cycler (Applied Biosystems) with Step One software (v.2.2.2; Applied Biosystems). The TaqMan gene expression assays (Invitrogen) for the reference gene Abt1 (Mm00803824_m1) and target genes Ctsd (Mm00515586_m1), Atp6v1h (Mm00505548_m1), ATP6V1D (Mm00445832_m1), LAMP1 (Mm00495262_m1) and Mcoln1 (Mm00522550_m1) were done in triplicate. Target gene expression was determined by relative quantification (ΔΔCt method) to Abt1 and the vehicle-treated control sample.
Endo/lysosomal labelling
Endo/lysosomes in RAW cells, HeLa cells and MEFs were labelled by incubating cells with either 200 µg/ml Alexa Fluor 488- or 546-conjugated dextran (ThermoFisher) or with 2.5 mg/ml Lucifer Yellow (ThermoFisher) in cell-specific complete medium for 2 h at 37°C in 5% CO2, followed by washing in phosphate-buffered saline and replenishing with fresh complete medium for 1 h before live-cell imaging. This method is likely to label a spectrum of late endosomes, lysosomes and endolysosomes, which are endosome-lysosome hybrids (Huotari and Helenius, 2011). Thus, we refer to dextran- or Lucifer Yellow-labelled structures as endo/lysosomes.
Epifluorescence and spinning disc confocal microscopy
Manual quantification of lysosome size and imaging of GFP-based transfections were imaged using a Leica DM5000B system outfitted with a DFC350FX camera and controlled by Leica application suite (Leica Microsystems Inc., Concord, ON). TFEB-GFP transfected into PIKfyve knockout cells and endogenous TFEB in RAW cells were imaged using an Olympus IX83 microscope with a Hamamatsu ORCA-Flash4.0 digital camera controlled by CellSens Dimensions software (Olympus Canada Inc., Richmond Hill, ON). Endogenous TFEB was further deconvolved using a constrained iterative in CellSens Dimensions. All other imaging of fixed cells was done with a Quorum DisKovery Spinning Disc confocal microscope system equipped with a Leica DMi8 microscope connected to Andor Zyla 4.2 Megapixel sCMOS camera and controlled by Quorum Wave FX Powered by MetaMorph software (Quorum Technologies, Guelph, ON). Live-cell imaging by spinning disc confocal microscopy was accomplished with either an Olympus IX81 inverted microscope equipped with a Hamamatsu C9100-13 EMCCD camera and a 60×1.35 N.A. objective and controlled with Volocity 6.3.0 (PerkinElmer, Bolton, ON). Alternatively, we employed the Quorum DisKovery Spinning Disc confocal microscope system above but using the iXON 897 EMCCD camera. Cells were imaged live using cell-specific complete medium in a 5% CO2 chamber at 37°C. All microscopes were equipped with standard filters appropriate to fluorophores used in this study.
3H-myo-inositol labelling of phosphoinositides and phosphoinositide measurement by HPLC-coupled flow scintillation
RAW cells were incubated for 24 h in inositol-free DMEM (MP Biomedical, CA) with 10 µCi/ml myo-[2-3H(N)] inositol (Perkin Elmer, MA), 10% dialyzed FBS (Gibco), 4 mM L-glutamine (Sigma Aldrich), 1× insulin-transferrin-selenium-ethanolamine (Gibco) and 20 mM HEPES (Gibco). Cells were then treated with torin1 or apilimod for 1 h. Reactions were arrested in 600 µl of 4.5% perchloric acid (v/v) on ice for 15 min, scraped, and pelleted at 12,000 g for 10 min. Pellets were washed with 1 ml ice-cold 0.1 M EDTA and resuspended in 50 µl water. Phospholipid deacylation was then done in 500 µl of methanol/40% methylamine/1-butanol [45.7% methanol: 10.7% methylamine: 11.4% 1-butanol (v/v)] for 50 min at 53°C. Samples were vacuum-dried and washed twice by resuspending in 300 µl water and drying. Dried samples were resuspended again in 450 µl water and extracted with 300 µl 1-butanol/ethyl ether/ethyl formate (20:4:1), vortexed for 5 min and centrifuged at 12,000 g for 2 min. The bottom aqueous layer was collected and extracted two more times. The aqueous layer was dried in vacuum and resuspended in 50 µl water. Equal counts of 3H were separated by HPLC (Agilent Technologies, Mississauga, ON) through an anion exchange 4.6×250-mm column (Phenomenex, Torrance, CA) with a flow rate of 1 ml/min and subjected to a gradient of water (buffer A) and 1 M (NH4)2HPO4, pH 3.8 (adjusted with phosphoric acid) (buffer B) as follows: 0% B for 5 min, 0 to 2% B for 15 min, 2% B for 80 min, 2 to 10% B for 20 min, 10% B for 30 min, 10 to 80% B for 10 min, 80% B for 5 min, 80 to 0% B for 5 min. The radiolabelled eluate was detected by β-RAM 4 (LabLogic, Brandon, FL) with a 1:2 ratio of eluate to scintillant (LabLogic) and analyzed by Laura 4 software. Phosphoinositides were normalized against the parent phosphatidylinositol peak.
Live-cell imaging with swept-field confocal microscopy
Cells were seeded into single glass-bottom micro-well dishes (MatTek, Ashland, MA) and grown to 70-80% confluency. Cells were then incubated for 2 h in 200 µl DMEM medium supplemented with 10% FBS with a 200 µg/ml Alexa Fluor 555-conjugated dextran (ThermoFisher). Samples were washed and incubation medium was replaced with fresh complete DMEM followed by a 90 min post-incubation period. Live-cell imaging was performed with a Nikon Ti inverted microscope equipped with a Prairie Technologies swept-field slit scanning confocal scan head and a 100×1.49 N.A. objective (Prairie Technologies, Sioux Falls, SD). Nikon NIS Elements software equipped with a Photometrics Prime 95B back illuminated sCMOS camera was used for image acquisition while high-speed triggered acquisition was controlled by a National Instruments DAQ card and Mad City Labs Piezo Z stage controller. Sample volumes were acquired at 100 ms per z-slice at 30 s interval over a 30 min observation period before adding 20 nM apilimod and then for 2 h in the presence of apilimod.
Immunofluorescence
Following treatment, cells were fixed with 4% paraformaldehyde (v/v) for 15 min. Cells were blocked and permeabilized with 0.2% (v/v) Triton X-100 and 2% (v/v) BSA for 10 min, then subjected to rabbit polyclonal antibody against TFEB (1:250; Bethyl) and Dylight-conjugated donkey polyclonal antibody against rabbit (1:500; Bethyl). Nuclei were counterstained with 0.4 μg/ml DAPI. For LAMP1-specific immunostaining, cells were fixed and permeabilized with 100% ice-cold methanol for 5 min, followed by a 2% BSA block, incubation with Dylight-conjugated rat monoclonal antibody against LAMP1 (1:200, Clone 1D4B; Developmental Studies Hybridoma Bank) and polyclonal donkey anti-rat IgG antibodies (1:500; Bethyl).
Imaging analysis and volumetrics
To quantify the proportion of cells with nuclear TFEB-GFP (including TFE3-GFP or MiTF-GFP), RAW cells were visually scored as cytosolic if the cytosol had equal or greater staining intensity compared with the nucleus. Alternatively, the nuclear:cytosolic ratio of endogenous TFEB was measured as the ratio of the mean intensity in the nucleus over the mean intensity in the cytosol using ImageJ. To manually quantify vacuole diameter in DIC images, vacuoles were first defined as having a diameter greater than 1.5 µm and then the diameter was measuring using line plot in ImageJ.
To quantify average endo/lysosome volume and number per cells, we employed volumetric and particle detection tools in Volocity (Volocity 6.3.0) or Icy BioImage Analysis. Z-stack images were imported into the software and subject to signal thresholding. First, we tried thresholding at 1.5×, 2× and 2.5× average cytosolic fluorescence background. This was then subject to particle detection by defining particles as those with a volume >0.3 µm3, which helped eliminate noise-derived particles. Regions of interest were then drawn around each cell for analysis and particles split using the watershed function. Importantly, while increasing thresholding levels reduced absolute number of particles, particle size and total particle volume, (i) the reduction occurred at similar levels between control and apilimod-treated cells and (ii) the relative trends between control and apilimod-treated cells remained the same (Fig. S3A,B). Hence, we employed a threshold of 2× the average cytosol fluorescence intensity for the remaining analysis. Analysis of endo/lysosome splitting frequency was performed using Imaris (BitPlane, Concord, MA) using its ‘ImarisTrackLineage’ module, where endo/lysosome splitting was defined as the frequency of events producing two particles from a single apparent particle. Image adjustments such as enhancing contrast were done with ImageJ or Adobe Photoshop (Adobe Systems, San Jose, CA) without altering the relative signals within images. Figures were assembled with Adobe Illustrator (Adobe Systems).
Statistical analysis
All experiments were repeated at least three independent times. The respective figure legends indicate the total number of experiments and number of samples/cells assessed. The mean and measure of variation (s.d.) or error (s.e.m.) are indicated.
All experimental conditions were statistically analysed with either an unpaired Student's t-test when comparing two parameters only or with a one-way ANOVA test when comparing multiple conditions in non-normalized controls. ANOVA tests were coupled to Tukey's post hoc test to analyse pairwise conditions. For multiple comparisons with normalized controls, Student's t-tests were utilized with Bonferroni correction. Here, we accept P<0.05 as indication of statistical significance.
Supplementary Material
Acknowledgements
Plasmid constructs expressing the kinesin-1 dominant negative and of the p50 dynamitin subunit were obtained from Addgene (Cambridge, MA). Plasmids encoding GFP fusion proteins of TFEB, TFE3 and MITF were kind gifts from Dr Shawn Ferguson at Yale University. The Tfeb−/− Tfe3−/− Mitf−/− HeLa cells were a kind gift from Dr Richard Youle at the National Institutes of Health.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.J.B.; Methodology: C.H.C., G.S., M.A.G., Z.-Y.A.O., G.L., R.P.; Software: G.S.; Formal analysis: C.H.C., G.S., S.C.W., R.J.B.; Investigation: C.H.C., G.S., M.A.G., C.W., R.M.D., R.J.B.; Resources: G.L., R.P., S.C.W., R.J.B.; Data curation: C.H.C., G.S., M.A.G., C.W., R.M.D., Z.-Y.A.O.; Writing - original draft: C.H.C., G.S., R.J.B.; Writing - review & editing: R.J.B.; Visualization: C.H.C., G.S., M.A.G., C.W., R.M.D., Z.-Y.A.O., S.C.W.; Supervision: S.C.W., R.J.B.; Project administration: R.J.B.; Funding acquisition: R.J.B.
Funding
The research performed in the lab of R.J.B. was funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada, by the Early Researcher Award program from the Government of Ontario, by a Tier II Canada Research Chairs Award and by contributions from Ryerson University. Research by C.H.C. was made possible through support from an Ontario Graduate Scholarship and Queen Elizabeth II Graduate Scholarship. G.L. was funded through the National Institutes of Health (GM24872). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.213587.supplemental
References
- Bagshaw R. D., Callahan J. W. and Mahuran D. J. (2006). The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem. Biophys. Res. Commun. 344, 1186-1191. 10.1016/j.bbrc.2006.03.221 [DOI] [PubMed] [Google Scholar]
- Bissig C., Hurbain I., Raposo G. and van Niel G. (2017). PIKfyve activity regulates reformation of terminal storage lysosomes from endolysosomes. Traffic. 11, 747-757. 10.1111/tra.12525 [DOI] [PubMed] [Google Scholar]
- Bridges D., Ma J.-T., Park S., Inoki K., Weisman L. S. and Saltiel A. R. (2012). Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol. Biol. Cell 23, 2955-2962. 10.1091/mbc.E11-12-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright N. A., Gratian M. J. and Luzio J. P. (2005). Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr. Biol. 15, 360-365. 10.1016/j.cub.2005.01.049 [DOI] [PubMed] [Google Scholar]
- Bryant N. J., Piper R. C., Weisman L. S. and Stevens T. H. (1998). Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J. Cell Biol. 142, 651-663. 10.1083/jcb.142.3.651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Xu Y., Cheung A. K., Tomlinson R. C., Alcázar-Román A., Murphy L., Billich A., Zhang B., Feng Y., Klumpp M. et al. (2013). PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chem. Biol. 20, 912-921. 10.1016/j.chembiol.2013.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. H. M., Reyes L., Sohail S. M., Tran N. K. and Marshall W. F. (2016). Organelle size scaling of the budding yeast vacuole by relative growth and inheritance. Curr. Biol. 26, 1221-1228. 10.1016/j.cub.2016.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. Y., Zhang Y., Dowling J. J., Jin N., Adamska M., Shiga K., Szigeti K., Shy M. E., Li J., Zhang X. et al. (2007). Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature 448, 68-72. 10.1038/nature05876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currinn H., Guscott B., Balklava Z., Rothnie A. and Wassmer T. (2016). APP controls the formation of PI(3,5)P2 vesicles through its binding of the PIKfyve complex. Cell. Mol. Life Sci. 73, 393-408. 10.1007/s00018-015-1993-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lartigue J., Polson H., Feldman M., Shokat K., Tooze S. A., Urbé S. and Clague M. J. (2009). PIKfyve regulation of endosome-linked pathways. Traffic 10, 883-893. 10.1111/j.1600-0854.2009.00915.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delamarre L., Pack M., Chang H., Mellman I. and Trombetta E. S. (2005). Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307, 1630-1634. 10.1126/science.1108003 [DOI] [PubMed] [Google Scholar]
- Deng Y. P. and Storrie B. (1988). Animal cell lysosomes rapidly exchange membrane proteins. Proc. Natl. Acad. Sci. USA 85, 3860-3864. 10.1073/pnas.85.11.3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villen J., Beausoleil S. A., Bakalarski C. E., Elledge S. J. and Gygi S. P. (2008). A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 105, 10762-10767. 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L. S., Delling M. et al. (2010). PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat. Commun. 1, 38 10.1038/ncomms1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove S. K., Piper R. C., McEwen R. K., Yu J. W., King M. C., Hughes D. C., Thuring J., Holmes A. B., Cooke F. T., Michell R. H. et al. (2004). Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 23, 1922-1933. 10.1038/sj.emboj.7600203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclos S., Diez R., Garin J., Papadopoulou B., Descoteaux A., Stenmark H. and Desjardins M. (2000). Rab5 regulates the kiss and run fusion between phagosomes and endosomes and the acquisition of phagosome leishmanicidal properties in RAW 264.7 macrophages. J. Cell Sci. 113, 3531-3541. [DOI] [PubMed] [Google Scholar]
- Duclos S., Corsini R. and Desjardins M. (2003). Remodeling of endosomes during lysosome biogenesis involves “kiss and run” fusion events regulated by rab5. J. Cell Sci. 116, 907-918. 10.1242/jcs.00259 [DOI] [PubMed] [Google Scholar]
- Ferguson C. J., Lenk G. M. and Meisler M. H. (2009). Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum. Mol. Genet. 18, 4868-4878. 10.1093/hmg/ddp460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson C. J., Lenk G. M. and Meisler M. H. (2010). PtdIns(3,5)P2 and autophagy in mouse models of neurodegeneration. Autophagy 6, 170-171. 10.4161/auto.6.1.10626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. R., DiBenedetto J. R., West M., Rowland A. A. and Voeltz G. K. (2013). Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol. Biol. Cell 24, 1030-1040. 10.1091/mbc.E12-10-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayle S., Landrette S., Beeharry N., Conrad C., Hernandez M., Beckett P., Ferguson S. M., Xu T., Rothberg J. and Lichenstein H. (2017). B-cell non-Hodgkin lymphoma: Selective vulnerability to PIKFYVE inhibition. Autophagy. 13, 1768-1778. 10.1080/15548627.2017.1304871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. A., Choy C. H., Dayam R. M., Ospina-Escobar E., Somerville A., Xiao X., Ferguson S. M. and Botelho R. J. (2016). Phagocytosis enhances lysosomal and bactericidal properties by activating the transcription factor TFEB. Curr. Biol. 26, 1955-1964. 10.1016/j.cub.2016.05.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B.-K. and Emr S. D. (2011). Phosphoinositide [PI(3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev. 25, 984-995. 10.1101/gad.1998611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. Y., Alghamdi T. A. and Botelho R. J. (2012). Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic 13, 1-8. 10.1111/j.1600-0854.2011.01246.x [DOI] [PubMed] [Google Scholar]
- Huotari J. and Helenius A. (2011). Endosome maturation. EMBO J. 30, 3481-3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov O. C., Sbrissa D. and Shisheva A. (2001). Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J. Biol. Chem. 276, 26141-26147. 10.1074/jbc.M101722200 [DOI] [PubMed] [Google Scholar]
- Ikonomov O. C., Fligger J., Sbrissa D., Dondapati R., Mlak K., Deeb R. and Shisheva A. (2009a). Kinesin adapter JLP links PIKfyve to microtubule-based endosome-to-trans-Golgi network traffic of furin. J. Biol. Chem. 284, 3750-3761. 10.1074/jbc.M806539200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov O. C., Sbrissa D. and Shisheva A. (2009b). YM201636, an inhibitor of retroviral budding and PIKfyve-catalyzed PtdIns(3,5)P2 synthesis, halts glucose entry by insulin in adipocytes. Biochem. Biophys. Res. Commun. 382, 566-570. 10.1016/j.bbrc.2009.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov O. C., Sbrissa D., Delvecchio K., Xie Y., Jin J.-P., Rappolee D. and Shisheva A. (2011). The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve-/- embryos but normality of PIKfyve+/- mice. J. Biol. Chem. 286, 13404-13413. 10.1074/jbc.M111.222364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N., Chow C. Y., Liu L., Zolov S. N., Bronson R., Davisson M., Petersen J. L., Zhang Y., Park S., Duex J. E. et al. (2008). VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 27, 3221-3234. 10.1038/emboj.2008.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. E., Ostrowski P., Jaumouillé V. and Grinstein S. (2016). The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 212, 677-692. 10.1083/jcb.201507112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R. and Neefjes J. (2001). The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11, 1680-1685. 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Kim G. H. E., Dayam R. M., Prashar A., Terebiznik M. and Botelho R. J. (2014). PIKfyve inhibition interferes with phagosome and endosome maturation in macrophages. Traffic 15, 1143-1163. 10.1111/tra.12199 [DOI] [PubMed] [Google Scholar]
- Kim S. M., Roy S. G., Chen B., Nguyen T. M., McMonigle R. J., McCracken A. N., Zhang Y., Kofuji S., Hou J., Selwan E. et al. (2016). Targeting cancer metabolism by simultaneously disrupting parallel nutrient access pathways. J. Clin. Invest. 126, 4088-4102. 10.1172/JCI87148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Palm W., Lee Y., Yang W., Bandyopadhyay U., Xu H., Florey O., Thompson C. B. and Overholtzer M. (2016). PIKfyve regulates vacuole maturation and nutrient recovery following engulfment. Dev. Cell 38, 536-547. 10.1016/j.devcel.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk G. M., Frei C. M., Miller A. C., Wallen R. C., Mironova Y. A., Giger R. J. and Meisler M. H. (2016). Rescue of neurodegeneration in the Fig4 null mouse by a catalytically inactive FIG4 transgene. Hum. Mol. Genet. 25, 340-347. 10.1093/hmg/ddv480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Rydzewski N., Hider A., Zhang X., Yang J., Wang W., Gao Q., Cheng X. and Xu H. (2016a). A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 18, 404-417. 10.1038/ncb3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xu M., Ding X., Yan C., Song Z., Chen L., Huang X., Wang X., Jian Y., Tang G. et al. (2016b). Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol. 18, 1065-1077. 10.1038/ncb3407 [DOI] [PubMed] [Google Scholar]
- Luzio J. P., Pryor P. R. and Bright N. A. (2007). Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8, 622-632. 10.1038/nrm2217 [DOI] [PubMed] [Google Scholar]
- Luzio J. P., Parkinson M. D. J., Gray S. R. and Bright N. A. (2009). The delivery of endocytosed cargo to lysosomes. Biochem. Soc. Trans. 37, 1019-1021. 10.1042/BST0371019 [DOI] [PubMed] [Google Scholar]
- Marchand B., Arsenault D., Raymond-Fleury A., Boisvert F.-M. and Boucher M.-J. (2015). Glycogen Synthase Kinase-3 (GSK3) inhibition induces prosurvival autophagic signals in human pancreatic cancer cells. J. Biol. Chem. 290, 5592-5605. 10.1074/jbc.M114.616714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., Harper C. B., May L. M., Coulson E. J., Meunier F. A. and Osborne S. L. (2013). Inhibition of PIKfyve by YM-201636 dysregulates autophagy and leads to apoptosis-independent neuronal cell death. PLoS ONE 8, e60152 10.1371/journal.pone.0060152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J. A., Chen Y., Gucek M. and Puertollano R. (2012). MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903-914. 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J. A., Diab H. I., Lishu L., Jeong-A L., Patange S., Raben N. and Puertollano R. (2014). The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 7, ra9 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J. A., Diab H. I., Brady O. A. and Puertollano R. (2016). TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 35, 479-495. 10.15252/embj.201593428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayya V., Lundgren D. H., Hwang S.-I., Rezaul K., Wu L., Eng J. K., Rodionov V. and Han D. K. (2009). Quantitative Phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2, ra46 10.1126/scisignal.2000007 [DOI] [PubMed] [Google Scholar]
- Mccartney A. J., Zhang Y. and Weisman L. S. (2014). Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. BioEssays 36, 52-64. 10.1002/bies.201300012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D. L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J. A., Sardiello M. et al. (2011). Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell 21, 421-430. 10.1016/j.devcel.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D. L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A. et al. (2015). Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17, 288-299. 10.1038/ncb3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S. H., Suzuki A., Stalker T. J., Zhao L., Wang Y., McKennan C., Riese M. J., Guzman J. F., Zhang S., Lian L. et al. (2014). Loss of PIKfyve in platelets causes a lysosomal disease leading to inflammation and thrombosis in mice. Nat. Commun. 5, 4691 10.1038/ncomms5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell J. A. (2012). Lysosomal acidification mechanisms. Annu. Rev. Physiol. 74, 69-86. 10.1146/annurev-physiol-012110-142317 [DOI] [PubMed] [Google Scholar]
- Najibi M., Labed S. A., Visvikis O. and Irazoqui J. E. (2016). An evolutionarily conserved PLC-PKD-TFEB pathway for host defense. Cell Rep. 15, 1728-1742. 10.1016/j.celrep.2016.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezich C. L., Wang C., Fogel A. I. and Youle R. J. (2015). MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell. Biol. 210, 435-450. 10.1083/jcb.201501002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore N., Brady O. A., Diab H. I., Martina J. A., Sun L., Huynh T., Lim J.-A. A., Zare H., Raben N., Ballabio A. et al. (2016). TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy 12, 1240-1258. 10.1080/15548627.2016.1179405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Llopis S., Vega-Rubin-de-Celis S., Schwartz J. C., Wolff N. C., Tran T. A. T., Zou L., Xie X.-J., Corey D. R. and Brugarolas J. (2011). Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30, 3242-3258. 10.1038/emboj.2011.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan J. P., Millson S. H., Parker P. J., Piper P. W. and Cooke F. T. (2006). Fab1p and AP-1 are required for trafficking of endogenously ubiquitylated cargoes to the vacuole lumen in S. cerevisiae. J. Cell Sci. 119, 4225-4234. 10.1242/jcs.03188 [DOI] [PubMed] [Google Scholar]
- Ploper D. and De Robertis E. M. (2015). The MITF family of transcription factors: Role in endolysosomal biogenesis, Wnt signaling, and oncogenesis. Pharmacol. Res. 99, 36-43. 10.1016/j.phrs.2015.04.006 [DOI] [PubMed] [Google Scholar]
- Ploper D., Taelman V. F., Robert L., Perez B. S., Titz B., Chen H.-W., Graeber T. G., von Euw E., Ribas A. and De Robertis E. M. (2015). MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. USA 112, E420-E429. 10.1073/pnas.1424576112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor P. R., Mullock B. M., Bright N. A., Gray S. R. and Luzio J. P. (2000). The role of intraorganellar Ca(2+) in late endosome-lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J. Cell Biol. 149, 1053-1062. 10.1083/jcb.149.5.1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Schindler C., Jia R., Jarnik M., Backlund P. and Bonifacino J. S. (2015). BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell 33, 176-188. 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N. and Puertollano R. (2016). TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu. Rev. Cell Dev. Biol. 32, 255-278. 10.1146/annurev-cellbio-111315-125407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A., Caler E. V. and Andrews N. W. (2001). Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell 106, 157-169. 10.1016/S0092-8674(01)00421-4 [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C. S., Froehlich F., Qian S., Ky J., Angarola B., Walther T. C. and Ferguson S. M. (2012). The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Ferreira C. and Munro S. (2011). Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell 21, 1171-1178. 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland A. A., Chitwood P. J., Phillips M. J. and Voeltz G. K. (2014). ER contact sites define the position and timing of endosome fission. Cell 159, 1027-1041. 10.1016/j.cell.2014.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford A. C., Traer C., Wassmer T., Pattni K., Bujny M. V., Carlton J. G., Stenmark H. and Cullen P. J. (2006). The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J. Cell Sci. 119, 3944-3957. 10.1242/jcs.03153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S. and Sabatini D. M. (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290-303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S. et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473-477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Saric A., Hipolito V. E. B., Kay J. G., Canton J., Antonescu C. N. and Botelho R. J. (2016). mTOR controls lysosome tubulation and antigen presentation in macrophages and dendritic cells. Mol. Biol. Cell 27, 321-333. 10.1091/mbc.E15-05-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O. C. and Shisheva A. (1999). PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J. Biol. Chem. 274, 21589-21597. 10.1074/jbc.274.31.21589 [DOI] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O. C., Fu Z., Ijuin T., Gruenberg J., Takenawa T. and Shisheva A. (2007). Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport: Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J. Biol. Chem. 282, 23878-23891. 10.1074/jbc.M611678200 [DOI] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O. C., Filios C., Delvecchio K. and Shisheva A. (2012). Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636. Am. J. Physiol. Cell Physiol. 303, C436-C446. 10.1152/ajpcell.00105.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwake M., Schröder B. and Saftig P. (2013). Lysosomal membrane proteins and their central role in physiology. Traffic 14, 739-748. 10.1111/tra.12056 [DOI] [PubMed] [Google Scholar]
- Settembre C. and Ballabio A. (2014). Lysosomal adaptation: how the lysosome responds to external cues. Cold Spring Harb. Perspect. Biol. 6, a016907 10.1101/cshperspect.a016907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D. L., Vetrini F., Erdin S., Erdin S. U., Huynh T., Ferron M., Karsenty G., Vellard M. C. et al. (2012). A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Eur. Mol. Biol. Organ. J. 31, 1095-1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J., Burke E., Silverstein S. C., Bushnell A. and Silverstein S. C. (1987). Tubular lysosome morphology and distribution within macrophages depend on the integrity of cytoplasmic microtubules. Proc. Natl. Acad. Sci. USA 84, 1921-1925. 10.1073/pnas.84.7.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T., Ashe T. D., Morrison B. E., Soriano K. R., Au J., Roque R. A. V., Lazarowski E. R., Damian V. A., Masliah E. and La Spada A. R. (2012). PGC-1α rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 4, 142ra97 10.1126/scitranslmed.3003799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas J. M., Kim Y.-M., Artavanis-Tsakonas K., Love J. C., Van der Veen A. G. and Ploegh H. L. (2007). Tubulation of class II MHC compartments is microtubule dependent and involves multiple endolysosomal membrane proteins in primary dendritic cells. J. Immunol. 178, 7199-7210. 10.4049/jimmunol.178.11.7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Gao Q., Yang M., Zhang X., Yu L., Lawas M., Li X., Bryant-Genevier M., Southall N. T., Marugan J. et al. (2015). Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc. Natl. Acad. Sci. USA 112, E1373-E1381. 10.1073/pnas.1419669112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J. and Zhu M. X. (2016). Regulation of lysosomal ion homeostasis by channels and transporters. Sci. China Life Sci. 59, 777-791. 10.1007/s11427-016-5090-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zolov S. N., Chow C. Y., Slutsky S. G., Richardson S. C., Piper R. C., Yang B., Nau J. J., Westrick R. J., Morrison S. J., Meisler M. H. and Weisman L. S. (2007). Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc. Natl. Acad. Sci. USA. 104, 17518-17523. 10.1073/pnas.0702275104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolov S. N., Bridges D., Zhang Y., Lee W.-W., Riehle E., Verma R., Lenk G. M., Converso-Baran K., Weide T., Albin R. L. et al. (2012). In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl. Acad. Sci. USA 109, 17472-17477. 10.1073/pnas.1203106109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.