Summary
The development of cell therapy for repairing damaged or diseased skeletal muscle has been hindered by the inability to significantly expand immature, transplantable myogenic stem cells (MuSCs) in culture. To overcome this limitation, a deeper understanding of the mechanisms regulating the transition between activated, proliferating MuSCs and differentiation-primed, poorly engrafting progenitors is needed. Here, we show that methyltransferase Setd7 facilitates such transition by regulating the nuclear accumulation of β-catenin in proliferating MuSCs. Genetic or pharmacological inhibition of Setd7 promotes in vitro expansion of MuSCs and increases the yield of primary myogenic cell cultures. Upon transplantation, both mouse and human MuSCs expanded with a Setd7 small molecule inhibitor are better able to repopulate the satellite cell niche, and treated mouse MuSCs show enhanced therapeutic potential in preclinical models of muscular dystrophy. Thus, Setd7 inhibition may help bypass a key obstacle in the translation of cell therapy for muscle disease.
Introduction
The functional unit of skeletal muscle is the myofiber, a large syncytial structure containing hundreds of nuclei generated by fusion of myogenic progenitors with each other. Gene products are able to diffuse freely within these structures and it has been estimated that replacing a fraction of nuclei in a fiber would be enough to restore sufficient expression of genes mutated in congenital diseases. Thus, skeletal muscle is an ideal target for cell therapy, and this notion fuelled the characterization of adult myogenic progenitors (skeletal muscle stem cells, MuSCs) which are today among the best understood adult stem cells.
Defects in MuSC function have been shown to contribute to the etiology of muscle diseases (Morgan and Zammit, 2010). Age related declines in muscle mass (sarcopenia) and regenerative potential are associated with MuSC senescence (García-Prat et al., 2016; Sousa-Victor et al., 2014) and improper cell cycle kinetics (Chakkalakal et al., 2012; Cosgrove et al., 2014). In muscular dystrophy, MuSCs have been shown to undergo exhaustion (Sacco et al., 2010) and have impaired self-renewal mechanisms (Dumont et al., 2015b). Thus, in addition to the use of myogenic cells as gene delivery vehicles to myofibers, the rejuvenation of the stem cell population by transplantation of expanded MuSCs also represent a promising therapeutic avenue (Marg et al., 2014).
However, the translation of the field’s findings into an efficient cellular therapy has been hampered by our inability to mimic the environment that supports MuSC self-renewal, making in vitro cultivation of ‘transplantable’ MuSCs that retain their potency following in vivo engraftment a significant hurdle (Montarras et al., 2005; Rinaldi and Perlingeiro, 2014).
Skeletal muscle stem cells, also called satellite cells, are identified by the expression of transcription factor Pax7 (Seale et al., 2000) and lie beneath the basal lamina of myofibers (Mauro, 1961). In response to tissue injury, MuSCs progress along a stepwise process to generate MyoD-positive proliferating myoblasts and eventually differentiation-committed myocytes. Myocytes “donate” their nuclei by fusing into damaged myofibers, thus playing an essential role in restoring myofiber function. As a population, MuSCs are capable of returning to their niche and replenishing the stem cell pool, although following damage-induced activation most of their progeny lose this potential and eventually commit to differentiation (Kuang et al., 2007; Montarras et al., 2005; Rocheteau et al., 2012; Sacco et al., 2008). Loss of self-renewal potential is thought to take place shortly following activation, consistent with asymmetric division playing an early role in the maintenance of MuSCs (Dumont et al., 2015a), and has been associated with lower levels of Pax7 expression (Rocheteau et al., 2012).
Recent efforts to provide sufficient numbers of cells for successful therapy have focused on optimizing in vitro conditions that permit propagation of MuSCs whilst maintaining an undifferentiated state. Strategies aimed at “rejuvenating” aged myogenic MuSCs have included culturing cells on substrates that mimic the in vivo muscle niche (Gilbert et al., 2010; Quarta et al., 2016) and using small molecules to target signaling pathways involved in differentiation (Bernet et al., 2014; Cosgrove et al., 2014; Tierney et al., 2014). These strategies represent attempts to restore the function of old MuSC to the level observed in younger cells. However, even young MuSCs cannot be expanded efficiently enough for use in cellular therapies under current conditions. Progress towards this goal has been recently obtained by mimicking the inflammatory milieu present in regenerating skeletal muscle (Fu et al., 2015; Ho et al., 2017) or by favouring the maintenance of quiescence in culture, which on the other hand limits in vitro expansion (Quarta et al., 2016; Zismanov et al., 2016). Although these studies have provided encouraging results, the identification of druggable targets that can be manipulated to enhance the therapeutic efficacy of expanded MuSCs while favouring their expansion remains an important goal in the development of stem cell therapies for muscle.
Although conventionally known for their role in chromatin remodeling, methyltransferases can hold much broader substrate specificity to include receptors, signaling molecules and transcription factors (Del Rizzo and Trievel, 2011). The su(var)3–9, enhancer-of-zeste, trithorax (SET) domain-containing protein 7 (Setd7), a lysine methyltransferase, provides one of the best examples of the wide-ranging targets such enzymes can have. Studies have found Setd7 can methylate an array of non-histone proteins, including ERα (Subramanian et al., 2008), YAP (Oudhoff et al., 2013), TAF-10 (Kouskouti et al., 2004) and STAT3 (Yang et al., 2010), fine tuning various cellular pathways and functions in a cell type specific manner. To date however, little is known about the role of Setd7 in skeletal muscle biology. Data from the C2C12 myoblast cell line suggested a role for Setd7 in regulating myogenesis in vitro, and in particular in modulating differentiation (Tao et al., 2011).
In this paper we explore the role of Setd7 in MuSC biology, finding that its expression is critical for effective skeletal muscle regeneration, and for modulating MuSC cell fate both in vitro and in vivo. We identify the underlying mechanism, showing that Setd7 binds to β-catenin and primes its nuclear translocation and interaction with transcription factor BCL-9 (Brack et al., 2009) which is required for progression along the myogenic differentiation cascade (Jones et al., 2015; Rudolf et al., 2016). Finally, we validate that Setd7 can be targeted pharmacologically to expand immature murine and human MuSCs in culture leading to enhanced engraftment, self-renewal and ultimately therapeutic efficiency following transplantation.
Results
Setd7 is absent in quiescent MuSCs and its expression increases with myogenic progression
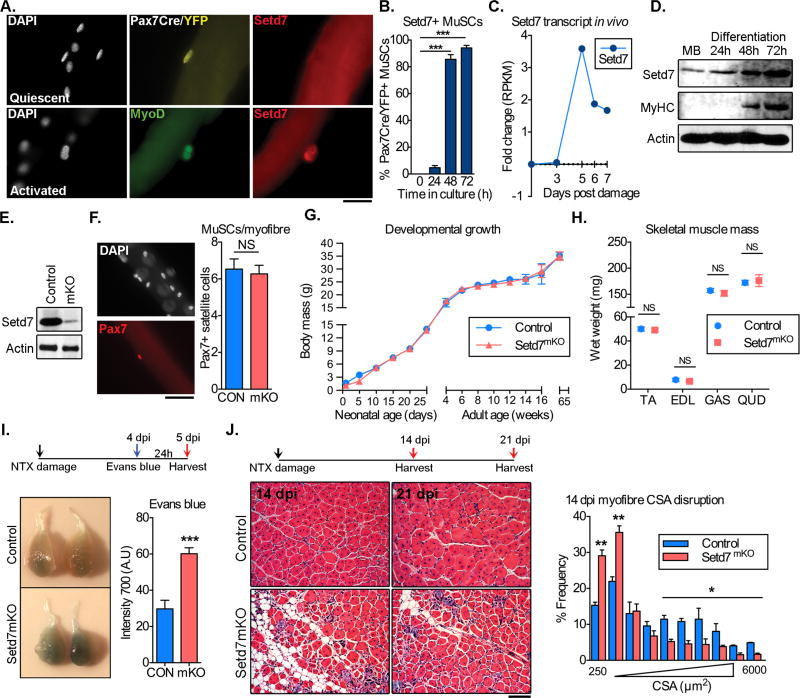
To examine the expression of Setd7 in myogenesis, we cultured isolated myofibers from Pax7-Cre/YFP mice. No Setd7 expression was detected in Pax7+ quiescent MuSCs upon isolation (Figure 1A, top panel). However, robust expression of Setd7 protein was detected in >90% of MyoD+ cells after 48 and 72 hours in culture, concomitantly with progression along the myogenic continuum (Figure 1A, bottom panel. Quantified in Figure 1B). In agreement with these ex vivo findings, analysis of Setd7 transcript levels from MuSCs isolated over a time course of skeletal muscle regeneration in vivo shows an elevation of Setd7 mRNA between day 3–5 after notexin (NTX) damage (Figure 1C). Finally, we found that Setd7 protein increased during induction of primary myoblast differentiation into myotubes (Figure 1D and S1, A). In summary, Setd7 expression is upregulated with progression of myogenesis.
Figure 1. Setd7 is expressed in activated MuSCs and is required for skeletal muscle regeneration.
(A) Setd7 expression in myofiber-associated MuSCs fixed immediately after isolation (quiescent, top panel) or after 48 hours ex vivo culture (activated, lower panel). Scale bar = 50µm
(B) Percentage of Setd7+ myofiber associated MuSCs (quantification from 5 mice).
(C) Setd7 expression in FACS isolated MuSCs following acute damage.
(D) Setd7 protein expression in MuSC during in vitro differentiation.
(E) Western blot analysis of Setd7 protein expression in whole muscle lysates.
(F) Representative images and quantification of quiescent Pax7+ MuSCs from myofibers isolated from control (CON - MyoDi-Cre/Setd7WT) and Setd7 mKO (mKO - MyoDi-Cre/Setd7FL/FL) mice. (n=3 per group). Scale bar = 50µm
(G) Body weights of control and Setd7 mKO mice during development, adulthood and aging (n≥15 per group).
(H) Skeletal muscle wet weights at 15 weeks of age. EDL – extensor digitorum longus, TA – tibialis anterior, GAS – gastrocnemius, QUD – quadriceps. (n≥15 per group)
(I) Evan’s blue incorporation in TA muscle of NTX damaged control and Setd7 mKO mice (n≥5 per group).
(J) TA muscles of control and Setd7 mKO mice following NTX damage. Quantification of myofiber cross sectional area (CSA) (n≥5 per group). Scale bar = 100µm
Data represented as mean ± SEM. NS = p > 0.05, *p<0.05, **p<0.01, ***p<0.005
Deletion of Setd7 in the myogenic lineage impairs adult skeletal muscle regeneration
The elevation of Setd7 observed during myogenic differentiation suggests this event may be important for skeletal muscle development and/or regeneration in vivo. To test this, we generated a mouse model that allows deletion of Setd7 in the myogenic lineage by crossing mice harboring a MyoD-iCre (Chen et al., 2005) allele with Setd7 floxed (Lehnertz et al., 2011) mice (Setd7 mKO). In this strain, Setd7 is deleted in both skeletal muscle fibers (Figure 1E) and MuSCs (Figure 2A). These mice had no overt developmental phenotype. Loss of Setd7 had no effect on adult quiescent MuSC numbers (Figure 1F) or on whole body growth rates (Figure 1G). Finally, adult skeletal muscles from Setd7 mKO and control mice (MyoDiCre/Setd7WT) were histologically similar (Figure S1B) and not significantly different in weight (Figure 1H) or myofiber cross-sectional area (CSA) (Figure S1C). Thus, Setd7 does not appear to be a critical factor for muscle development, although thorough analysis of discrete developmental stages would be required to rule out possible compensatory mechanisms at play.
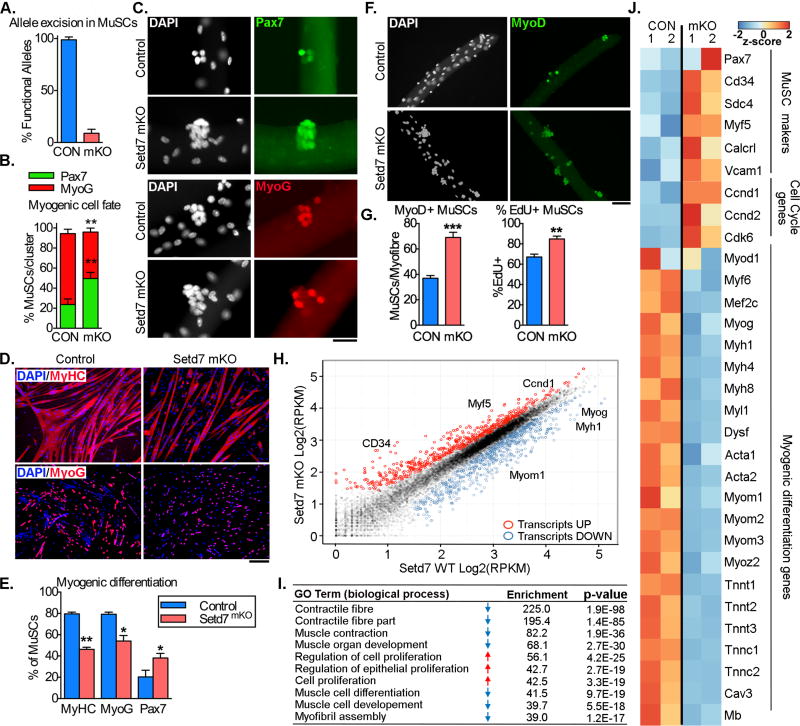
Figure 2. Loss of Setd7 impairs myogenic differentiation and enhances proliferation of MuSCs ex vivo.
(A) Allele excision efficiency in FACS sorted MuSCs (CD45/31-, Sca1-, alpha7-integrin/Vcam1+) from MyoDiCre/Setd7FL/FL mice (mKO) and controls (CON) (n=3 per group).
(B) Frequency of Pax7+ and MyoG+ myofiber-associated MuSCs after 72 hours in culture (n=3 per group).
(C) Pax7 and MyoG immuno-staining of myofiber-associated MuSCs after 72 hours in culture. Scale bar = 50µm.
(D) Quantification of the percentage of cells positive for MyHC, MyoG and Pax7 in cultured primary MuSCs following differentiation for 72 hours (n=3 per group).
(E) MyHC, MyoG and Pax7 immuno-staining of cultured primary MuSCs following differentiation for 72 hours. Scale bar = 100µm
(F) Quantification of total MyoD+ and percentage of EdU+ myofiber-associated MuSCs after 72 hours in culture. (n=3 per group).
(G) MyoD and EdU immunostaining of myofiber-associated MuSCs after 72 hours in culture. Scale bar = 100µm
(H) RNA-Seq scatter plot with key myogenic regulatory genes indicated. Each data point represents the mean Log2(RPKM) from two independent biological replicates. Red and blue indicate upregulated and downregulated genes respectively.
(I) Gene Ontology analysis of RNA-Seq data highlighting top 10 associated biological processes.
(J) Heat map of key differentially expressed genes associated with MuSC identity, cell cycle and myogenic differentiation.
Data represented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.005
We next tested if Setd7 is involved in adult skeletal muscle regeneration. Compared to control mice, Setd7 mKO mice showed an increase in Evans Blue dye incorporation over controls 5 days after acute injury (Figure 1I), a sign of reduced myofiber integrity (Hamer et al., 2002). These findings were supported by histological analysis 14 and 21 days after damage, showing signs of fibrofatty degeneration and an increased frequency of small regenerating myofibers in Setd7 mKO animals (Figure 1J). Together, these findings indicate that depletion of Setd7 in the myogenic lineage leads to an impaired regenerative response.
Setd7 regulates MuSC proliferation and differentiation ex vivo
Impairment in the regenerative capacity of Setd7 mKO mice suggests a defect in MuSC function. As expected, we observed deletion of >90% of Setd7 loci in FACS-isolated MuSCs from Setd7 mKO mice (Figure 2A). We assessed the proliferation and differentiation potential of MuSCs in vitro in single myofibers cultured for 72 hours (Figure 2B). We found an increase in cells expressing stem/progenitor marker Pax7 and a reduction in differentiating, Myogenin+ (MyoG)+ cells on Setd7 mKO fibers (Figure 2C), suggesting a delay in myogenic progression. Consistent with this finding, after 72 hours in differentiation conditions, myoblast cultures from Setd7 mKO mice contained fewer MyoG+ cells and nuclei within myosin heavy chain+ (MHC) structures, but more Pax7+ cells than controls (Figure 2D,E). Consistent with a higher frequency of immature cells, Setd7mKO cultures also expanded at a greater rate. Indeed, the absolute number of MyoD+ cells on cultured fibres was nearly double in Setd7 mKO samples compared to controls, with an increase in the frequency of KO cells incorporating EdU (Figure 2F,G). Together, these results show that deletion of Setd7 affects MuSC cell fate by impairing maturation along the myogenic lineage.
To gain further insight into the effects of Setd7 deletion, we performed RNASeq on cultured MuSCs from Setd7 mKO mice. We identified 1789 transcripts differentially expressed (768 upregulated and 1021 downregulated) between Setd7 KO and controls (Figure 2H). Gene Ontology (GO) analysis revealed a reduction in gene sets linked with muscle differentiation, development and sarcomere assembly and an enrichment of transcripts related to cell proliferation, cell division and mitosis in mKO cells (Figure 2I). In agreement with our in vitro assays, Setd7 KO myoblasts appeared to maintain higher expression levels of early MuSC markers such as Myf5 (Crist et al., 2012), CD34 (Beauchamp et al., 2000) and Calcitonin receptor (Yamaguchi et al., 2015) as well as of cell cycle genes such as CyclinD1 and Cdk6 (Figure 2J). In addition, Setd7 mKO myoblasts had lower expression levels of myogenic regulatory factors MRF4 (Myf6), Myogenin and Mef2c as well as of markers of myofibril assembly such as MyHC, Troponin and Actin genes (Figure 2J). These transcriptomic data provide further evidence that Setd7 is necessary for progression of MuSCs towards differentiation.
MuSC specific deletion of Setd7 impairs myogenic maturation ex vivo and blunts adult skeletal muscle regeneration in vivo
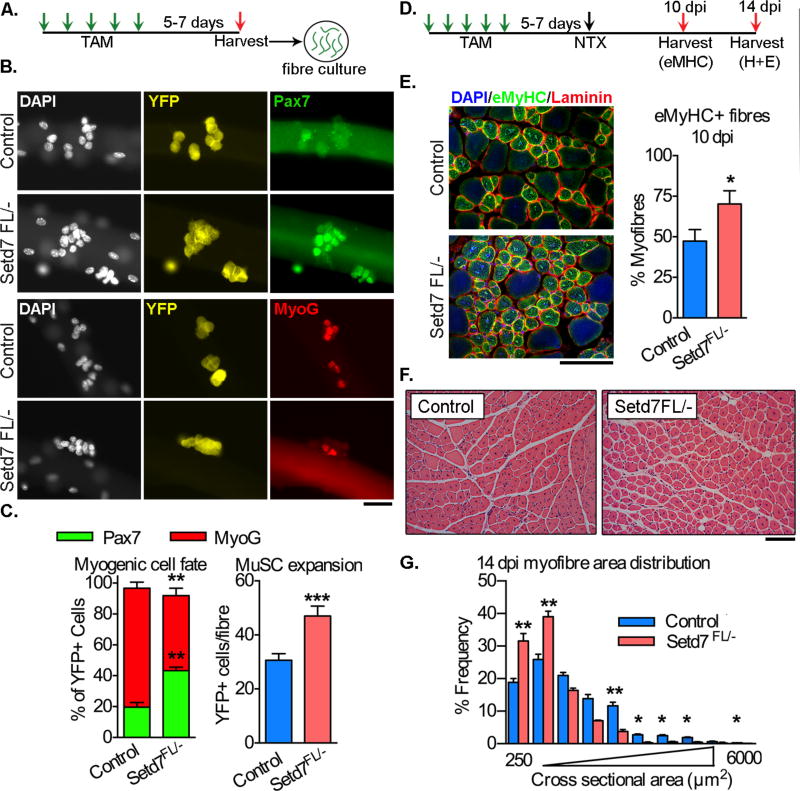
The MyoDiCre driver acts early in development, raising the possibility that compensation mechanisms may be activated mitigating the Setd7 KO phenotype. To address this concern, we conditionally deleted Setd7 in adult MuSCs. To this end, we bred mice carrying a Pax7-CreERT2 allele (Lepper et al., 2009), a CRE-inducible YFP and Setd7FL/− or Setd7+/+ (control) alleles, leading to deletion of Setd7 and expression of YFP in >80% of MuSCs upon tamoxifen administration (Figure S3, A). This strain closely recapitulated the phenotype observed in the developmental KO animals. Myofiber cultures from induced Pax7CrERT2/YFP/Setd7FL/− mice displayed a lower frequency of MyoG+/YFP+ cells and an increase in Pax7+/YFP+ MuSCs compared to controls (Figure 3A,B,C). In addition, the total number of YFP+ cells per fiber increased, consistent with Setd7 deletion imparting a proliferative advantage (Figure 3C).
Figure 3. Inducible deletion of Setd7 in adult MuSCs impairs myogenic commitment ex vivo and skeletal muscle regeneration in vivo.
(A) Schematic experimental design for ex vivo experiments.
(B) Quantification of total YFP+ and frequency of Pax7+ and MyoG+ myofiber-associated MuSCs from EDL of control and Pax7CreERT2/YFP/Setd7FL/− mice cultured for 72 hours (n=3 per group).
(G) Pax7, MyoG and YFP immunostaining of myofiber-associated MuSCs from control and Pax7CreERT2/YFP/Setd7FL/− mice cultured for 72 hours. Scale bar = 50µm
(D) Schematic experimental design for in vivo injury experiments.
(E) Quantification of frequency of eMyHC+ myofibers 10 days post injury (n=3 per group). Scale bar = 100µm
(F) H+E staining of cross sections from TA muscles of control and Pax7CreERT2/YFP/Setd7FL/− mice 14 days following NTX damage. Scale bar = 50µm
(G) Quantification of myofiber cross sectional area (CSA) from control and Pax7CreERT2/YFP/Setd7FL/− mice 14 days following NTX damage (n≥5 per group).
Data represented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.005
In these mice, impairment in the regenerative response to damage was similar to that observed in Setd7 mKO mice, including an increase in Evans Blue incorporation at 3 and 5 days post NTX injury (Figure S3, B,C) and in the number of eMYH+ myofibers 10 days post injury (Figure 3D,E). Finally, quantification of myofiber CSA 14 days post damage revealed an increase in the frequency of small regenerating myofibers in Pax7CrERT2/Setd7FL/− mice over controls, indicating a delayed/impaired regenerative process (Figure 3F+G). Thus, Setd7 expression in MuSCs is required for effective myogenic progression for regeneration following acute damage.
Little evidence of Setd7 acting as a histone methyltransferase or regulating MyoD in MuSCs
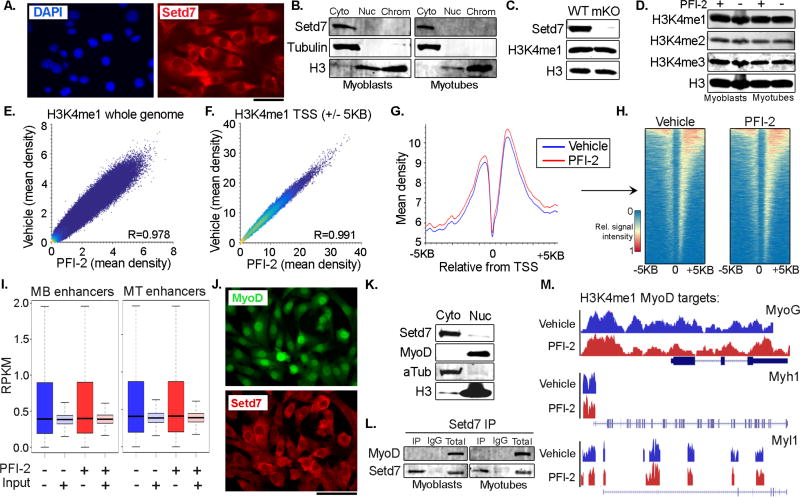
We next investigated the molecular mechanisms underlying regulation of MuSC behavior by Setd7. Previous data from immortalized C2C12 myoblasts suggests Setd7 binds MyoD and regulates transcription of myogenic genes by catalyzing the formation of H3K4me1 on target promoters (Tao et al., 2011). Surprisingly however, our experiments do not support this mechanism in primary MuSCs. Instead, immunostaining (Figure 4A) and protein fractionation (Figure 4B) revealed that Setd7 is predominantly localized to the cytoplasm in MuSCs cultured on isolated fibers (Figure 1A), bulk myoblast cultures (Figure 4A) and differentiated myotubes (Figure S4, C), suggesting a major role in chromatin regulation is unlikely. In addition, no changes in total H3K4me1 status were observed between WT and Setd7 mKO myoblasts (Figure 4C) or following acute inhibition of Setd7 by PFI-2 (Figure 4D and S4, A), a selective and potent small molecule inhibitor of the catalytic activity of Setd7 (Barsyte-Lovejoy et al., 2014).
Figure 4. Little evidence of Setd7 interacting with MyoD or acting as an H3K4me1 histone methyltransferase in MuSCs.
(A) Immunostaining of Setd7 in plated MuSCs highlighting cytoplasmic localization. Scale bar = 50µm.
(B) Western blot analysis of Setd7 in Cytoplasmic (cyto), nuclear (nuc) and chromatin (chrom) fractions from plated MuSCs and myotubes.
(C) Western blot analysis of H3K4me1 in WT and Sets7KO plated MuSCs.
(D) Western blot of H3K4me1, me2, me3 in plated MuSCs and myotubes treated with or without a Setd7 inhibitor (PFI-2).
(E+F) H3K4me1 ChIPSeq scatter plot correlating mean peak density in plated MuSCs treated with or without PFI-2 across whole genome (E) or at TSS (+/− 5KB) sites (F).
(G) Mean signal of normalized peak density surrounding TSSs for H3K4me1 ChIPSeq for control treated MuSCs (blue line) and PFI-2 treated MuSCs (red line)
(H) Heatmap displaying single gene resolution of H3K4me1 ChIP-Seq TSS enrichment data in (G).
(I) Comparisons in H3K4me1 enrichment (RPKM) at myoblast (MB) and myotube (MT) enhancer regions in MuSCs treated with or without PFI-2.
(J) Setd7 and MyoD co-immunostaining in plated MuSCs highlighting lack of co-localization. Scale bar = 100µm.
(K) Western blot analysis of Setd7 and MyoD in cytoplasmic (cyto) and nuclear (nuc) fractions of plated MuSCs.
(L) Setd7 IP samples blotted and probed with an anti-MyoD antibody.
(M) H3K4me1 genome browser tracks in proximal promoters and gene bodies of MyoD targets (MyoG, Myh1 and Myl1). Control MuSCs (blue line) and PFI-2 treated MuSCs (red line)
R values calculated with Pearson’s correlation coefficient.
To examine if Setd7 could be acting on a limited subset of specific genomic loci, we performed chromatin immunoprecipitation sequencing (ChIPSeq) with a H3K4me1 antibody on primary myoblasts treated with PFI-2 in vitro. This analysis revealed a robust correlation (R=0.978) in H3K4me1 distribution between vehicle and PFI-2 treated myoblasts (Figure 4E), indicating no major alterations in the status of this mark at the whole genome level. As previously reported, we observed enrichment of H3K4me1 in genomic areas immediately flanking transcriptional start sites (TSS). However, no noticeable differences in mean peak density were seen between treated myoblast and controls (Figure 4F, G, H). Recent reports that H3K4me1 can also mark distal regulatory elements (Blum et al., 2012) led us to investigate H3K4me1 content in myoblast and myotube enhancers, identified through a published dataset (Blum et al., 2012). Again, inhibition of Setd7 failed to produce significant changes in peak density at these annotated enhancer regions (Figure 4I). Finally, using the same workflow, we again failed to observe any significant differences in H3K4me1 status between WT and mKO MuSCs (Figure S4, E-I).
We next examined the relationship between Setd7 and MyoD in primary myoblasts and myotubes. We found little evidence of their co-localization by immunofluorescence (Figure 4J), and we observed them to partition in different subcellular fractions (Figure 4K and S4, B). Immunoprecipitation (IP) experiments also failed to demonstrate a physical interaction between MyoD and endogenous Setd7 (Figure 4L), or between MyoD and Myc-tagged Setd7 overexpressed in primary myoblasts and myotubes (Figure S4, D). Finally, we found no major alterations in H3K4me1 density at the TSS of known MyoD target genes (MyoG, Myh1, Mly1) (Blum et al., 2012; Tao et al., 2011) upon loss of Setd7 activity (Figure 4M). Together these results strongly argue against a role for Setd7 in regulating histone methylation or as a direct partner of MyoD in MuSCs, and prompted us to investigate alternative mechanisms.
Setd7 is required for BCL-9 mediated β-catenin nuclear import in MuSCs
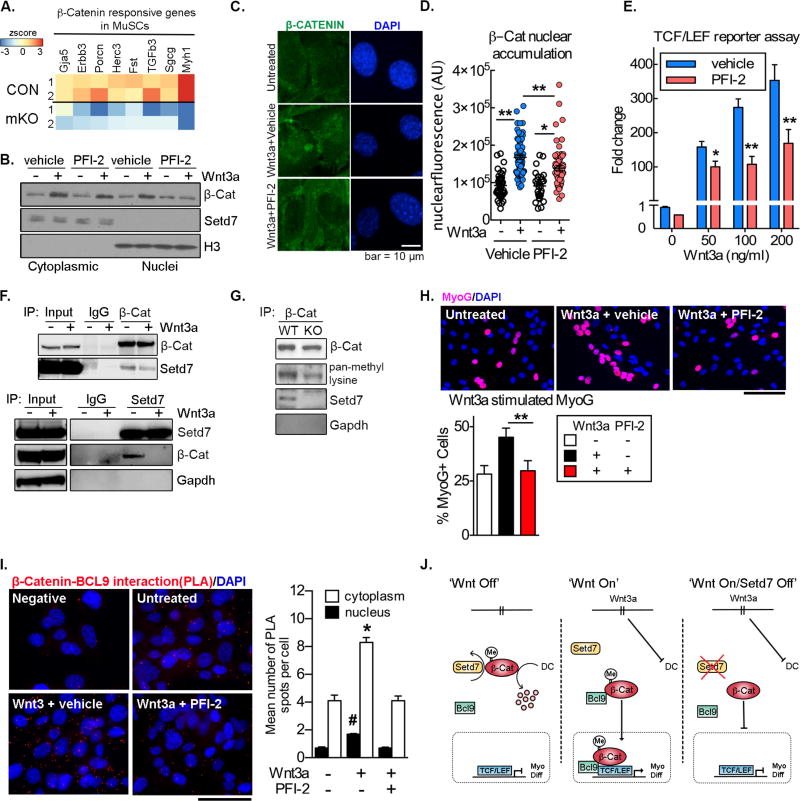
Recent studies have shown that Setd7 can regulate Wnt signaling in epithelial cells (Oudhoff et al., 2016). As canonical Wnt signaling is a major regulatory pathway in myogenesis, we investigated if it may be altered in cells lacking Setd7 activity. We compared our RNASeq data from Setd7 mKO MuSCs (Figure 2, H-J) with a published dataset (GEO: GSE72496) (Rudolf et al., 2016) in which β-catenin was constitutively activated (β-catenin-EX3loxP/+) in MuSCs. We observed a significant negative correlation (R=−0.624) (Figure S5B), with top ranked transcripts upregulated by β-catenin activation, down-regulated in Setd7 mKO MuSCs (Figure S5, A). These transcripts included Gja5, Erbb3, Porcn, Herc3 - key Wnt responsive genes in MuSCs (Jones et al., 2015; Parisi et al., 2015; Rudolf et al., 2016), as well as TGF-β pathway members, Fst and TGFb3 - regulators of Wnt/β-catenin mediated myogenic differentiation (Rudolf et al., 2016) (Figure 5A). These data prompted further investigation of the interplay between Setd7 and Wnt/β-catenin signalling.
Figure 5. Setd7 regulates β-catenin nuclear accumulation and transcriptional output via BCL-9 to control myogenic differentiation of MuSCs.
(A) Heat map of key β-catenin responsive genes in MuSCs and their differential expression in Control (CON) vs Setd7 mKO cells.
(B) β-catenin and Setd7 subcellular localization in C2C12 cells following stimulation with Wnt3a in the presence or absence of PFI-2.
(C) Confocal microscopy images of β-catenin localization in C2C12 cells following treatment with Wnt3a in the presence of absence of PFI-2.
(D) Quantification of β-catenin localization in C2C12 cells following treatment with Wnt3a in the presence of absence of PFI-2 (n=3).
(E) TopFLASH reporter assay of β-catenin transcriptional output (TCF/LEF) by plated MuSCs in response to Wnt3a stimulation and in the presence or absence of PFI-2 (n=3).
(F) β-catenin IP samples blotted and probed with anti-Setd7 antibody (top panel) and Setd7 IP samples blotted and probed with anti-β-catenin antibody (bottom panel) in the presence and absence of Wnt3a.
(G) β-catenin IP samples blotted and probed with an anti-pan-methyl lysine antibody from cultured wild type (WT) and Setd7 null (KO) MEFs.
(H) Frequency of MyoG+ plated MuSCs following 24 treatment with Wnt3a with or without PFI-2. Results from 3 independent experiments (n=3). Scale bar = 100µm.
(I) BCL-9 and β-catenin protein-protein interactions evaluated using in-situ proximity ligation assay (PLA). Complexes visualized as red dots. Scale bar = 100µm.
Quantification of BCL-9/β-catenin PLA assay. Red dots quantified in cytoplasm and nucleus from at least 800 cells per condition (n=4). *p<0.05, Wnt3a treated and untreated cells in cytoplasm. #p<0.05, Wnt3a treated and untreated cells in nucleus.
(J) Schematic summary of interplay between Setd7 and Wnt signaling.
Data represented as mean ± SEM. *p<0.05, **p<0.01
Stimulation of C2C12 myoblasts with Wnt3a yielded the expected upregulation of β-catenin in both PFI-2 and vehicle treated cells (Figure S5, C). However, inactivation of Setd7 significantly impaired translocation of β-catenin to the nucleus (Figure 5B–D). In addition, Wnt3a treatment and GSK3-β inhibition of transfected myoblasts both triggered an increase in TCF/LEF reporter (TOPFlash) output that was significantly blunted upon treatment with PFI-2 (Figure 5E + S5D).
We next investigated if Setd7 was physically interacting with β-catenin. Indeed, Setd7 co-immunoprecipitated with β-catenin (Figure 5F) and this interaction was reduced upon Wnt3a stimulation, consistent with the translocation of β-catenin, but not Setd7, into the nucleus (Figure 5F). Using MEFs to gain sufficient protein material, we also observed lysine methylation of β-catenin was reduced in cells lacking Setd7 (Figure 5G), suggesting β-catenin may act as a substrate for Setd7 in myogenic cells, echoing observations made in other cell types (Shen et al., 2015).
Myogenin is a critical mediator of canonical Wnt signaling in promoting myogenic differentiation (Jones et al., 2015). We found that inhibition of Setd7 significantly reduced the appearance of MyoG+ cells in response to Wnt3a stimulation (Figure 5H). Thus, inhibition of Setd7 impairs the nuclear accumulation and transcriptional function of β-catenin, preventing the up-regulation of myogenic genes including Myogenin.
Our data suggest that Setd7 regulates a checkpoint in canonical Wnt signaling, namely the nuclear translocation of β-catenin following its cytoplasmic upregulation. In myogenic cells, nuclear import of β-catenin is dependent on the formation of a complex with legless/BCL-9 (Brack et al., 2009). To assess whether Setd7 activity is required for the formation of this complex, we quantified the interaction between β-catenin and BCL-9 in myogenic cells using proximity ligation assays (PLA). We observed the expected increase in this complex in response to Wnt stimulation, which was completely blocked by both PFI-2 treatment in C2C12s and genetic deletion of Setd7 in MuSCs (Figure 5I and S5E).
In summary, we show that Setd7 interacts with β-catenin, regulates its subcellular localization and is required for transcription of Wnt responsive genes in MuSCs.
PFI-2 inhibition of Setd7 enhances engraftment, self-renewal and therapeutic potential of cultured MuSCs
A critical hurdle in the development of stem cell therapies for muscle is the inability to manipulate stem cells in vitro without loss of potency. Ideally, expanded MuSCs should remain capable of both engraftment into myofibers as well as replenishment of the stem cell niche to maintain homeostasis. As Setd7 inhibition maintains cultured cells in an immature state, it could provide a strategy to improve engraftment in an experimental model of autologous muscle stem cell transplantation.
We cultured MuSCs in the presence or absence of Setd7 inhibitor (Figure 6A) and compared their expansion. PFI-2 treatment significantly increased the yield of these cultures (Figure 6B,C). As expected, Pax7/MyoD co-staining revealed PFI-2 decreased the proportion of cells undergoing differentiation (Pax7-/MyoD+ cells) (Figure 6B,D). Similar to Setd7 mKO cells, PFI-2 treated cells also displayed impaired myotube formation under differentiation conditions (Figure S6A).
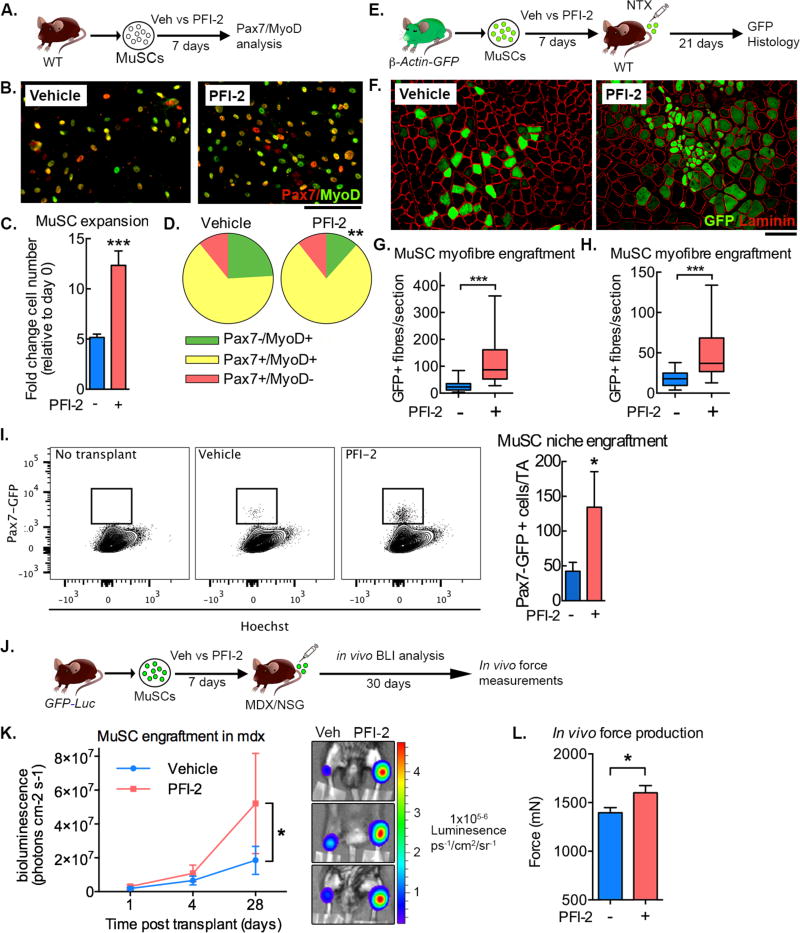
Figure 6. Setd7 inhibition enhances MuSC expansion, maintains MuSC potency and improves therapeutic efficacy.
(A) Schematic of experimental design.
(B) Pax7/MyoD co-immunostaining of plated MuSCs treated with or without PFI-2 for 7 days. Scale bar = 100µm.
(C+D) Quantification of expansion (C) and frequency of Pax7/MyoD+ (D) MuSCs after seven days of culture with or with without PFI-2. (n=3).
(E) Schematic of β-Actin-GFP transplantation experimental design.
(F) GFP+ myofibers following transplantation of MuSCs expanded with or without PFI2. Scale bar = 100µm.
(G) Quantification of GFP+ myofibers following transplantation of MuSCs expanded from 10,000 cells in the presence or absence of PFI-2. (n=5. Data from 5 mice each)
(H) Quantification of GFP+ myofibers following transplantation of 10,000 MuSCs expanded in the presence or absence of PFI-2. (n=6. Data from 6 mice each)
(I) Representative FACS plots and quantification of GFP+ MuSCs recovered from recipient mice transplanted with MuSCs expanded in vitro for 7 days with or without PFI-2. (n=8. Data from 8 mice each)
(J) Schematic of GFP-Luc transplantation experimental design.
(K) Quantification of bioluminescence over 28 days following transplantation of GFP-Luc labelled MuSCs expanded in vitro with or without PFI-2. (n=8. Data from 8 mice each)
(L) Quantification of specific tetanic force production in TA muscle of NSG-MDX mice 28 days following transplantation of GFP-Luc labelled MuSCs expanded in vitro with or without PFI-2. (n=8. Data from 8 mice each)
Data represented as mean ± SEM. *p≤0.05, **p<0.01, ***p<0.005.
Next we investigated if PFI-2 treated cells had an enhanced capacity to engraft in vivo. MuSCs isolated from mice expressing ubiquitous GFP (β-Actin-GFP) were expanded in the presence or absence of PFI-2 for 7 days, and the whole resulting cultures were transplanted into injured TA muscles (Figure 6E). Twenty-one days later, TAs injected with PFI-2 treated cells contained more GFP+ myofibers, indicating enhanced engraftment (Figure 6F,G). This could simply rest on the larger yield obtained from PFI-treated cultures or could also rely on a higher propensity of the treated cells to engraft on a per cell basis. To distinguish between these two possibilities, we repeated the experiment, injecting equal numbers (10,000 MuSCs/TA) of vehicle and PFI-2-expanded MuSCs. Again we observed that cells cultured in the presence of PFI-2 yielded more GFP+ myofibers (Figure 6, H), supporting the notion that inhibition of Setd7 during in vitro expansion both increases MuSC yield as well as improving their engraftment potency.
We then tested if PFI-2 could improve the capacity of cultured cells to re-enter the MuSC niche. GFP+ cells were FACS isolated from a Pax7-GFP reporter mouse (Rocheteau et al., 2012) and expanded in the presence or absence of PFI-2. Expanded cells (10,000 MuSCs/TA) were then transplanted into injured TA muscles of irradiated NSG mice. Under these conditions, only cells engrafting in the satellite cell niche retain Pax7 expression and GFP positivity. Twenty-one days later, we found a significant increase in the number of GFP+ cells in muscles transplanted with PFI-2-treated MuSCs (Figure 6I), indicating enhanced capacity to return to the MuSC niche.
We next asked whether the improved engraftment potency of PFI-2-treated MuSCs could translate into functional improvements in muscle strength in mdx-NSG mice (an immunocompromised mouse model of muscular dystrophy). To this end, expanded MuSCs from GFP-Luc mice were transplanted, allowing longitudinal quantification of their engraftment by non-invasive bioluminescence imaging (BLI) (Figure 6J). BLI confirmed that MuSCs cultured with PFI-2 engrafted better than controls (Figure 6K). Importantly, this enhanced engraftment also corresponded to a functional increase in specific tetanic force production compared to muscles transplanted with vehicle-treated MuSCs (Figure 6L). Thus, Setd7 can be targeted pharmacologically to increase the yield and the potency of cultured MuSCs boosting their engraftment, self-renewal and therapeutic efficacy.
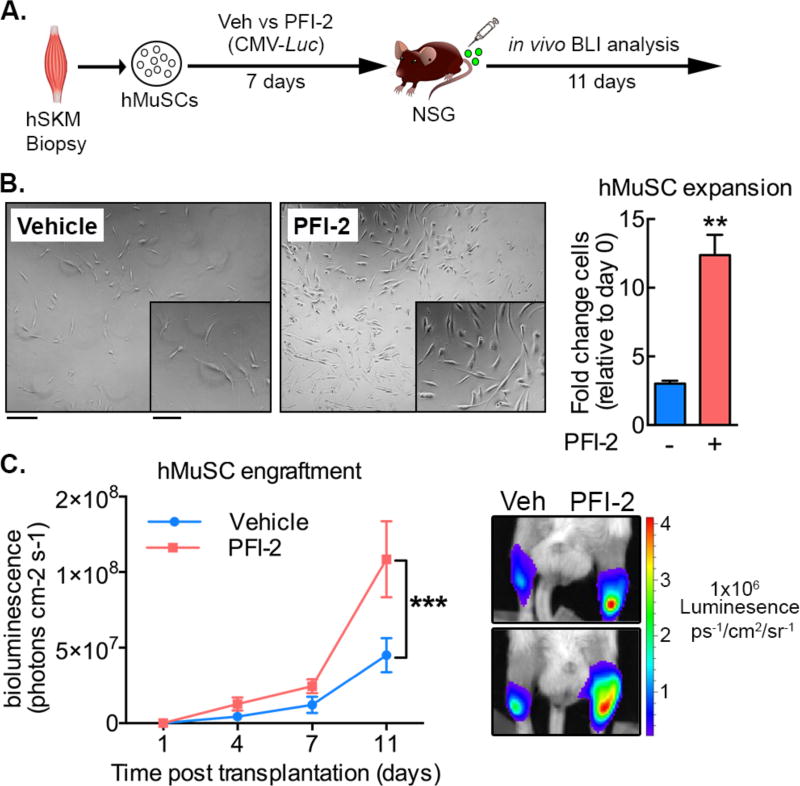
PFI-2 inhibition of Setd7 enhances expansion of transplant-competent human MuSCs
Finally, we investigated if PFI-2 could enhance the expansion of human MuSCs (hMuSCs). Using a similar experimental workflow (Figure 7A), hMuSCs were FACS isolated from human skeletal muscle operative samples, expanded in the presence of PFI-2, infected with a lentivirus expressing luciferase protein and then transplanted into the injured TA of immunocompromised mice. Consistent with murine MuSCs, we observed PFI-2 induced a significant increase in hMuSC expansion (Figure 7B). Upon transplantation, in vivo BLI analysis revealed enhanced engraftment of hMuSCs previously exposed to PFI-2 (Figure 7C). Thus SETD7 inhibition is a viable strategy to enhance the therapeutic potential of human MuSCs.
Figure 7. Setd7 inhibition enhances expansion of transplantable human MuSCs.
(A) Schematic of experimental design. hSKM (Human skeletal muscle).
(B) Freshly isolated hMuSCs culture-expanded with/without PFI-2 for 7 days (n=3 donor samples). Scale bars = 20µm (main image) and 10µm (inset).
(C) Quantification of bioluminescence over 11 days following transplantation of GFP-Luc transduced hMuSCs expanded in vitro with or without PFI-2. (Data from 6 mice (n=6) transplanted with donor cells from three operative samples).
Data represented as mean ± SEM. **p≤0.01, ***p<0.005.
Discussion
Here we provide evidence that methyltransferase Setd7 is important for MuSCs progression towards differentiation. Mechanistically, Setd7 influences canonical Wnt signalling by interacting with β-catenin and modulating its methylation status. This primes the response to Wnts by enabling β-catenin to interact with BCL-9, accumulate in the nucleus and induce myogenic gene expression. We also demonstrate that pharmacological inhibition of Setd7 boosts in vitro expansion of both murine and human MuSCs whilst maintaining their stemness, thus enhancing their therapeutic potential.
MuSCs that lack Setd7 displayed enhanced proliferation and maintained a phenotypically immature state when expanded in vitro. These data are consistent with experiments in C2C12 cells (Tao et al., 2011), showing that Setd7 knock down blocks the expression of myogenic genes such as Myogenin and MyHC. Tao et al., also provided evidence that Setd7 interacts with MyoD and regulates H3K4me1 status. Despite agreement on the role of Setd7 in delaying progress towards differentiation, our data do not support this molecular mechanism. We found little evidence of Setd7 interacting with MyoD in MuSCs and ChIPSeq analysis failed to detected changes in H3K4me1 status in Setd7 KO cells. These discrepancies are perhaps unsurprising given cultured primary MuSCs and C2C12, while both myogenic, are fundamentally different in their biology – C2C12 cells are immortalized, have shorter doubling times, minimal engraftment capacity, altered differentiation potential and abnormal karyotype.
Instead, we show that Setd7 is a key component of the Wnt signaling pathway, and it is required for the nuclear functions of activated β-catenin. A recent report in cancer cells demonstrated that β-catenin is directly methylated by Setd7 at K180, facilitating its degradation (Shen et al., 2015). Although we confirm this interaction in muscle cells and also find that Setd7 likely methylates β-catenin, we did not observe significant changes in total β-catenin protein levels when we blocked its activity. Future work is required to determine if Setd7 acts on the same substrates in MuSCs as in cancer cells, and what the exact mechanistic consequences are. Our previously published data show that in epithelial cells a complex containing Setd7 and β-catenin exists and that Setd7 activity is required for β-catenin nuclear localization following stimulation of the Wnt pathway (Oudhoff et al., 2016). However, this regulation of β-catenin by Setd7 was dependent on the presence of YAP (Oudhoff et al., 2016). Whether YAP plays a similar mechanistic role in MuSCs remains unclear, however, we did not observe changes in YAP localization or transcriptional output in response to Setd7 deletion/inactivation (data not shown). Taken together these findings highlight a general interaction between Setd7 and Wnt signaling, with further work required to more completely define their relationship in MuSCs.
The function of canonical Wnt signaling and β-catenin in regulating myogenesis and MuSCs has been studied extensively over the past decade, but controversy still exists on its precise role. In vitro, studies have shown both positive (Figeac and Zammit, 2015; Han et al., 2011; Jones et al., 2015) and negative (Tanaka et al., 2011) effects of activating the Wnt/β-catenin pathway on myogenic differentiation. In vivo, Wnt signaling appears critical for developmental myogenesis (Hutcheson et al., 2009; Ikeya and Takada, 1998) whereas activation or deletion of β-catenin in MuSCs in regenerative myogenesis has produced inconsistent findings from several independent groups (Murphy et al., 2014; Parisi et al., 2015; Rudolf et al., 2016). The most recent study however, employing inducible transgenic strategies to modulate β-catenin specifically in MuSCs, convincingly demonstrated a reduction in regenerative potential following loss of β-catenin and conversely, precocious differentiation upon its activation (Rudolf et al., 2016). Our data is in support of this observation, as in the absence of Setd7, β-catenin fails to accumulate in the nucleus upon Wnt stimulation, blunting its transcriptional output and leading to impaired differentiation in vitro and inhibited regenerative potential in vivo.
In an experimental model of autologous MuSC cell therapy, we demonstrate that treatment of MuSCs with PFI-2 during their in vitro expansion leads to enhanced cell yields and superior engraftment both overall as well as on a per cell basis. We detected an obvious improvement in engraftment both into regenerating fibres and the MuSC niche, indicating that inhibition of Setd7 allows the maintenance of some degree of self-renewal in vitro. Interestingly, among the earliest events in MuSC activation is the downregulation of stem cell marker CD34 (Alfaro et al., 2011). Indeed, CD34 is highly expressed by efficiently engrafting quiescent MuSCs but absent from poorly engrafting cultured myogenic cells (Montarras et al., 2005). Our data as well as recent reports (Alfaro et al., 2011; Fu et al., 2015) by others showing continued CD34 expression in cultures retaining self-renewal potential extend this correlation and suggest that this marker may help identify the least immature proliferative progeny of quiescent MuSCs know to date.
Our work also has translational implications. Early clinical trials involving transplantation of cultured myoblasts into muscular dystrophy patients yielded poor results and highlighted several problems such as poor myoblast survival and engraftment, limited migration into the muscle tissue and importantly, negligible capacity to self-renewal (Wilschut et al., 2012). Since these trials it’s become clear that culturing MuSCs following isolation significantly impairs their engraftment and self-renewal potential (Montarras et al., 2005) - making in vitro manipulation to, for example, genetically correct disease mutations extremely challenging. As a result, defining culture conditions that permit propagation of MuSCs whilst maintaining potency is a major goal. PFI-2 treatment allowed us to expand greater numbers of higher quality MuSCs, which when transplanted in dystrophic muscle produced significant improvement in muscle function. Importantly, Setd7 inhibition also allowed the enhanced expansion of human MuSCs, providing, a significant step towards refining optimal MuSC culture conditions achieving this goal and making cell therapy for muscle disease a reality.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for reagents and resources should be directed to the lead contact, Fabio Rossi (fabio@brc.ubc.ca)
Experimental Model and Subject Details
In vivo mouse models
Housing, husbandry and experimental protocols were conducted following institutional and national guidelines at the University of British Columbia, Canada. Setd7FL/FL mice were generated as previously described (Lehnertz et al., 2011; Oudhoff et al., 2013). Mice harbouring the MyoDiCre allele were a kind gift from Prof. David Goldhammer (Chen et al., 2005). Pax7-CreERT2 mice were obtained from The Jackson Laboratory and originally generated by (Lepper et al., 2009). Allelic recombination under the Pax7-CreERT2 allele was induced by daily injections of 0.1mg/g Tamoxifen (TMX) for 5 days, administered either intraperitoneally or by oral gavage. To control for tamoxifen toxicity, all mice were administered with tamoxifen. Pax7-GFP reporter mice were a kind gift from Prof. Shahragim Tajbakhsh (Rocheteau et al., 2012). Mdx (C57BL/10ScSn-Dmdmdx/J), GFP-Luc reporter mice (FVB-Tg(CAG-luc,-GFP)L2G85Chco/J), NSG (NOD-scid IL2Rgammanull), NSG-mdx4Cv (B6Ros.Cg-Dmdmdx-4Cv/J) and β-Actin-GFP (C57BL/6-Tg(CAG-EGFP)1Osb/J) mice were obtained from The Jackson Laboratory. All experimental mice used were 2−6 months old unless stated and randomly assigned into experimental groups of matching sex. Skeletal muscle injury was induced by a single injection of 0.15µg Notexin (NTX) into the tibialis anterior (TA) muscle. Evans Blue Dye (FW 960.82 g mol−1) solution (25mg/kg) was injected intraperitoneally 24 hours before sacrifice. Evans Blue dye incorporation was quantified on whole TA muscles measuring fluorescence at 700nm using a LI-COR® machine.
Murine primary cultures and cell lines
For plated murine MuSC cultures, cells were FACs isolated from male mice and seeded onto Matrigel (BD Biosciences) coated dishes and expanded in MuSC media consisting of high glucose (4500mg/L) DMEM (ThermoFisher) supplemented with 20% (v/v) fetal bovine serum, 10 ng/ml bFGF (Peprotech) and 1% (v/v) Penicillin Streptomycin (ThermoFisher). Differentiation into myotubes was stimulated by switching to high glucose DMEM supplemented with 2% (v/v) horse serum (differentiation medium) for up to 96 hours. For single fiber cultures, myofibers were cultured in high glucose DMEM supplemented with 20% (v/v) fetal bovine serum, 0.5% (v/v) chick embryo extract and 1% (v/v) Penicillin Streptomycin (ThermoFisher) for upto 72 hours. All single fibre experiments were conducted using male mice. C2C12 myoblasts were maintained in high glucose DMEM supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) Penicillin Streptomycin (ThermoFisher).
Human primary cultures
Human skeletal muscle tissue was obtained from latissimus dorsi muscles of operative patients (aged 15–53 years old) following informed consent and institutional and national guidelines. Sorted human MuSCs were immediately counted and plated in growth media (DMEM + MCDB 131 (1:1) supplemented with 10% (v/v) fetal bovine serum, 1% (v/v) ITS (Gibco) and 1% (v/v) Penicillin Streptomycin (ThermoFisher) into plates precoated with extracellular matrix (1:100, Sigma).
Method Details
Flow cytometry analysis and FACs
Skeletal muscle tissue from mouse hindlimbs were dissected, gently minced with scissors and then digested with Collagenase II (250 CDU/mL) solution for 30 minutes at 37°C. Digested tissue was then washed once with cold PBS and centrifuged at 130g for 10 minutes. Second digestion was then performed by adding Collagenase D (1.5 CDU/mL) and Dispase II (2.4 CDU/mL) solution for 1 hour at 37°C. Digested tissue was then passed through a 45uM cell strainer to generate a single cell suspension ready for a antibody staining. Digested tissue was stained using antibodies: 1:500 FITC-conjugated rat anti-mouse CD31 (eBioscience, clone: 390), 1:500 FITC-conjugated rat anti-mouse CD45 (AbLab, clone: I3/2), 1: 5000 PeCy7-conjugated rat anti-mouse Sca-1 (eBioscience, clone: D7), 1:1000 Alexa Fluor 647-conjugated rat anti-mouse integrin alpha7 (AbLab, clone: R2F2) and 1:2000 anti-Vcam1-Biotin (AbLab, clone: MK1.9). Cells were incubated with primary antibodies for 20 minutes on ice, washed once with cold FACs buffer (2% FBS, 2mM EDTA) and then incubated with 1:1000 streptavidin-PE (ThermoFisher) for a further 15 minutes on ice. Cells were analyzed using Flow Cytometry or MuSCs FACs isolated into complete MuSC media based on cell surface antigen markers: CD31-/CD45-/Sca1-/integrin-α7+/Vcam1+ using a BD LSRII Flow Cytometer (BD Biosciences) or FACsAria™ III (BD Biosciences) respectively.
For isolation of human MuSCs, skeletal muscle biopsies were immediately placed on ice and prepared for sorting. Muscles were minced using fine surgical scissors and then digested for 60 minutes in collagenase II solution (700 U/mL) at 37°C in a 60 rpm shaking water bath. Samples were then washed with PBS and placed in 100U/mL collagenase II and 1U/mL dispase solution for an additional 30 minutes. To further disassociate MuSCs from the muscle fibers, the digested sample was subjected to mechanical agitation via multiple passes through 20 gauge needle and syringe. Muscle samples were then washed in PBS and passed through a 40 µm nylon cell strainer to generate a single cell suspension. Cells were stained with the following antibodies for 45 minutes on ice: 1:100 anti-CD31-Alexa Fluor 488, (BioLegend, clone WM59), 1:100 anti-CD45-Alexa Fluor 488 (Invitrogen, clone HI30), 1:100 anti-CD34-FITC (BioLegend, clone 581), 1:100 anti-CD29-APC (BioLegend, clone TS2/16) and 1:100 anti-CD56-APC (Biolegend, clone 5.1H11). Human MuSCs were isolated by FACs based on the following cell surface antigen markers: CD45-/CD31-/CD34-/CD29+/CD56+ using a FACsAria™ III (BD Biosciences).
Myofiber ex vivo culture
Viable single myofibers were isolated from the extensor digitorum longus (EDL) muscle of 8-week-old mice following dissociation with collagenase I solution (0.2%) for 1.5 hours at 37°C. Myofibers and their associated MuSCs were cultured for up to 72 hours, before being fixed with 4% (v/v) PFA and processed for immunofluorescent staining. For proliferation assays, myofibers were incubated with 10µM EdU for 4 hours and then processed using Click-iT® EdU imaging kit (ThermoFisher) following the manufacturer’s instructions. Quantitation of % MuSCs expressing Pax7 and MyoG was performed by counting MuSC colonies (3 or more MuSCs) associated with myofibers.
Immunofluorescent staining and microscopy
For plated MuSCs, 8 well Permanox® chamber slides (Nunc® Lab-Tek®) or black-walled 96 well plates were used and cells fixed with 4% (v/v) paraformaldehyde (PFA) for 10 minutes. Single myofibers were fixed in suspension with 4% (v/v) PFA for 10 minutes. Following fixation, material was permeabilized with 0.5% (v/v) Triton X-100 solution for 5 minutes and then blocked with 10% (v/v) goat serum and 10% (v/v) horse serum solution for 30 minutes to reduce nonspecific antibody binding. Cells were then incubated with the following antibodies overnight at 4°C, 1:150 mouse anti-Setd7 (Abcam, clone 2D10), 1:10 mouse anti-Pax7 (DSHB, clone PAX7 supernatant), 1:50 mouse anti-MyoD (Dako, clone 5.8A), 1:150 rabbit anti-MyoD (Santa Cruz, clone C-20), 1:15 mouse anti-Myogenin (DSHB, clone FD5 supernatant), 1:500 mouse anti-MyHC (DSHB, clone MF20 concentrate), 1:400 mouse anti-β-catenin (BD Biosciences, clone 14/β-catenin) and 1:1000 chicken anti-GFP (Abcam). Species-specific fluorochrome-conjugated secondary antibodies (ThermoFisher) were then applied for 2 hours at room temperature, before being mounted with 100 ng/ml−1 of DAPI (4,6-diamidino-2-phenylindole; Vector Laboratories).
Quantification of nuclear β-Catenin was performed using Fiji software where masks for individual nuclei were drawn using the DAPI signal and the β-Catenin signal within these masks was determined as nuclear β-Catenin. We subtracted the background signal that was measured in parts of the well where no cells were present. This was performed for 3–5 fields from 3 independent experiments with total numbers of cells analyzed from these pooled experiments ranging from 30 to 45 for untreated to 68 to 78 for Wnt3a treated cells (Barsyte-Lovejoy et al., 2014).
Muscle histology and immunohistochemistry
Mice were sacrificed and perfused transcardially with 25 mL PBS/2 mM EDTA, followed by 25 mL 4% (v/v) PFA. TA muscles were dissected and postfixed in 2% (v/v) PFA for 1 hour. For immunohistochemistry, whole muscles were embedded in OCT, frozen and cryosectioned onto glass slides. Preceding embryonic myosin staining, slides underwent antigen retrieval consisting of incubation in sodium citrate buffer (10mM Sodium Citrate, 0.05% Tween 20, pH 6.0) at 85°C for 20 minutes. Tissue sections were then blocked with a 0.25% (v/v) Triton X-100 and 5% (v/v) goat serum PBS solution and then incubated with the following antibodies overnight at 4°C, 1:7.5 mouse anti-eMyHC (DSHB, clone F1.652 supernatant) and 1:500 rabbit anti-Laminin (Abcam, ab11575). Species-specific fluorochrome-conjugated secondary antibodies (ThermoFisher) were then applied for 2 h at room temperature, before being mounted with 100 ng/ml−1 of DAPI (4,6-diamidino-2-phenylindole; Vector Laboratories).
For histochemical stains, whole muscles were placed in 70% (v/v) ethanol overnight before being paraffin embedding and microtome sectioned. Hematoxylin and eosin (H+E) staining was performed following standard methods.
PFI-2 treatment
Small molecule inhibition of Setd7 was achieved by incubation of cells with 4 µM (R)-PFI-2 for at least 2 hours and replaced every 24 hours for extended culturing (Barsyte-Lovejoy et al., 2014). Dose matched DMSO (vehicle) or enantiomer (S)-PFI-2 were used as controls as indicated.
Cell fractionation, immunoprecipitation and Western blotting
Preparation of chromatin, nuclear and cytoplasmic fractions was performed as previously described (Mendez and Stillman, 2000). For cytoplasmic and nuclear fractionation cells were lysed in a hypotonic buffer (20 mM Hepes-KOH [pH 8.0], 5 mM KCl, 1.5 mM MgCl2, 5 mM Na butyrate, 0.1 mM dithiothreitol [DTT]) for 10 minutes on ice. Lysates were then centrifuged (10 minutes at 16,000g, 4°C) and the resulting supernatant maintained as the cytoplasmic fraction. Pelleted material was then resuspensed in nuclear extraction buffer (15 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.4 M NaCl, 10% sucrose, 1 mM DTT). After 30 minutes on ice nuclear extracts were collected by final centrification (10 minutes at 16,000g, 4°C) and resuspended in Laemmli buffer. To isolate chromatin associated proteins, cells were resuspended in buffer A (10 mM HEPES, [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT) supplemented with protease inhibitor cocktail (ThermoFisher). Triton X-100 (0.1%) was added and cells incubated on ice for 5 minutes. Nuclei were then collected by low-speed centrifugation (4 minutes, 1,300g, 4°C) and washed once in buffer A and then lysed in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT) supplemented with protease inhibitor cocktail (ThermoFisher). Insoluble chromatin was collected by centrifugation (4 minutes, 1,700g, 4°C), washed once in buffer B, and centrifuged again under the same conditions. The final chromatin pellet was resuspended in Laemmli buffer and sonicated.
For immunoprecipitation (IP), cells were lysed in IP lysis buffer (50 mM Tris HCl, 150 mM NaCl, 50 mM NaF, 1% Triton X-100, 1mM sodium orthovanadate, 0.5 mM EDTA) and protease inhibitor cocktail (ThermoFisher), homogenized and centrifuged at 13,000g for 15 minutes at 4°C. IP lysates were incubated with 5 µg of anti-Setd7 (Abcam, clone EPR5574) and anti-Myc-Tag (Cell Signalling, #2276) antibody or an equivalent amount of species matched IgG. IP was performed using 50 µl protein A+G Dynabeads™ (ThermoFisher) following the manufacturer’s instructions.
For whole cell lysates, cells were lysed in RIPA buffer (25mM Tris•HCl pH 7.6, 150mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS supplemented protease inhibitor cocktail).
Protein lysates were separated via SDS-PAGE electrophoresis and transferred to nitrocellulose membranes using standard Western blot methods. Membranes were probed with the following primary antibodies overnight at 4°C: 1:500 mouse anti-Setd7 (Abcam, clone 2D10), 1:500 rabbit anti-Setd7 (Cell Signalling Technologies), 1:200 rabbit anti-MyoD (Santa-Cruz, clone C-20), 1:2000 mouse anti-MyHC (DSHB, clone MF20 concentrate), 1:1000 rabbit anti-alpha Tubulin (Abcam), 1:1000 rabbit anti-β-Actin (Abcam), rabbit anti-histone 3 (Cell Signalling Technologies), 1:2000 rabbit anti-H3K4me1 (Cell Signalling Technologies, Clone D1H2), 1:1000 rabbit anti-H3K4me2 (Cell Signalling Technologies, clone D1A9), 1:1000 rabbit anti-H3K4me3 (Cell Signalling Technologies), 1:200 mouse anti-β-catenin (Santa Cruz, clone E5). Primary antibodies were then visualised using species-specific Alexa Fluor conjugated secondary antibodies (ThermoFisher) and scanned using a LI-COR Odyssey infrared imager.
gDNA allele excision assay
Efficiency of Cre mediated excision under MyoDiCre and Pax7CreERT2 alleles was assessed using the methods previously described (Zhang et al., 2016). Genomic DNA was extracted using a PureLink® Genomic Mini Kit (ThermoFisher) from 15,000 MuSCs purified by FACS. Digital droplet PCR (Bio-Rad QX200) was used to measure absolute number of copies of the wildtype Setd7 allele, with a probe set targeted to a region within exon 7 of the Setd7 locus (catalog #4400291, assay Mm00100479_cn, ThermoFisher); exon 7 is deleted in the knockout model (Lehnertz et al., 2011). The number of wild type copies is normalized to a locus reference that is not deleted in the knockout model, which we employed a probe set designed to target a region overlapping intron 1 and exon 2 of the Setd7 locus (catalog #4442487, assay Mm00100458_cn, ThermoFisher). Signals are fitted to a Poisson distribution, corrected by a standard curve specifically made for this locus by mixing known amounts of Setd7-wt and Setd7-null genomic DNA. This measure represents the absolute frequency of a functional allele of Setd7 in the population and its reciprocal is therefore the percent efficiency of the knockout system.
TCF/LEF reporter assay
After overnight treatment with PFI-2 (5 µM) , expanding MuSCs were transfected with TOPFlash plasmid (Addgene #12456) using Lipofectamine 3000 (Life Technologies) following the manufacturer’s instructions. MuSCs were then treated with Wnt3a (Peprotech) overnight at indicated concentrations before being lysed and Firefly/Renilla luciferase activity quantified by luminescence using a Dual-Luciferase® Reporter Assay System (Promega) following the manufacturer’s instructions.
RNASeq methods and analysis
Total RNA was isolated from cultured and FACs sorted cells using an RNeasy Mini Kit (Qiagen). Sample quality control was performed using the Agilent 2100 Bioanalyzer to check for RNA integrity with RNA Integrity Numbers (RIN) over 8. Qualifying samples were then prepped following the standard protocol for the TruSeq stranded mRNA library kit (Illumina). Sequencing was performed on the Illumina Miseq v3 with Paired End 75bp × 75bp reads. De-multiplexed read sequences were then aligned to the mus musculus mm10 reference sequence using TopHat2 splice junction mapper (Kim et al., 2013). Mapping and Reads Per Kilobase Per Million (RPKM) were generated using Genomic Suite (ParTek Inc). Significant differentially expressed genes were called using EdgeR analysis (Robinson et al., 2009) and then scatter plots and heatmaps generated by open source R and VisRseq platforms (Younesy et al., 2015). Heatmaps generating using z-score values (Pocock, 2006). Gene Ontology (GO) analysis of biological functions was performed using online bioinformatics resource NextBio (Illumina).
RNA-seq data sets prepared for this manuscript have been deposited to the Sequence Read Archive (SRA) under accession number SRP118943.
ChIP-Seq methods and analysis
FACs isolated MuSCs were culture expanded for 7 days before either being lysed or where indicated, treated with vehicle (DMSO) or PFI-2 overnight. ChIPSeq was performed using methods described and validated previously (Lorzadeh et al., 2016). Cell lysis was performed using non-inonic detergents (0.1% Triton X 100 and Deoxycholate) supplemented with protease cocktail inhibitor (Calbiochem). Chromatin was sheared by incubation with Micrococcal nuclease (MNase) at room temperature for 5 minutes. For ChIP, anti-H3K4me1 antibody (Diagenode, Cat# C15410037 ) was incubated with anti-IgA magnetic beads (Dynabeads, ThemoFisher) for 2 hours. This antibody-bead mix was then incubated with the digested chromatin overnight at 4° in IP buffer (20mM Tris-HCl pH 7.5, 2mM EDTA, 150mM NaCl, 0.1% Triton X-100, 0.1% Deoxycholate). IPs were then sequentially washed twice in Low Salt (20mM Tris-HCl pH 8.0, 2mM EDTA, 150mM NaCl, 1% Triton X-100, 0.1% SDS) and High Salt (20 mM Tris-HCl pH 8.0, 2 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.1% SDS) wash buffers before being eluted in elution buffer (1% SDS, 100mM Sodium Bicarbonate) for 1.5 hours at 65°. Histones were digested by Protease (ThermoFisher) for 30 minutes at 50° and DNA fragments were purified using Sera Mag magnetic beads in 30% PEG.
Illumina sequencing libraries were generated by end repair, 3’ A-addition, and Illumina sequencing adaptor ligation (New England BioLabs, E6000B-10). Libraries were then indexed and PCR amplified (10 cycles) and sequenced using a NextSeq 500 High-Output Kit (Illumina) on a Illumina NextSeq 500 sequencing platform following the manufacturer’s protocols (Illumina).
Sequence reads were mapped to mm10 (GRCm38) using Burrows-Wheeler Aligner (BWA) (Li and Durbin, 2009). Regions enriched for H3K4me1 were determined using MACS2 (Feng et al., 2012) peak callers on non-duplicate, uniquely aligned reads. For H3K4me1 peaks, broad regions were called using MACS2 broadpeaks (--qvalue 0.1) (Feng et al., 2012). The MACS output BedGraph files with filtered peak islands were converted to Bigwig format and were loaded in the UCSC genome browser for visualization of the ChIP libraries. Normalization of relative ChIP enrichment from raw data (bam files) was calculated as reads per kilobase per million mapped reads (RPKM) for enhancer Boxplots using VisRseq (Younesy et al., 2015). For scatter plots and read counts profile genome-wide and within 5 kb of TSS regions, we used SeqMonk. The ChIP-seq heatmaps within 5 kb of TSS were generated by ChAsE (Younesy et al., 2016, 2013).
ChIP-seq data sets prepared for this manuscript have been deposited to the Sequence Read Archive (SRA) under accession numbers: SRP118552 (ChIPSeq: H3K4me1 PFI-2 treated myoblasts) and SRP118947 (ChIPSeq: H3K4me1 Setd7 mKO cultured myoblasts).
RNA-seq data sets prepared for this manuscript have been deposited to the Sequence Read Archive (SRA) under accession number SRP118943.
Detection of β-catenin interaction with BCL9 using in-situ proximity ligation assay (PLA)
Duolink® in situ PLA reagents (Sigma-Aldrich Inc.) were used to detect the interaction between β-catenin and BCL9 and the manufacturer’s protocol was followed. Briefly, C2C12 cells, cultured on chamber slides to 60–70% confluence, were treated with Wnt3a (100ng/mL for 4 hours) ± SetD7 inhibitor PFI-2 (5µM for the last 2 hours). Cells were then fixed, permeabilized and incubated overnight with mouse anti-β-catenin (Santa Cruz) and rabbit anti-BCL9 (Thermo Fisher). The following day, cells were incubated with secondary antibodies conjugated to oligonucleotides (PLA probe PLUS and PLA probe MINUS) for 1 hour at 37°C. Afterwards, ligation solution containing 2 oligonucleotides (that hybridize to the PLA probes) and Ligase was added for 30 minutes at 37°C. A closed circle is only formed if the 2 PLA probes are in close proximity. Finally, the closed circle was amplified using rolling-circle amplification reaction and the product was hybridized to fluorescently-labelled oligonucleotides. The fluorescent spots generated from positive interactions were quantified using a Nikon eclipse Ti (Nikon Instruments, Inc) epifluorescence microscope.
Murine MuSC transplantation experiments
MuSCs were freshly isolated from donor mice and expanded in 12 well dishes in the presence of PFI-2 or DMSO (vehicle). After 7 days, expanded MuSCs were pelleted, resuspended in 30–50µl PBS and counted using a haemocytometer. Recipient mice received randomly via intramuscular injection, MuSCs into the TA. Where stated recipient mice also received a single injection of 0.15µg Notexin (NTX) and/or a non-lethal dose of 20Gy irradiation to the hindlimb legs 24 hours prior to transplantation. The injection was performed either by hand or with a Hamilton syringe controlled by an automated micropump (SP220I syringe pump, World Precision Instruments; Sarasota, FL).
Human MuSC transplantation experiments and lentiviral GFP-Luc transduction
Human MuSCs (hMuSCs) were isolated from operative samples of consented patients (age range: 15 to 53 years old) and 15,000 cells were cultured in an 8 well chamber slide with 4 µM PFI-2 or vehicle. At day 1 and day 7, hMuSCs were counted in each well. After 7 days in culture, hMuSCs were transduced with a dual promoter lentivirus (CD51X DPS, SystemBio) expressing Luciferase and GFP (Luc-GFP) proteins. MuSCs were transduced by spinoculation for 5 minutes at 3200g and then 1 hour at 2500g at 25°C with 8µg/mL polybrene. Cells were then dissociated, spun down, re-suspended in Barium Chloride solution (1.2% in PBS) and transplanted into TA muscles of immunocompromised NSG mice. Bioluminescence was then imaged non-invasively over the course of 11 days.
Bioluminescence Imaging and Analysis
Bioluminescent imaging was performed using the Xenogen IVIS-Spectrum System (Caliper Life Sciences). Mice were anesthetized using 2% isoflurane at a flow rate of 2.5 l/min. Intraperitoneal injection of D-Luciferin (50 mg/ml, Biosynth International Inc.) dissolved in sterile PBS was administered. Immediately following the injection, mice were imaged for 30 seconds at maximum sensitivity (f-stop 1) at the highest resolution (small binning). Every minute a 30 second exposure was taken, until the peak intensity of the bioluminescent signal began to diminish. Each image was saved for subsequent analysis. Imaging was performed in bind: the investigators performing the imaging did not know the identity of the experimental conditions for the transplanted cells. Analysis of each image was performed using Living Image Software, version 4.0 (Caliper Life Sciences). A manually-generated circle was placed on top of the region of interest and resized to completely surround the limb or the specified region on the recipient mouse. Similarly a background region of interest was placed on a region of a mouse outside the transplanted leg.
In vivo muscle function measurements
Methods used to measure in vivo muscle function were adapted from (Quarta et al., 2017). Experimental mice were placed under anesthesia for TA isolation surgery. The sciatic nerve adjacent to the cranial thigh muscle was exposed and a small, two wire, bipolar stimulatory nerve cuff was then positioned around the sciatic nerve. The TA muscle was then exposed and gently separated from surround muscles using a blunt dissection technique. Saline was added throughout the procedure to ensure the tissue remained hydrated. The TA tendon was then cut distal to free up the muscle for subsequent force production. An Aurora Scientific 1300-A Whole Mouse Test System was used to gather force production data. The mouse was placed on the stage and a pin was placed through an L bracket behind the patellar tendon to secure the knee joint and ensure isometric muscle contractions. The ankle of the mouse was also secured using tape to ensure the leg remained in the same position throughout the testing procedure.
Quantification and Statistical Analysis
All results are presented as mean ± standard error of the mean (SEM) with the exception of measurements of GFP+ myofibers which are expressed as box-and-whisker plots. Statistical analysis was performed using either a student’s t-tests to calculate differences between two groups and either one-way or two-way ANOVA with post-hoc test for multiple comparisons (Graphpad Prism®; version 6). For genomic and transcriptomic correlation analysis, R values were calculated by Pearson’s correlation coefficient (RStudio®, open source software). Sample size and/or replicate number for each experiment are indicated in the figure legends. Results with p values of less than 0.05 were considered statistically significant.
Data and Software Availability
RNA sequencing data from this study has been submitted to the NCBI Sequence Read Archive (SRA: http://www.ncbi.nlm.nih.gov/sra) repository under the accession number SRP118943. ChIPSeq data has also been deposited to the SRA under accession numbers: SRP118552 (Vehicle vs PFI-2 treated MuSCs) and SRP118947 (Control vs Setd7 mKO MuSCs).
Supplementary Material
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Setd7 (clone 2D10) | Abcam | Cat# ab119400; RRID: AB_11126988 |
| Rabbit polyclonal anti-Set7/Set9 | Cell Signalling Technology | Cat# 2813; RRID: AB_823636 |
| Rabbit monoclonal anti-Setd7 (clone EPR5574) | Abcam | Cat# 124708; RRID:A B_10975432 |
| Rat monoclonal anti-CD45-FITC (clone I3/2) | AbLab | N/A |
| Rat monoclonal anti-CD31-FITC (clone 390) | eBiosciences | Cat# 11-0311-81; RRID: AB_465011 |
| Rat monoclonal anti-Integrinα7-APC (clone R2F2) | AbLab | N/A |
| Rat monoclonal anti-Ly6A/E(Sca-1)-PE-Cy7 (clone D7) | eBiosciences | Cat# 25-5981; RRID: AB_469669 |
| Anti-mouse Vcam1(CD106)-Biotin (clone MK1.9) | AbLab | N/A |
| Mouse monoclonal anti-human CD31-Alexa® Fluor 488 (clone WM59) | Biolegend | Cat# 303110; RRID: AB_493074 |
| Mouse monoclonal anti-human CD34-FITC (clone 581) | Biolegend | Cat# 343503; RRID: AB_1731923 |
| Mouse monoclonal anti-human CD45-Alexa® Fluor 488 (clone HI30) | ThermoFisher | Cat# MHCD4520; RRID: AB_10392555 |
| Mouse monoclonal anti-human CD29-APC (clone TS2/16) | Biolegend | Cat# 303008; RRID: AB_314324 |
| Mouse monoclonal anti-human CD56-APC (clone 5.1H11) | Biolegend | Cat# 362503; RRIS: AB_2563912 |
| Mouse monoclonal anti-Pax7 (clone PAX7) | DSHB | AB_528428; RRID: AB_528428 |
| Mouse monoclonal anti-MyoD (clone 5.8A) | BD Biosciences | Cat# 554130; RRID: AB_395255 |
| Rabbit polyclonal anti-MyoD (clone C-20) | Santa Cruz | Cat# sc-304; RRID: AB_631992 |
| Mouse monoclonal anti-Myogenin (clone F5D) | DSHB | AB_2146602; RRID: 2146602 |
| Mouse monoclonal anti-MyHC (clone MF20) | DSHB | AB_2147781; RRID: AB_2147781 |
| Mouse monoclonal anti-eMyHC (clone F1.652) | DSHB | AB_528358; RRID: AB_528358 |
| Mouse monoclonal anti-β-catenin (clone 14/β-catenin) | BD Biosciences | Cat# 610153; RRID: AB_397554 |
| Mouse monoclonal anti-β-catenin (clone E5) | Santa Cruz | Cat# sc-7963; RRID: AB_626807 |
| Rabbit polyclonal anti-BCL9 | ThermoFisher | Cat# PA5-49466; RRID: AB_2634920 |
| Mouse monoclonal anti-Myc-Tag (9B11) | Cell Signalling Technology | Cat# 2276; RRID: AB_331783 |
| Chicken polyclonal anti-GFP | Abcam | Cat# ab13970; RRID: AB_300798; RRID: AB_300798 |
| Rabbit polyclonal anti-Laminin | Abcam | Cat# ab11575; RRID: AB_298179 |
| Rabbit polyclonal anti-alpha tubulin | Abcam | Cat# ab4074; RRID: AB_2288001 |
| Rabbit polyclonal anti-beta actin | Abcam | Cat# ab8227; RRID: AB_2305186 |
| Rabbit monoclonal anti-H3K4me1 (clone D1A9) | Cell Signalling Technology | Cat# 5362; RRID: AB_10695148 |
| Rabbit polyclonal anti-H3K4me1 | Diagenode | Cat# C15410037; RRID: AB_2637078 |
| Rabbit monoclonal anti-H3K4me2 (clone C64G9) | Cell Signalling Technology | Cat# 9725; RRID: AB_10205451 |
| Rabbit monoclonal anti-H3K4me3 (clone C42D8) | Cell Signalling Technology | Cat# 9751; RRID: AB_2616028 |
| Rabbit monoclonal anti-Histone3 (clone D1H2) | Cell Signalling Technology | Cat# 4499; RRID: AB_10544537 |
| Goat anti-mouse IgG (H+L) Alexa® Fluor 488 | ThermoFisher | Cat# A32723; RRID: AB_2633275 |
| Goat anti-rabbit IgG (H+L) Alexa® Fluor 594 | ThermoFisher | Cat# R37117; RRID: AB_2556545 |
| Goat anti-mouse IgG (H+L) Alexa® Fluor 594 | ThermoFisher | Cat# A-11032; RRID: AB_141672 |
| Goat anti-chicken IgY (H+L) Alexa® Fluor 488 | ThermoFisher | Cat# A-11039; RRID: AB_2534096 |
| Streptavidin-PE | ThermoFisher | Cat# SA1004 |
| Goat anti-mouse IgG (H+L) IRDye® 680LT | LI-COR | Cat# 925-68022; RRID: AB_2687826 |
| Goat anti-rabbit IgG (H+L) IRDye® 800CW | LI-COR | Cat# 925-32213; RRID: AB_2651127 |
| Biological Samples | ||
| Healthy human skeletal muscle tissue | Stanford University, California, USA | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Notexin venom | Latoxan | Cat# L8104 |
| Collagenase I | Worthington Biochemical | Cat# CLS-1 |
| Collagenase II | Worthington Biochemical | Cat# CLS-2 |
| Collagenase D | Roche | Cat# 11 088 858 001 |
| Dispase II | Sigma-Aldrich | Cat# D4693 |
| (R)-PFI-2 | TOCRIS | Cat# 4892 |
| (S)-PFI-2 (enantiomer) | TOCRIS | Cat# 5400 |
| CHIR99021 | TOCRIS | Cat# 4953 |
| Evans Blue Dye (FW 960.82 g mol−1) | Sigma-Aldrich | Cat# E2129 |
| Recombinant murine basic FGF | Peprotech | Cat# 450-33 |
| Recombinant murine Wnt-3a | Peprotech | Cat# 315-20 |
| Barium Chloride | Sigma | Cat# B0750 |
| Polybrene | Santa Cruz Biotechnology | Cat# sc-134220 |
| Critical Commercial Assays | ||
| Dual-Luciferase® Reporter Assay System | Promega | Cat# E1910 |
| RNeasy Plus Micro Kit | QIAGEN | Cat#: 74034 |
| TruSeq stranded mRNA library kit | Illumina | Cat# RS-122-2101 |
| PureLink® Genomic Mini Kit | ThermoFisher | Cat# K182001 |
| NextSeq 500 High-Output Kit | Illumina | Cat# FC-404-1002 |
| Click-iT® EdU imaging kit | ThermoFisher | Cat# C10337 |
| Lipofactamine 3000 reagent | ThermoFisher | Cat# L3000015 |
| DuoLink® in situ PLA® Kit | Sigma | Cat# DUO92101 |
| Deposited Data | ||
| RNAseq: mKO cultured myoblasts | This paper | SRA: SRP118943 |
| ChIPSeq: H3K4me1 PFI-2 treated myoblasts | This paper | SRA: SRP118552 |
| ChIPSeq: H3K4me1 mKO cultured myoblasts | This paper | SRA: SRP118947 |
| Experimental Models: Cell Lines | ||
| Mouse: C2C12 myoblasts | Laboratory of Prof. Helen Blau | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Setd7FL/FL | Lehnertz et al., 2011 | N/A |
| Mouse: Pax7tm1(Cre)Mrc | The Jackson Laboratory | JAX: 01530 |
| Mouse: MyoDiCre | Chen et al., 2005 | N/A |
| Mouse: B6;129-Pax7tm2.1(cre/ERT2)Fan/J | The Jackson Laboratory | JAX: 012476 |
| Mouse: Pax7-GFP | Rocheteau et al., 2012 | N/A |
| Mouse: C57BL/10ScSn-Dmdmdx/J | The Jackson Laboratory | JAX: 001801 |
| Mouse: NOD-scid IL2gmmanull | The Jackson Laboratory | JAX: 005557 |
| Mouse: B6Ros.Cg-Dmdmdx-4Cv/J | The Jackson Laboratory | JAX: 002378 |
| Mouse: C57BL/6-Tg(CAG-EGFP)1Osb/J | The Jackson Laboratory | JAX: 003291 |
| Mouse: FVB-Tg(CAG-Luc,-GFP)L2G85Chco/J | The Jackson Laboratory | JAX: 008450 |
| Oligonucleotides | ||
| Setd7 WT primers (exon 7) assay Mm00100479_cn | ThermoFisher | Cat# 4400291 |
| Setd7 reference primers (exon 1–2) assay Mm00100458_cn modified with VIC dye | ThermoFisher | Cat# 4442487 |
| Recombinant DNA | ||
| M50 super 8xTOPFLASH | Addgene | Cat# 12456 |
| pMSCV-Setd7-3xmyc-IRES-GFP | This paper | N/A |
| CD51X–GFP-Luc | This paper | N/A |
| Software and Algorithms | ||
| TopHat2 | Kim et al., 2013 | http://ccb.jhu.edu/software/tophat/index.shtml |
| EdgeR | (Robinson et al., 2009) | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Genomic Suite | ParTek® Inc | N/A |
| Burrows-Wheeler Aligner | (Li and Durbin, 2009) | http://bio-bwa.sourceforge.net/ |
| MACS2 | Feng et al., 2012 | https://github.com/taoliu/MACS |
| ChAsE | (Younesy et al., 2016, 2013) | http://chase.cs.univie.ac.at/overview |
| Seqmonk | Bioinformatics Group, Babraham Institute | http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/ |
| Correlation Engine (GO Analysis) | NextBio, Ilummina | http://www.nextbio.com/b/nextbio.nb |
| VisRSeq | (Younesy et al., 2015) | http://visrseq.github.io/ |
| Graphpad Prism 6 | GraphPad Software | https://www.graphpad.com/ |
| Fiji | Fiji | https://fiji.sc/ |
| FlowJo V10 | FlowJo LLC | https://www.flowjo.com/solutions/flowjo |
Acknowledgments
We acknowledge the animal unit staff and genotyping core facility at the Biomedical Research Centre especially Mr Taka Muakami, Ms Tara Stach, Mrs Krista Ranta and Mr Wei Yuan. We thank Mr Andy Johnson and Mr Justin Wong of UBC Flow Cytometry. Mr Alireza Lorzadeh, Ms Michelle Moska and Dr. Martin Hirst from the Centre of High-Throughput Biology are sincerely acknowledged for their ChIPSeq services. Dr. Cheryl Arrowsmith is acknowledged for helping acquire PFI-2 molecule. For administrative assistance we thank Ms Vittoria Canale. This work was supported by Canadian Institutes of Health Research (CIHR) grants MOP-97856 to F.M.R. M.J.O was supported by the Research Council of Norway (Project 223255/F50) and the Norwegian Cancer Society (182767-2016). C.Z. was supported by CIHR’s Canadian Epigenetics, Environment and Health Research Consortium (CEEHRC) [funding reference number 128090], CIHR operating grants (MOP-89773 and MOP-106623), Michael Smith Foundation for Health Research, and Australian National Health and Medical Research Council (NHMRC) project grants (APP1104433 and APP1104466 to C. Zaph). C.Z. is a veski innovation fellow. Grants from the NIH (P01 AG036695) and the VA (RR&D Merit Review) to T.A.R also supported this work.
Footnotes
Author contributions:
Conceptualization, R.N.J, M.J.O, M.Q, T.A.R and F.M.R; Methodology, R.H.Z; Validation, M.Q, M.H and J.B; Formal analysis, R.N.J, M.J.O, M.Q, H.S, M.H, J.B, M.M.K, A.C and R.V.W; Investigation, R.N.J, M.Q, M.J.O, H.S, L.Y, C.K.C, G.L, M.H, J.B, P.P, L.T.N.D, E.G, W.H, L.S, R.H.Z, P.X, A.C, R.V.W, I.B, C.E C.B.L and M.L; Writing - original draft, R.N.J; Writing - review & editing, R.N.J, M.Q, M.J.O, L.S, T.A.R and F.M.R; Supervision, T.A.R, C.Z and F.M.R; Funding acquisition, T.A.R, C.Z and F.M.R.
Declarations of interest:
The authors declare no competing interests.
References
- Alfaro LAS, Dick SA, Siegel AL, Anonuevo AS, McNagny KM, Megeney LA, Cornelison DDW, Rossi FMV. CD34 Promotes Satellite Cell Motility and Entry into Proliferation to Facilitate Efficient Skeletal Muscle Regeneration. Stem Cells. 2011;29:2030–2041. doi: 10.1002/stem.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsyte-Lovejoy D, Li F, Oudhoff MJ, Tatlock JH, Dong A, Zeng H, Wu H, Freeman SA, Schapira M, Senisterra GA, Kuznetsova E, Marcellus R, Allali-Hassani A, Kennedy SJPL, Couzens AL, Aman AA-CG, Al-Awar R, Fish PV, Gerstenberger BS, Roberts L, Benn CL, Grimley RL, Braam MJS, Rossi FMV, Sudol M, Brown PJ, Bunnage ME, Owen DR, Zaph C, Vedadi M, Arrowsmith CH. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proceedings of the National Academy of Sciences. 2014;111:12853–12858. doi: 10.1073/pnas.1407358111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, Zammit PS. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J. Cell Biol. 2000;151:1221–1234. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernet JD, Doles JD, Hall JK, Kelly Tanaka K, Carter TA, Olwin BB. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat. Med. 2014;20:265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev. 2012;26:2763–2779. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Murphy-Seiler F, Hanifi J, Deka J, Eyckerman S, Keller C, Aguet M, Rando TA. BCL9 is an essential component of canonical Wnt signaling that mediates the differentiation of myogenic progenitors during muscle regeneration. Dev. Biol. 2009;335:93–105. doi: 10.1016/j.ydbio.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JCJ, Justin M, Jason M, Goldhamer DJ. MyoD-cre transgenic mice: A model for conditional mutagenesis and lineage tracing of skeletal muscle. Genesis. 2005;41:116–121. doi: 10.1002/gene.20104. [DOI] [PubMed] [Google Scholar]
- Cosgrove BD, Gilbert PM, Porpiglia E, Mourkioti F, Lee SP, Corbel SY, Llewellyn ME, Delp SL, Blau HM. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011;6:1059–1067. doi: 10.4161/epi.6.9.16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont NA, Bentzinger CF, Sincennes M-C, Rudnicki MA. Satellite Cells and Skeletal Muscle Regeneration. Compr. Physiol. 2015a;5:1027–1059. doi: 10.1002/cphy.c140068. [DOI] [PubMed] [Google Scholar]
- Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE, Rudnicki MA. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 2015b;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Jianxing F, Tao L, Bo Q, Yong Z, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figeac N, Zammit PS. Coordinated action of Axin1 and Axin2 suppresses β-catenin to regulate muscle stem cell function. Cell. Signal. 2015;27:1652–1665. doi: 10.1016/j.cellsig.2015.03.025. [DOI] [PubMed] [Google Scholar]
- Fu X, Xiao J, Wei Y, Li S, Liu Y, Yin J, Sun K, Sun H, Wang H, Zhang Z, Zhang B-T, Sheng C, Wang H, Hu P. Combination of inflammation-related cytokines promotes long-term muscle stem cell expansion. Cell Res. 2015;25:1082–1083. doi: 10.1038/cr.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Prat L, Martínez-Vicente M, Perdiguero E, Ortet L, Rodríguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, Sandri M, Muñoz-Cánoves P. Autophagy maintains stemness by preventing senescence. Nature. 2016;529:37–42. doi: 10.1038/nature16187. [DOI] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer PW, McGeachie JM, Davies MJ, Grounds MD. Evans Blue Dye as an in vivo marker of myofibre damage: optimising parameters for detecting initial myofibre membrane permeability. J. Anat. 2002;200:69–79. doi: 10.1046/j.0021-8782.2001.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han XH, Jin Y-R, Seto M, Yoon JK. A WNT/beta-catenin signaling activator, R-spondin, plays positive regulatory roles during skeletal myogenesis. J. Biol. Chem. 2011;286:10649–10659. doi: 10.1074/jbc.M110.169391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho ATV, Palla AR, Blake MR, Yucel ND, Wang YX, Magnusson KEG, Holbrook CA, Kraft PE, Delp SL, Blau HM. Prostaglandin E2 is essential for efficacious skeletal muscle stem-cell function, augmenting regeneration and strength. Proc. Natl. Acad. Sci. U. S. A. 2017 doi: 10.1073/pnas.1705420114. [DOI] [PMC free article] [PubMed]
- Hutcheson DA, Zhao J, Merrell A, Haldar M, Kardon G. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev. 2009;23:997–1013. doi: 10.1101/gad.1769009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya M, Takada S. Wnt signaling from the dorsal neural tube is required for the formation of the medial dermomyotome. Development. 1998;125:4969–4976. doi: 10.1242/dev.125.24.4969. [DOI] [PubMed] [Google Scholar]
- Jones AE, Price FD, Le Grand F, Soleimani VD, Dick SA, Megeney LA, Rudnicki MA. Wnt/β-catenin controls follistatin signalling to regulate satellite cell myogenic potential. Skelet. Muscle. 2015;5:14. doi: 10.1186/s13395-015-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Rogalski JC, Schulze FM, Yi L, Lin S, Kast J, Rossi FMV. p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Mol. Cell. 2011;43:673–680. doi: 10.1016/j.molcel.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan C-M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorzadeh A, Bilenky M, Hammond C, Knapp DJHF, Li L, Miller PH, Carles A, Heravi-Moussavi A, Gakkhar S, Moksa M, Eaves CJ, Hirst M. Nucleosome Density ChIP-Seq Identifies Distinct Chromatin Modification Signatures Associated with MNase Accessibility. Cell Rep. 2016;17:2112–2124. doi: 10.1016/j.celrep.2016.10.055. [DOI] [PubMed] [Google Scholar]
- Marg A, Escobar H, Gloy S, Kufeld M, Zacher J, Spuler A, Birchmeier C, Izsvák Z, Spuler S. Human satellite cells have regenerative capacity and are genetically manipulable. J. Clin. Invest. 2014;124:4257–4265. doi: 10.1172/JCI63992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin Association of Human Origin Recognition Complex, Cdc6, and Minichromosome Maintenance Proteins during the Cell Cycle: Assembly of Prereplication Complexes in Late Mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Morgan JE, Zammit PS. Direct effects of the pathogenic mutation on satellite cell function in muscular dystrophy. Exp. Cell Res. 2010;316:3100–3108. doi: 10.1016/j.yexcr.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Murphy MM, Keefe AC, Lawson JA, Flygare SD, Mark Y, Gabrielle K. Transiently Active Wnt/β-Catenin Signaling Is Not Required but Must Be Silenced for Stem Cell Function during Muscle Regeneration. Stem Cell Reports. 2014;3:475–488. doi: 10.1016/j.stemcr.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudhoff MJ, Braam MJS, Freeman SA, Wong D, Rattray DG, Wang J, Antignano F, Snyder K, Refaeli I, Hughes MR, McNagny KM, Gold MR, Arrowsmith CH, Sato T, Rossi FMV, Tatlock JH, Owen DR, Brown PJ, Zaph C. SETD7 Controls Intestinal Regeneration and Tumorigenesis by Regulating Wnt/β-Catenin and Hippo/YAP Signaling. Dev. Cell. 2016;37:47–57. doi: 10.1016/j.devcel.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Oudhoff MJ, Freeman SA, Couzens AL, Antignano F, Kuznetsova E, Min PH, Northrop JP, Lehnertz B, Barsyte-Lovejoy D, Vedadi M, Arrowsmith CH, Nishina H, Gold MR, Rossi FMV, Gingras A-C, Zaph C. Control of the hippo pathway by Set7-dependent methylation of Yap. Dev. Cell. 2013;26:188–194. doi: 10.1016/j.devcel.2013.05.025. [DOI] [PubMed] [Google Scholar]
- Parisi A, Lacour F, Giordani L, Colnot S, Maire P, Le Grand F. APC is required for muscle stem cell proliferation and skeletal muscle tissue repair. J. Cell Biol. 2015;210:717–726. doi: 10.1083/jcb.201501053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock SJ. The simplest statistical test: how to check for a difference between treatments. BMJ. 2006;332:1256–1258. doi: 10.1136/bmj.332.7552.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarta M, Brett JO, DiMarco R, De Morree A, Boutet SC, Chacon R, Gibbons MC, Garcia VA, Su J, Shrager JB, Heilshorn S, Rando TA. An artificial niche preserves the quiescence of muscle stem cells and enhances their therapeutic efficacy. Nat. Biotechnol. 2016;34:752–759. doi: 10.1038/nbt.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarta M, Cromie M, Chacon R, Blonigan J, Garcia V, Akimenko I, Hamer M, Paine P, Stok M, Shrager JB, Rando TA. Bioengineered constructs combined with exercise enhance stem cell-mediated treatment of volumetric muscle loss. Nat. Commun. 2017;8:15613. doi: 10.1038/ncomms15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi F, Perlingeiro RCR. Stem cells for skeletal muscle regeneration: therapeutic potential and roadblocks. Transl. Res. 2014;163:409–417. doi: 10.1016/j.trsl.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- Rudolf A, Schirwis E, Giordani L, Parisi A, Lepper C, Taketo MM, Le Grand F. β-Catenin Activation in Muscle Progenitor Cells Regulates Tissue Repair. Cell Rep. 2016;15:1277–1290. doi: 10.1016/j.celrep.2016.04.022. [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, Shkreli M, Delp S, Pomerantz JH, Artandi SE, Blau HM. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Shen C, Wang D, Liu X, Gu B, Du YFZWLLC, Song B, Lu X, Yang Q, Zhu Q, Hou T, Li M, Wang L, Wang H, Zhao Y, Yang YWGZ. SET7/9 regulates cancer cell proliferation by influencing -catenin stability. The FASEB Journal. 2015;29:4313–4323. doi: 10.1096/fj.15-273540. [DOI] [PubMed] [Google Scholar]
- Sousa-Victor P, Gutarra S, García-Prat L, Rodriguez-Ubreva J, Ortet L, Ruiz-Bonilla V, Jardí M, Ballestar E, González S, Serrano AL, Perdiguero E, Muñoz-Cánoves P. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Terada K, Nohno T. Canonical Wnt signaling is involved in switching from cell proliferation to myogenic differentiation of mouse myoblast cells. J. Mol. Signal. 2011;6:12. doi: 10.1186/1750-2187-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Neppl RL, Huang Z-P, Chen J, Tang R-H, Cao R, Zhang Y, Jin S-W, Wang D-Z. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J. Cell Biol. 2011;194:551–565. doi: 10.1083/jcb.201010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney MT, Aydogdu T, Sala D, Malecova B, Gatto S, Puri PL, Latella L, Sacco A. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat. Med. 2014;20:1182–1186. doi: 10.1038/nm.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilschut KJ, Ling VB, Bernstein HS. Concise review: stem cell therapy for muscular dystrophies. Stem Cells Transl. Med. 2012;1:833–842. doi: 10.5966/sctm.2012-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Watanabe Y, Ohtani T, Uezumi A, Mikami N, Nakamura M, Sato T, Ikawa M, Hoshino M, Tsuchida K, Miyagoe-Suzuki Y, Tsujikawa K, Takeda S‘ichi, Yamamoto H, Fukada S-I. Calcitonin Receptor Signaling Inhibits Muscle Stem Cells from Escaping the Quiescent State and the Niche. Cell Rep. 2015;13:302–314. doi: 10.1016/j.celrep.2015.08.083. [DOI] [PubMed] [Google Scholar]
- Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, An L, Wang Q, Lu T, Zhang X, Wang Z, Stark GR. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younesy H, Möller T, Lorincz MC, Karimi MM, Jones SJM. VisRseq: R-based visual framework for analysis of sequencing data. BMC Bioinformatics. 2015;(16 Suppl 11):S2. doi: 10.1186/1471-2105-16-S11-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younesy H, Nielsen CB, Lorincz MC, Jones SJM, Karimi MM, Möller T. ChAsE: chromatin analysis and exploration tool. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw382. [DOI] [PMC free article] [PubMed]
- Younesy H, Nielsen CB, Möller T, Alder O, Cullum R, Lorincz MC, Karimi MM, Jones SJM. An Interactive Analysis and Exploration Tool for Epigenomic Data. Comput. Graph. Forum. 2013;32:91–100. [Google Scholar]
- Zhang R-H, Judson RN, Liu DY, Kast J, Rossi FMV. The lysine methyltransferase Ehmt2/G9a is dispensable for skeletal muscle development and regeneration. Skelet. Muscle. 2016;6:22. doi: 10.1186/s13395-016-0093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zismanov V, Chichkov V, Colangelo V, Jamet S, Wang S, Syme A, Koromilas AE, Crist C. Phosphorylation of eIF2α Is a Translational Control Mechanism Regulating Muscle Stem Cell Quiescence and Self-Renewal. Cell Stem Cell. 2016;18:79–90. doi: 10.1016/j.stem.2015.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.