ABSTRACT
Force generation within cells, mediated by motor proteins along cytoskeletal networks, maintains the function of multicellular structures during homeostasis and when generating collective forces. Here, we describe the use of chromatin dynamics to detect cellular force propagation [a technique termed SINK (sensors from intranuclear kinetics)] and investigate the force response of cells to disruption of the monolayer and changes in substrate stiffness. We find that chromatin dynamics change in a substrate stiffness-dependent manner within epithelial monolayers. We also investigate point defects within monolayers to map the impact on the strain field of a heterogeneous monolayer. We find that cell monolayers behave as a colloidal assembly rather than as a continuum since the data fit an exponential decay; the lateral characteristic length of recovery from the mechanical defect is ∼50 µm for cells with a 10 µm spacing. At distances greater than this characteristic length, cells behave similarly to those in a fully intact monolayer. This work demonstrates the power of SINK to investigate diseases including cancer and atherosclerosis that result from single cells or heterogeneities in monolayers.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Mechanobiology, Cell mechanics, Cell rheology, Cell structure, Substrate stiffness
Summary: Factors that affect cellular force propagation through cells lead to changes in chromatin dynamics. The SINK technique can be used to characterize mechanical aspects of cell monolayers.
INTRODUCTION
Generation and propagation of forces by cells in monolayers is critical in development and disease (Saw et al., 2017). The mechanobiology of monolayers and collective cellular behavior is of increasing interest since cells integrate forces for communication in addition to chemical signaling (Ladoux and Mège, 2017). Important aspects of collective cellular behaviors occur in response to mechanical differences within an intact monolayer environment. Current methods to measure cellular force typically require an external, physiologically deformable probe or substrate, as in traction force microscopy (Polacheck and Chen, 2016), or require drastic alterations to cell architecture, as in stress fiber ablation (Kassianidou and Kumar, 2015). Additionally, many of the current techniques are more easily performed on isolated cells, as opposed to cell monolayers.
Previously, we developed a particle-tracking technique in which fluctuations of probes bound to chromatin are measured as readouts of cell force propagation (Spagnol and Dahl, 2014). Details on previous applications of the technique are provided in the Materials and Methods. Here, we describe the use of chromatin dynamics to detect cellular force propagation [a technique termed sensors from intranuclear kinetics (SINK)] and investigate the force response of cells to disruption of the monolayer and changes in substrate stiffness. SINK provides previously unattainable information about monolayer force dynamics. Unlike traction force microscopy in which actin-myosin forces are transmitted to focal adhesions and ultimately the extracellular matrix (Rape et al., 2011), SINK is based on actin-myosin forces transmitted to the linker of the nucleoskeleton and cytoskeleton (LINC) complex and into the chromatin of the nucleus (Alam et al., 2015). Thus, SINK can be utilized to investigate systems previously unreachable by current force measurement techniques or in similar systems to further understand the extent that forces impact the cell nucleus, and ultimately, gene expression.
Here, we use SINK to compare measurements in intact cell monolayers with either modulated substrate stiffness or single cells mechanically decoupled from the monolayer. Using this intracellular technique, we can determine the maximum effective stiffness that the monolayer can sense and the lateral length scale that the cells can mechanically perturb.
RESULTS AND DISCUSSION
Active cellular forces increase intranuclear dynamics
SINK does not require isolated cells, and we present studies here on epithelial cell (NRK-52E) monolayers. We tracked GFP-tagged upstream binding factor (GFP-UBF) and quantified intranuclear motion as mean square displacement (MSD). With extensive processing and nuclear alignment, punctate regions of GFP-UBF within the nucleus were tracked over time (Fig. 1A-D). The MSD of these tracks were then averaged and power-law fits (of the form of Eqn 2 in the Materials and Methods) of the MSD versus lag time (τ) yielded β, the force generation exponent (Fig. 1E,F). Details of this equation and fitting are provided in the Materials and Methods. We have shown previously that the fitting parameters β and Deff, indicative of the force propagation through cells and related to the chromatin condensation state, respectively, are affected by independent cellular phenomena (Spagnol and Noel Dahl, 2014), and we have successfully performed SINK on a variety of cell types (Armiger et al., 2016; Booth-Gauthier et al., 2012). Here, we reduced force generation in NRK-52E monolayers with the Rho-associated protein kinase (ROCK) inhibitor Y-27632 (Fig. S1 for characterization) and physically decoupled the nucleus from the cytoskeleton using a dominant-negative Klarsicht, ANC-1, Syne homology (DN-KASH) domain construct to reduce LINC complex connectivity (Figs S2 and S3 for characterization), via displacement of nesprins (Luxton et al., 2010). Both Y-27632-treated and DN-KASH-transfected cells showed reduced intranuclear movement, and the force generation exponent β was similar despite the independent mechanisms used to reduce the propagation of cell force (Fig. 1F). Increased agitation within cells from active forces has been observed in the cytoplasm based on stochastic motion of tracer particles, driven by molecular motor activity on the cytoskeleton (Guo et al., 2014). Additionally, as a result of connections through the LINC complex, motor forces propagate to the nucleus and affect subnuclear movements (Tajik et al., 2016). In addition to the work described here, we previously demonstrated this propagation of active force in endothelial cells by the inhibition of myosin II with blebbistatin, ATP depletion and physical disruption of the LINC complex using DN-KASH, wherein all cases showed a reduction of β (Spagnol and Noel Dahl, 2014). While a quantitative force value cannot currently be extracted using this technique, we have shown that chromatin motion and the parameter β are responsive to changes in force propagation. We demonstrate the utility of SINK for probing complex systems in the following sections.
Fig. 1.
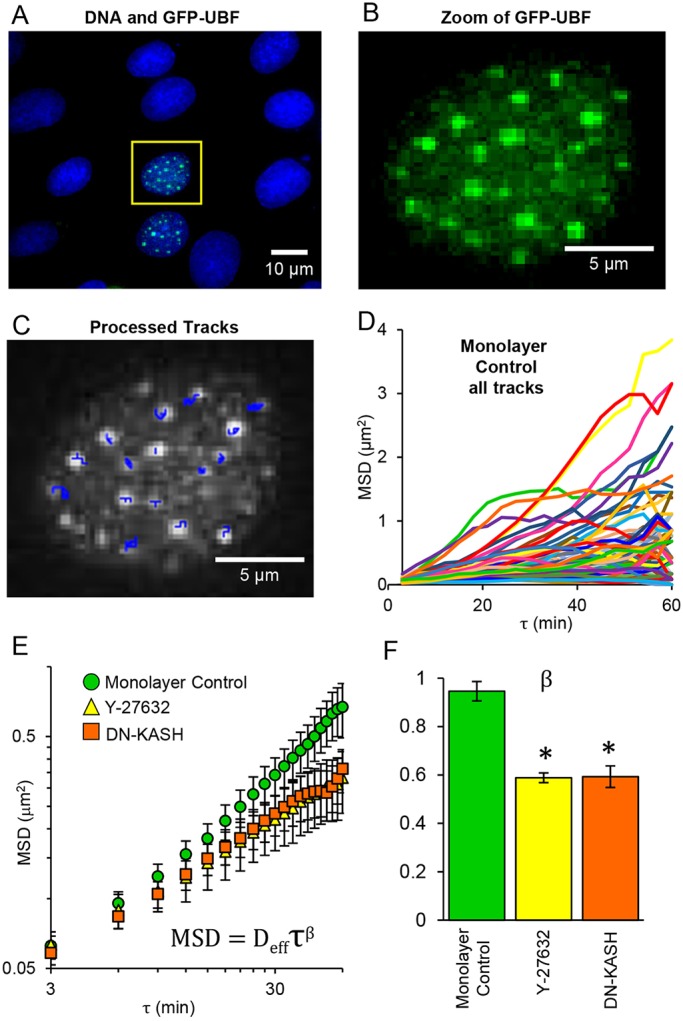
SINK method and mechanisms of intracellular force propagation reduction. (A) Monolayer of NRK-52E epithelial cells showing two cells expressing GFP-UBF (green); DNA is stained blue with Hoechst 33342. (B) Magnified image of outlined nucleus in A showing only the GFP-UBF channel. (C) Processed frame of B after imaging nucleus at 3 min time intervals for 60 min. Tracks of points are shown as blue overlay. Nuclei were aligned prior to tracking of points. (D) Tracks from all points in all nuclei for the control monolayer data shown on linear coordinates. (E) MSD average of intranuclear movement versus lag time (τ) plotted on log-log coordinates for control cells in a monolayer (green), cells in a monolayer treated with Y-27632 (yellow), and cells in a monolayer transfected with DN-KASH (orange). Error bars represent s.e.m. Data are fitted to a power law of the form shown. (F) Comparison of the force generation exponent (β) for control cells in a monolayer, Y-27632 and DN-KASH monolayers. Reduction of either cell force generation (Y-27632) or nuclear connectivity (DN-KASH) resulted in decreased β. Asterisk denotes significant difference based on the curve fit (P<0.05). Error bars represent 95% confidence interval for the fitting of β. Monolayer control, n=84; Y-27632, n=101; DN-KASH, n=64.
Subconfluent cells demonstrate reduced intranuclear motion compared with monolayer cells
SINK can be used to investigate epithelial cells in monolayers, which can be challenging using other biophysical techniques. We first considered the differences in intracellular force generation between subconfluent and monolayer cells. Previous studies using traction force microscopy on multicellular systems reported that the traction force generation of two adjoined cells was not the sum of forces of the two isolated cells on the substrate (Sim et al., 2015). We define monolayer cells as being in contact with adjacent cells on all sides, with actin at the midline of the cell (Fig. 2A-C) and with developed adherens junctions, as confirmed using immunofluorescence (Fig. S2). Isolated cells were imaged at sub-confluence where a portion of the cell was not in contact with adjacent cells. In these cells, actin distribution was observed primarily in the basal region of the cell (Fig. 2A-C). We used SINK to compare changes in intranuclear movement of GFP-UBF in isolated and monolayer cells; intranuclear movement was increased in a monolayer compared to isolated cells (Fig. 2D). When these MSD curves were fitted to a power law, the force generation exponent, β, was significantly increased in monolayer cells (Fig. 2E). We speculate that the increased force propagation to the nucleus in monolayer cells compared with isolated cells may be due to both the redistribution of force within cells due to changes in actin distribution, as well as increased net force resulting from increased cell-cell contact.
Fig. 2.
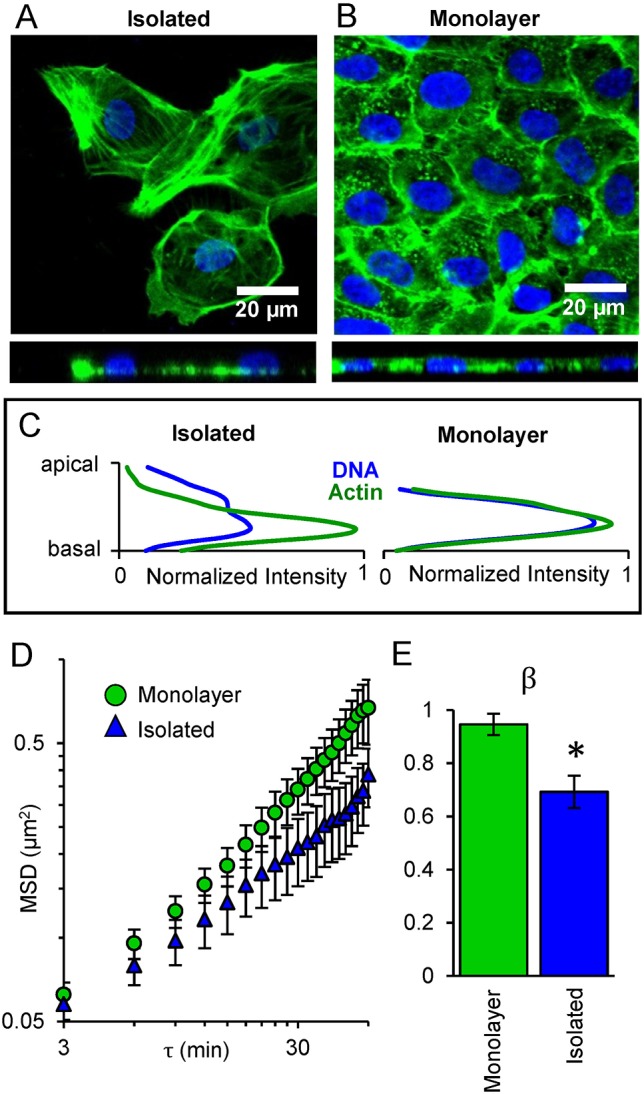
Reduction of intranuclear movement and β in isolated cells versus monolayers. (A) Confocal image of isolated cells and (B) cells in a monolayer, with DNA (blue), and actin (green, phalloidin). Top view (top) and side view (bottom) shown. (C) Image analysis of apical-basal (z-direction) distribution of actin and DNA shows colocalization in the monolayer but separation in the isolated cells. In isolated cells actin lies primarily along the basal region of cells, while in monolayer cells actin is primarily coplanar with the nucleus. (D) MSD of intranuclear movement versus lag time (τ) plotted on log-log coordinates, comparing monolayer (green) versus isolated (blue) cells. Error bars represent s.e.m. (E) Comparison of the force generation (β) for monolayer and isolated cells. *P<0.05 based on the curve fit; error bars represent 95% confidence interval for the fitting of β. Monolayer, n=84; Isolated, n=55.
We observed increased coplanar actin leading to the nucleus in monolayers (Fig. 2A-C). This is consistent with previous observations of monolayers wherein cell nuclei were uniform and flatter, with actin throughout the height of the nucleus compared with isolated cells, which had less-uniform nuclei with basal actin (Neelam et al., 2016). Cell-cell contacts in monolayers provide force generation through adherens junctions rather than only through focal adhesions. Previous studies have shown that the overall contractile moment in clusters of endothelial cells decreased after blocking VE-cadherin, (Krishnan et al., 2011), suggesting that cells increase their force generation upon forming cadherin junctions. Our results suggest that a larger amount of cell-generated force is propagated to the nucleus in monolayer systems compared with the nuclei of isolated cells.
Intranuclear motion changes with substrate stiffness in monolayer systems
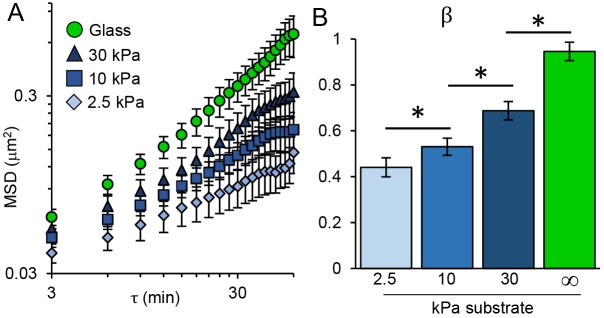
For more than a decade, traction force measurements and analogous studies have shown that individual cells generate increased force on stiffer substrates (Discher et al., 2005). We use SINK with NRK-52E monolayers on collagen coated polyacrylamide gels of varying elastic moduli. As the stiffness of the underlying substrate was increased from 2.5 to 10 and then to 30 kPa, the intranuclear movement of GFP-UBF and the force-generation exponent β increased, indicating a dose-dependent increase in force propagation through monolayer cells as their substrate stiffness increases (Fig. 3A,B). Additionally, these data were compared with that for cell monolayers grown on glass (not coated with collagen), with an elastic modulus several orders of magnitude greater than that of a 30 kPa gel, where a further increase in MSD and β was observed (Fig. 3). Each of these regimes were statistically different, suggesting that SINK has sufficient precision to measure substrate stiffness-dependent changes in cellular force generation. This result was in slight contrast to traction force microscopy data, which often plateaus on stiffer substrates (Ghibaudo et al., 2008). In contrast to SINK measurements, traction forces become difficult to measure at high stiffness, as substrate displacements decrease, and cannot be measured on glass substrates. There is evidence to suggest that epithelial monolayers are more responsive to substrate stiffness than sparsely seeded cells (Ng et al., 2012). Generally, the trend of increased force generation with increased substrate stiffness is consistent with traction force microscopy data (Andresen Eguiluz et al., 2017; Krishnan et al., 2011). From exponential fitting of our data, we found that cells would generate 90% of the force generated on glass (glass set to 50 GPa, essentially infinite stiffness) on a substrate with an elastic modulus of about 60 kPa. To our knowledge, this is the first example of measuring the effects of substrate stiffness on cellular force propagation to the nucleus in monolayer systems.
Fig. 3.
Effect of substrate stiffness on intranuclear movement via SINK in monolayers. (A) MSD of intranuclear movement versus lag time (τ) plotted on log-log coordinates for cells in monolayers seeded on glass (green), or collagen coated polyacrylamide gels with elastic moduli of 30, 10 or 2.5 kPa. Intranuclear movement increases as substrate stiffness increases. Error bars represent s.e.m. (B) Comparison of the force generation exponent (β) for monolayers of varying elastic moduli for curves shown in A. *P<0.05 based on the curve fit; error bars represent 95% confidence interval for the fitting of β. Glass, n=84; 30 kPa, n=235; 10 kPa, n=287; 2.5 kPa, n=173.
The SINK method can be used to characterize monolayers with single-cell defects
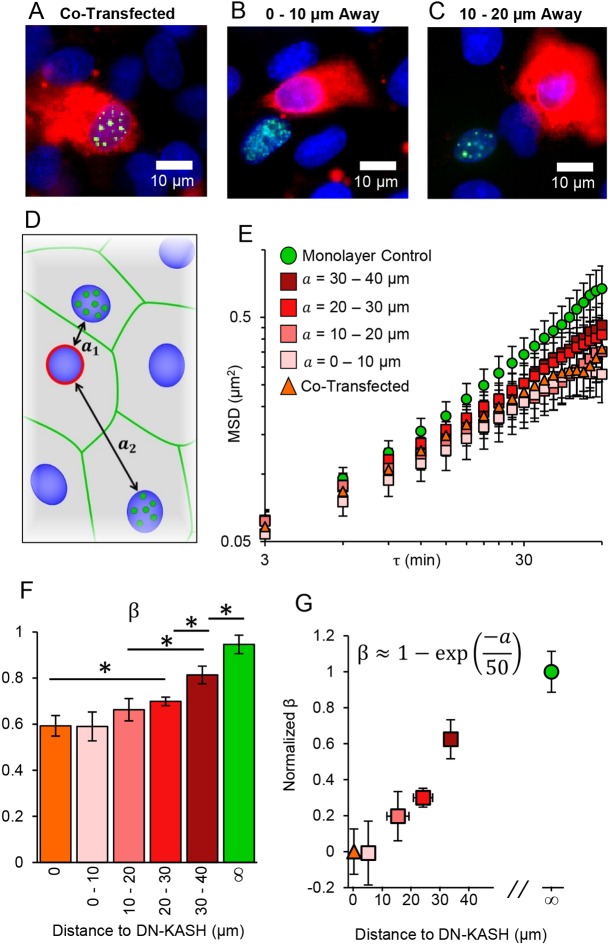
Cancers, cardiovascular plaques and other diseases often stem from the dysfunction of a single cell or cell cluster within a relatively homogeneous monolayer (Lodish et al., 2000; Mehta and Tzima, 2016). We used SINK to investigate spatial propagation of force from a single-cell defect through a monolayer. We decreased cellular force propagation through a cell by expressing DN-KASH, which has previously been shown to reduce force propagation from one side of a cell to the other (Alam et al., 2015). For these experiments, monolayers were initially transfected with DN-KASH, the medium was changed after several hours, and the cell monolayer was transfected again with GFP-UBF (Fig. 4A-C). We examined cells co-transfected with DN-KASH and GFP-UBF (Fig. 4A) or at various distances from DN-KASH-expressing cells (Fig. 4B,C) for SINK analysis. With the addition of the untransfected monolayer data and the co-transfected cells (from Fig. 1E,F), a trend in β was observed: β (and MSD) increased in nuclei further from the DN-KASH-expressing cell (Fig. 4D-F). We saw no significant difference in nuclear height or actin distribution between DN-KASH-expressing cells, either nearby or distant (Figs S2C,D and S3), suggesting that altered β is from force propagation through cells, not changes in cell architecture. Next, β was normalized and plotted versus distance between the DN-KASH-expressing nucleus and the cell in which the SINK analysis was performed. We found that the data can be fitted by an exponential, β≈1–exp(–a/50 µm), consistent with a two-dimensional strain field, where a is the distance between the two nuclei (Fig. 4G). These results suggest that mechanically compromised cells can affect additional cells in a monolayer, perhaps increasing the probability for disease propagation. It has been shown that cells can sense rigidity in fibrous protein matrices at distances >65 µm as compared with synthetic gel matrices where this distance is only ∼5 µm (Rudnicki et al., 2013). We have shown here that cells have a mechanical interaction distance greater than 50 µm and, based on this fit, would return to ∼90% of their control level force propagation at a distance of 100 µm from the compromised cell nucleus. These data are consistent with literature reports that suggest cells within a monolayer respond to physical perturbation (via magnetic twisting cytometer) several cells away from the applied force (Barry et al., 2015). Within the context of wound healing, a refined strain field allows for regulation and control of physical properties such as coordinated mechanotransduction (Sunyer et al., 2016) as a function of distance from the wound site.
Fig. 4.
SINK method to measure changes in force in heterogeneous monolayers. (A-C) GFP-UBF (green)-expressing nuclei (blue) with DN-KASH (red) being expressed in the same cell (A), a cell 0-10 µm away (B) or a cell 10-20 µm away from the GFP-UBF-expressing cell (C). Distances measured represent nearest nucleus to nucleus distance to a DN-KASH-expressing nucleus. (D) Schematic of target cells expressing GFP-UBF (green dots) at various distances (a) from a DN-KASH-expressing cell (red outline). (E) MSD versus lag time for DN-KASH-expressing cells, cells of varying distances from DN-KASH and control monolayer cells. Nearby cells have similar intranuclear motion to co-transfected cells. MSD increases as cells are further from a DN-KASH-expressing cell. Error bars represent s.e.m. (F) Comparison of the force generation exponent (β) for nuclei transfected with DN-KASH (orange), or at different distances away from a DN-KASH-expressing cell (shades of red) as well as monolayers not transfected with DN-KASH (green). Error bars represent 95% confidence interval for the fitting of β. (G) Plot of the normalized β value versus distance away from DN-KASH. The data were fitted as an exponential of the form β=1–exp(−a/n), where a is the distance (in µm) away from a DN-KASH-expressing nucleus. The parameter n is a spatial parameter such that forces at a distance n (in µm) no longer feel the majority of the effects of the DN-KASH-expressing cell, ∼50 µm based on the fit. The R2 for the fit was 0.93. Y error bars are 95% confidence intervals of β after normalization. X error bars are the s.d. of distance away from DN-KASH for the non-adjacent cells. Monolayer control, n=84; a=30-40 um, n=50; a=20-30 um, n=132; a=10-20 um, n=93; a=0-10 um, n=185; co-transfected, n=64.
The fitting of spatial data to an exponential suggests that these cells are integrated as colloidal crystals rather than as a continuum material (Alsayed et al., 2005). Based on theory, continuum materials with defects would scale as 1/a2 whereas colloidal crystal materials would scale as an exponential, as shown (Lipowsky, 1986). There are numerous analogies of cell monolayers to colloidal crystals: colloidal crystals have weak attraction energy between individual particles and assembly is driven primarily by volume exclusion rather than strong attraction. Thus, fluctuations allow for reorganization and redistribution in void sites and at edges, similar to the self-healing of monolayers. Also, cells can undergo geometric-sensitive changes in structure (Chanet et al., 2017), which resemble colloidal crystal phase transitions (Weeks et al., 2000). Intercellular forces calculated with traction force microscopy of cell monolayers utilizes an assumption of a continuum of the monolayer (Tambe et al., 2011). Our results suggest that this assumption should be modified to include the fact that cells are mechanically communicating as colloids rather than as a continuum.
In conclusion, we demonstrate that bulk intranuclear movement is responsive to intercellular forces and that the force generation exponent β can serve as a relative output of intracellular force propagation through modulated monolayer structures. Our results suggest that monolayer cells generate increased force as substrate stiffness increases, consistent with results in single cells (Califano and Reinhart-King, 2010; Discher et al., 2005; Han et al., 2012) and that this increased force leads to increased chromatin motion. In these monolayer systems, the data suggest that cells have a maximum stiffness-sensing threshold of ∼60 kPa and would respond similarly to 60 kPa and glass (GPa) substrates. Finally, heterogeneous monolayers arising from aberrant single-cell defects integrate as colloidal crystals with a length scale of ∼50 µm.
MATERIALS AND METHODS
Cell culture, transfection and chemical treatment
NRK-52E cells (a gift from Yu-Li Wang, Carnegie Mellon University, Pittsburgh, PA) were used in all experiments and cultured in DMEM high-glucose medium (Thermo Fisher), supplemented with 5% FBS (Thermo Fisher) and 1% penicillin-streptomycin (Thermo Fisher) at 37°C and 5% CO2. Live-cell imaging with Hoechst 33342 allowed us to confirm that contamination was not present during experiments. All cells were cultured on uncoated glass for imaging applications with the exception of cells grown on polyacrylamide gels to examine changes in substrate stiffness. To visualize chromatin fluctuations, cells were transfected with the GFP-UBF plasmid (Booth-Gauthier et al., 2012). To disrupt the LINC complex, cells were transfected with a DN-KASH plasmid tagged with red fluorescent protein (Spagnol and Noel Dahl, 2014) (a gift from Gant Luxton, University of Minnesota, Minneapolis, MN). Lipofectamine 3000 (Thermo Fisher) was used for transfection according to the manufacturer's protocol; cells were imaged 24 h after transfection. For experiments that required cells transfected with GFP-UBF to be either adjacent, or distant, to DN-KASH-expressing cells, monolayers were first transfected with DN-KASH. After 4 h, cells were washed with PBS, medium was changed and cells were re-transfected with the GFP-UBF. For isolated, monolayer control, and Y-27632-treated monolayers, cells were co-transfected with LifeAct-Tag RFP to visualize actin filaments in live cells (Fig. S1). For Y-27632 experiments, cells were treated for 1 h prior to imaging, at a concentration of 50 µM.
Cell imaging and fluorescent labeling
For SINK analysis, live cells were imaged in a live-cell imaging chamber (PeCON), maintained at 37°C and 5% CO2. Cells were imaged using a 63×1.4 NA oil immersion inverted wide-field fluorescence microscope (DMI6000, Leica) with a DFC350 camera. All cells were treated with Hoechst 33342 (Life Technologies) to visualize nuclei. When applicable, cells were fixed using 4% formaldehyde solution (Thermo Fisher) and permeabilized using 0.2% Triton X-100. For actin visualization in fixed cells, cells were stained using Oregon Green or Rhodamine-phalloidin (Thermo Fisher). Immunofluorescence against E-cadherin was performed by blocking with a 0.2% BSA (Sigma-Aldrich) solution in PBS for 20 min, followed by incubation with the primary mouse E-cadherin antibody (BD Biosciences, 610181) at a 1:100 dilution in 0.2% BSA for 1 h. The secondary antibody used was either donkey anti-mouse Alexa Fluorophore 647 (Thermo Fisher, A31571), or goat anti-mouse Alexa Fluor 488 (Thermo Fisher, R37120), at a 1:200 dilution in 0.2% BSA and incubated for 1 h. Confocal images were acquired on a Zeiss LSM700, with a 63×, 1.4 NA, oil immersion objective.
Image processing and SINK method
Live-cell time series images were acquired at 3 min time intervals for 1 h. Only the green (GFP-UBF) channel, and bright-field channel were acquired for this time to minimize phototoxicity. Bright-field images were obtained to assess viability of target, and adjacent cells throughout the length of imaging. Additionally, if gaps between cells in monolayers became apparent during imaging, these cells were not analyzed. Prior to time series imaging, single images were taken with all necessary channels for analysis (i.e. to confirm co-transfection, etc.). Additionally, images were taken 1 h after the hour-long time series was acquired to assure cells remained viable during and after imaging. After image acquisition, images were processed using custom MATLAB software, which is available from Ge Yang at Carnegie Mellon University (Yang et al., 2008). Briefly, nuclei were aligned to remove rigid body motion (Movies 1-4); such that only intranuclear motion of GFP-UBF punctate regions was captured. In order to ensure single unique points were tracked and to minimize artifacts of z-drift or particles moving in and out of focus in the z-direction, only particles which persisted throughout the entire movie were tracked and used for analysis. No changes in size of particles was observed during imaging. For the particles tracked for a given condition, the ensemble average MSD was calculated according to:
| (1) |
in which τ is the lag time, xt and yt are the x and y coordinates of the particle at a given time t, and xt+τ and yt+τ are the x and y coordinates of the particle after a given lag time τ. The ensemble average of the data for each condition was taken, after outliers were removed. Data in which the MSD was three standard deviations above the mean was considered an outlier. Outliers were removed to minimize the potential artifact in which a different punctate region was tracked between the beginning and end of imaging, and thus would obtain a significantly increased MSD. Outliers accounted for at most 6% of the total points tracked, as seen in Table S1. Curve fitting was performed using the MATLAB curve fitting toolbox (using the Trust-Region Algorithm), in which mean squared displacements were fitted to a power law of the form shown in Eqn 2, and the inset of Fig. 1E:
| (2) |
where MSD is the mean squared displacement, τ is the lag time, and the fitting parameters Deff and β are the effective diffusivity and diffusive exponent, respectively.
In previous work, we demonstrate that the parameters Deff and β appear to be modulated by independent phenomena (Spagnol and Noel Dahl, 2014). In live cell nuclei, the diffusive exponent β is altered by active forces. This was demonstrated by reducing myosin II activity with blebbistatin, in which a reduction in β was observed. Additionally, force propagation to the nucleus was reduced by disrupting the LINC complex using DN-KASH and again a decrease in β was observed. In cases in which chromatin was decondensed (via daunomycin or trichostatin A treatment), a decrease in Deff was observed with no significant change in β. In this work, we further confirm the force responsiveness of β by treating cells with Y-27632 (a ROCK inhibitor) or decoupling the LINC complex with DN-KASH. In both cases we note a decrease in the parameter β.
For heterogeneous DN-KASH experiments, the closest distance (edge to edge) between a DN-KASH-expressing nucleus and the nucleus of interest was taken to be the distance to a DN-KASH-expressing cell. These distances were measured for every cell of interest, and the data were binned into 10 µm intervals. Distances were measured using the Leica Application Suite Advanced Fluorescence software. For reference, in these systems we find that cells are on average 7.7±1.1 µm apart (schematically, average of a1, a2, etc. from Fig. 4D). Nuclear area was measured for cells in all conditions, and no significant difference between nuclear area for any condition was detected using a one-way ANOVA (P>0.05). Additionally, no correlation between nuclear area and β was seen (Fig. S4). All data showing number of cells, number of tracks, nuclear area, and β for each condition is presented in Table S1.
Polyacrylamide gel synthesis
Polyacrylamide (PA) gels were synthesized similar to previously published methods (Kraning-Rush et al., 2012; Lampi et al., 2016), with Sulfo-SANPAH used as the protein-substrate linker. Activated glass coverslips were coated with PA gels of varying elastic moduli, using acrylamide to bisacrylamide ratios of 5%:0.1%, 7.5%:0.35% and 12%:0.28% for a stiffness of 2.5, 10 and 30 kPa, respectively. Gels were then coated with Sulfo-SANPAH at 0.2 mg/ml (BioVision), and UV treated for 30 min, followed by coating with 0.1 mg/ml rat tail type 1 collagen (Corning) for cell adhesion. For comparison we also present the monolayer data on an uncoated glass substrate. While extracellular matrix coating probably influences cellular forces, we found it interesting that uncoated glass (orders of magnitude stiffer than 30 kPa) demonstrated the largest MSD within the nucleus, and β. While glass can be coated in collagen, the differences in surface chemistry between polyacrylamide and glass likely influence interfacial interactions with collagen, and the cells would, in turn, experience a different binding surface regardless. Nevertheless, the main finding presented for this section is the stiffness response of motion within the nucleus, which is clearly demonstrated with the different stiffness PA gels.
Statistics
Error bars shown on MSD versus lag time plots are s.e.m. of the population of tracks for a given experimental group. If a given MSD data point was greater than three standard deviations from the mean of that population at 60 min, the track was removed from analysis. Error bars on the bar graphs of β are the 95% confidence intervals based on the fit of this parameter. Values were considered significantly different if the 95% confidence intervals did not overlap.
Supplementary Material
Acknowledgements
We would like to acknowledge Ge Yang (Carnegie Mellon University) for providing the particle tracking software as well as Stephen Spagnol, Adam Feinberg, Rachelle Palchesko, Yu-Li Wang, Stephanie Wong-Noonan and David Li for their experimental insight.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.J.A., M.C.L., C.A.R., K.N.D.; Methodology: T.J.A., M.C.L., C.A.R., K.N.D.; Formal analysis: T.J.A., K.N.D.; Investigation: T.J.A., M.C.L.; Resources: T.J.A., C.A.R.; Data curation: T.J.A.; Writing - original draft: T.J.A.; Writing - review & editing: T.J.A., M.C.L., C.A.R., K.N.D.; Supervision: K.N.D.; Project administration: C.A.R.; Funding acquisition: T.J.A., M.C.L., C.A.R., K.N.D.
Funding
This work was funded by the National Science Foundation [NSF-CMMI-1634888 to K.N.D., NSF-CMMI-1435755 to C.A.R.-K., graduate research fellowship 201315546 to M.C.L.], the National Institutes of Health [NIH-EB003392 to T.J.A.], Cornell Sloan Foundation Fellowship and a Ford Foundation Fellowship to M.C.L. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.216010.supplemental
References
- Alam S. G., Lovett D., Kim D. I., Roux K. J., Dickinson R. B. and Lele T. P. (2015). The nucleus is an intracellular propagator of tensile forces in NIH 3T3 fibroblasts. J. Cell Sci. 128, 1901-1911. 10.1242/jcs.161703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsayed A. M., Islam M. F., Zhang J., Collings P. J. and Yodh A. G. (2005). Premelting at defects within bulk colloidal crystals. Science 309, 1207-1210. 10.1126/science.1112399 [DOI] [PubMed] [Google Scholar]
- Andresen Eguiluz R. C., Kaylan K. B., Underhill G. H. and Leckband D. E. (2017). Substrate stiffness and VE-cadherin mechano-transduction coordinate to regulate endothelial monolayer integrity. Biomaterials 140, 45-57. 10.1016/j.biomaterials.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armiger T. J., Spagnol S. T. and Dahl K. N. (2016). Nuclear mechanical resilience but not stiffness is modulated by αII-spectrin. J. Biomech. 49, 3983-3989. 10.1016/j.jbiomech.2016.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry A. K., Wang N. and Leckband D. E. (2015). Local VE-cadherin mechanotransduction triggers long-ranged remodeling of endothelial monolayers. J. Cell Sci. 128, 1341-1351. 10.1242/jcs.159954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth-Gauthier E. A., Alcoser T. A., Yang G. and Dahl K. N. (2012). Force-induced changes in subnuclear movement and rheology. Biophys. J. 103, 2423-2431. 10.1016/j.bpj.2012.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano J. and Reinhart-King C. (2010). Substrate stiffness and cell area predict cellular traction stresses in single cells and cells in contact. Cell Mol. Bioeng. 3, 68-75. 10.1007/s12195-010-0102-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanet S., Miller C. J., Vaishnav E. D., Ermentrout B., Davidson L. A. and Martin A. C. (2017). Actomyosin meshwork mechanosensing enables tissue shape to orient cell force. Nat. Commun. 8, 15014 10.1038/ncomms15014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D. E., Janmey P. and Wang Y.-L. (2005). Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139-1143. 10.1126/science.1116995 [DOI] [PubMed] [Google Scholar]
- Ghibaudo M., Saez A., Trichet L., Xayaphoummine A., Browaeys J., Silberzan P., Buguin A. and Ladoux B. (2008). Traction forces and rigidity sensing regulate cell functions. Soft Mat. 4, 1836 10.1039/b804103b [DOI] [Google Scholar]
- Guo M., Ehrlicher A. J., Jensen M. H., Renz M., Moore J. R., Goldman R. D., Lippincott-Schwartz J., Mackintosh F. C. and Weitz D. A. (2014). Probing the stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy. Cell 158, 822-832. 10.1016/j.cell.2014.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. J., Bielawski K. S., Ting L. H., Rodriguez M. L. and Sniadecki N. J. (2012). Decoupling substrate stiffness, spread area, and micropost density: a close spatial relationship between traction forces and focal adhesions. Biophys. J. 103, 640-648. 10.1016/j.bpj.2012.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassianidou E. and Kumar S. (2015). A biomechanical perspective on stress fiber structure and function. Biochim. Biophys. Acta - Mol. Cell Res. 1853, 3065-3074. 10.1016/j.bbamcr.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraning-Rush C. M., Carey S. P., Califano J. P. and Reinhart-King C. A. (2012). Quantifying traction stresses in adherent cells. In Methods in Cell Biology (ed. Asthagiri A. R. and Arkin A. P.), pp. 139-178. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Krishnan R., Klumpers D. D., Park C. Y., Rajendran K., Trepat X., van Bezu J., van Hinsbergh V. W. M., Carman C. V., Brain J. D., Fredberg J. J. et al. (2011). Substrate stiffening promotes endothelial monolayer disruption through enhanced physical forces. Am. J. Physiol. Cell Physiol. 300, C146-C154. 10.1152/ajpcell.00195.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoux B. and Mège R.-M. (2017). Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 18, 743-757. 10.1038/nrm.2017.98 [DOI] [PubMed] [Google Scholar]
- Lampi M. C., Faber C. J., Huynh J., Bordeleau F., Zanotelli M. R. and Reinhart-King C. A. (2016). Simvastatin ameliorates matrix stiffness-mediated endothelial monolayer disruption. PLoS One 11, e0147033 10.1371/journal.pone.0147033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky R. (1986). Melting at grain boundaries and surfaces. Phys. Rev. Lett. 57, 2876 10.1103/PhysRevLett.57.2876 [DOI] [PubMed] [Google Scholar]
- Lodish H., Berk A., Zipursky S. L., Matsudaira P., Balimore D. and Darnell J. (2000). Tumor cells and the onset of cancer. In Molecular Cell Biology. 4th edn. New York: W. H. Freeman. [Google Scholar]
- Luxton G. W. G., Gomes E. R., Folker E. S., Vintinner E. and Gundersen G. G. (2010). Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329, 956-959. 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta V. and Tzima E. (2016). Cardiovascular disease: a turbulent path to plaque formation. Nature 540, 531-532. 10.1038/nature20489 [DOI] [PubMed] [Google Scholar]
- Neelam S., Hayes P. R., Zhang Q., Dickinson R. B. and Lele T. P. (2016). Vertical uniformity of cells and nuclei in epithelial monolayers. Sci. Rep. 6, 19689 10.1038/srep19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M. R., Besser A., Danuser G. and Brugge J. S. (2012). Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J. Cell Biol. 199, 545-563. 10.1083/jcb.201207148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck W. J. and Chen C. S. (2016). Measuring cell-generated forces: a guide to the available tools. Nat. Methods 13, 415-423. 10.1038/nmeth.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape A. D., Guo W. H. and Wang Y.-L. (2011). The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials 32, 2043-2051. 10.1016/j.biomaterials.2010.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki M. S., Cirka H. A., Aghvami M., Sander E. A., Wen Q. and Billiar K. L. (2013). Nonlinear strain stiffening is not sufficient to explain how far cells can feel on fibrous protein gels. Biophys. J. 105, 11-20. 10.1016/j.bpj.2013.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saw T. B., Doostmohammadi A., Nier V., Kocgozlu L., Thampi S., Toyama Y., Marcq P., Lim C. T., Yeomans J. M. and Ladoux B. (2017). Topological defects in epithelia govern cell death and extrusion. Nature 544, 212-216. 10.1038/nature21718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim J. Y., Moeller J., Hart K. C., Ramallo D., Vogel V., Dunn A. R., Nelson W. J. and Pruitt B. L. (2015). Spatial distribution of cell-cell and cell-ECM adhesions regulates force balance while main taining E-cadherin molecular tension in cell pairs. Mol. Biol. Cell 26, 2456-2465. 10.1091/mbc.e14-12-1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnol S. T. and Dahl K. N. (2014). Active cytoskeletal force and chromatin condensation independently modulate intranuclear network fluctuations. Integr. Biol. 6, 523-531. 10.1039/C3IB40226F [DOI] [PubMed] [Google Scholar]
- Sunyer R., Conte V., Escribano J., Elosegui-Artola A., Labernadie A., Valon L., Navajas D., Garcia-Aznar J. M., Munoz J., Roca-Cusachs P. et al. (2016). Collective cell durotaxis emerges from long-range intercellular force transmission. Science 353, 1157-1161. 10.1126/science.aaf7119 [DOI] [PubMed] [Google Scholar]
- Tajik A., Zhang Y., Wei F., Sun J., Jia Q., Zhou W., Singh R., Khanna N., Belmont A. S. and Wang N. (2016). Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 15, 1287-1296. 10.1038/nmat4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambe D. T., Corey Hardin C., Angelini T. E., Rajendran K., Park C. Y., Serra-Picamal X., Zhou E. H., Zaman M. H., Butler J. P., Weitz D. A. et al. (2011). Collective cell guidance by cooperative intercellular forces. Nat. Mater. 10, 469-475. 10.1038/nmat3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks E. R., Crocker J. C., Levitt A. C., Schofield A. and Weitz D. A. (2000). Three-dimensional direct imaging of structural relaxation near the colloidal glass transition. Science 287, 627-631. 10.1126/science.287.5453.627 [DOI] [PubMed] [Google Scholar]
- Yang G., Cameron L. A., Maddox P. S., Salmon E. D. and Danuser G. (2008). Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J. Cell Biol. 182, 631-639. 10.1083/jcb.200801105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.