ABSTRACT
Facial form depends on the precise positioning of cartilage, bone, and tooth fields in the embryonic pharyngeal arches. How complex signaling information is integrated to specify these cell types remains a mystery. We find that modular expression of Forkhead domain transcription factors (Fox proteins) in the zebrafish face arises through integration of Hh, Fgf, Bmp, Edn1 and Jagged-Notch pathways. Whereas loss of C-class Fox proteins results in reduced upper facial cartilages, loss of F-class Fox proteins results in distal jaw truncations and absent midline cartilages and teeth. We show that Fox proteins are required for Sox9a to promote chondrogenic gene expression. Fox proteins are sufficient in neural crest-derived cells for cartilage development, and neural crest-specific misexpression of Fox proteins expands the cartilage domain but inhibits bone. These results support a modular role for Fox proteins in establishing the competency of progenitors to form cartilage and teeth in the face.
KEY WORDS: Forkhead, Foxc1, Foxf1, Foxf2, Sox9, Cartilage, Teeth, Bone, Pharyngeal arches, Craniofacial, Zebrafish
Highlighted Article: Combinatorial mutagenesis studies reveal that a large family of Forkhead transcription factors integrate diverse signaling outputs to specify distinct cartilages and teeth in the developing zebrafish face.
INTRODUCTION
Development of the vertebrate head skeleton involves the specification of cartilage, bone, tooth and other connective tissue fates in reproducible, interconnected positions. The progenitors for these skeletal cells derive from the head paraxial mesoderm and a specialized population of cranial neural crest-derived cells (CNCs). In the pharyngeal arches, CNCs are exposed to diverse extracellular signals that regulate skeletal differentiation. Endothelin 1 (Edn1), Bone morphogenetic protein (Bmp) and Jagged-Notch signaling interact to generate nested gene expression patterns along the dorsoventral/proximodistal arch axis, which help direct particular morphologies of cartilages and bones in the upper versus lower face (Zuniga et al., 2010, 2011; Alexander et al., 2011). In contrast, Hedgehog (Hh) and Fibroblast growth factor (Fgf) signaling are largely dispensable for dorsoventral gene expression, instead playing crucial roles in CNC proliferation, survival and differentiation (Balczerski et al., 2012). Given that many of the target genes for these pathways are expressed in broader domains than the precursor regions for individual skeletal elements (Medeiros and Crump, 2012), a central mystery is how complex signaling information is integrated to specify individual cartilages, bones and teeth.
Fox genes are a large family of transcription factors that share a highly conserved winged-helix/forkhead DNA-binding domain yet little conservation otherwise (Hong et al., 1999). During murine craniofacial development, CNC expression of Foxc1, Foxc2, Foxd1, Foxd2, Foxf1 and Foxf2 is positively regulated by Hh signaling, with their distinct arch expression domains leading to the proposal that Fox genes might constitute a code for facial skeletal development (Yamagishi et al., 2003; Jeong et al., 2004). In support of this, Foxc1 mutant mice display loss of the calvarium and increased ossification of the mandible, squamosal and zygomatic bones, leading to jaw fusion (syngnathia) (Hong et al., 1999; Kume et al., 2001; Inman et al., 2013); Foxc2 mutant mice develop cleft palate, defective pterygoquadrate cartilage development, and fusions of the middle ear bones (Iida et al., 1997); and Foxf1 and Foxf2 mutants display cleft palate (Wang et al., 2003; Xu et al., 2016). In humans, mutations in FOXC1 have been linked to the craniofacial and ocular defects of Axenfeld–Rieger syndrome (Mears et al., 1998; Fang et al., 2000). Foxf1 and Foxf2 are also prominently expressed in developing tooth buds (Wang et al., 2003), though a genetic requirement has yet to be identified. Whether Fox proteins broadly control regional facial skeletal fates, as well as their mechanisms of action, remains unclear.
Although examination of Fox-C and Fox-F genes in the mammalian face point to requirements in bone and palate formation, clues to potential mechanisms come from analysis of their roles in the appendicular and axial skeletons. The spontaneous congenital hydrocephalus mouse mutant carries a point mutation in Foxc1 and has defects in sternal chondrogenesis. In particular, cultured mesenchymal cells from mutants fail to differentiate into cartilage, even upon exposure to BMP2 (Kume et al., 1998). Foxc1 mutants also display delayed endochondral ossification of the femur, in part owing to normal binding of a Foxc1-Gli2 complex to promoters of the chondrocyte genes Col10a1 and Pthrp (Pthlh – Mouse Genome Informatics) (Yoshida et al., 2015). Sox9 is an essential regulator of cartilage differentiation, with loss of Sox9 in mouse or sox9a in zebrafish resulting in a near-complete absence of cartilage (Bi et al., 1999; Akiyama et al., 2002; Mori-Akiyama et al., 2003; Yan et al., 2005). However, Sox9 also has many roles in non-cartilage tissues (Jo et al., 2014), raising the question of how activity in chondrocytes is specifically regulated. Intriguingly, the identification of Fox consensus binding motifs as highly enriched in Sox9-bound enhancers of chondrocyte genes suggests that Fox proteins might cooperate with Sox9 at chondrocyte enhancers (Ohba et al., 2015).
Here, we employ the zebrafish system to test the roles of discrete Fox genes in regulating cartilage, bone and tooth development in the embryonic face. We find that the distinct expression patterns of individual Fox genes arise through integration of Hh, Bmp, Fgf, Edn1 and Notch signaling. Combinatorial mutagenesis reveals requirements for Fox-C genes in formation of cartilage in the upper face, and for Fox-F genes in formation of the distal jaws, ventral midline cartilages, and pharyngeal teeth. These Fox genes appear to be largely dispensable for dorsoventral patterning, as well as CNC proliferation and survival. Instead, we find that Fox-C and Fox-F genes are required for Sox9a to activate effectively a number of known targets in chondrocytes, including col2a1a, matrilins 1 and 4, and acana. Rescue experiments also show that Foxc1a is sufficient in CNCs for facial cartilage formation. Further, CNC-specific misexpression of either Foxc1a or Foxf1 expands the chondrogenic domain and inhibits dermal bone formation. We propose that Fox genes function to integrate diverse signaling outputs to position cartilage and tooth competency fields precisely in the developing face.
RESULTS
Establishment of distinct arch expression domains of Fox genes
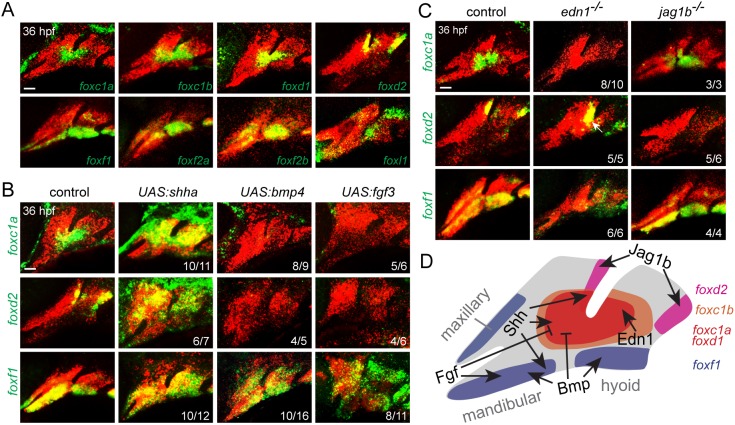
Examination of our published transcriptome data of arch CNCs (Askary et al., 2017) revealed the top eight expressed Fox genes to be foxc1b, foxd1, foxd2, foxc1a, foxf2a, foxl1, foxf1 and foxf2b [from highest to lowest, all above eight transcripts per million reads at 36 h post-fertilization (hpf); Fig. S1A]. Fluorescence in situ hybridization revealed distinct expression domains in arch CNCs (Fig. 1A). foxc1a, foxc1b and foxd1 were enriched in intermediate domains of the first and second arches, with strongest expression around the ventral limit of the first pharyngeal pouch, whereas foxc1b was expressed in broader domains than foxc1a and foxd1 (Fig. S1D). Although foxd2 showed similar expression at an earlier stage (30 hpf; Fig. S1B), by 36 hpf it became restricted to CNCs in the dorsal-posterior regions of the first and second arches. In contrast, foxf1, foxf2a and foxf2b were expressed in more ventral regions of the first and second arches, as well as in the maxillary domain. Expression of foxl1 was observed in the dorsal and ventral poles of the first two arches. In the more posterior gill-forming arches, we detected expression of foxf1 in ventral domains at 36 and 48 hpf, foxc1b in intermediate domains at 48 hpf, and foxd2 in dorsal domains at 48 hpf (Fig. S1C,E). We also observed exclusion of foxc1a, foxc1b, foxd2 and foxf1 from developing sox9a+ chondrocytes at 48 hpf, with the exception of foxc1a and foxc1b in some nascent sox9a+ cells in the hyoid joint region (Fig. S1E,F). Furthermore, foxc1b and foxf1 were expressed in distinct domains in and around the developing neurocranium, a structure analogous to the mammalian palate, and foxf1 was expressed in tooth-forming mesenchyme of the fifth branchial arch (Fig. S1G,H). These patterns are reminiscent of the expression of their mouse orthologs at embryonic day 10.5, such as the reported ‘first pouch adjacent’ expression of Foxc2, Foxd1 and Foxd2 and the mandibular and maxillary expression of Foxf1 and Foxf2 (Jeong et al., 2004).
Fig. 1.
Combinatorial signaling establishes distinct Fox expression domains. (A) In situ hybridization at 36 hpf shows expression of Fox genes (green) relative to dlx2a+ CNCs (red) of the first two arches. (B) Expression of foxc1a, foxd2 and foxf1 (green) relative to dlx2a+ CNCs (red) upon misexpression of Shha, Bmp4 or Fgf3. Embryos doubly transgenic for hsp70l:Gal4 and UAS:shha, UAS:bmp4 or UAS:fgf3 were subjected to heat shock induction from 20 to 24 hpf and then fixed at 36 hpf for analysis. (C) Expression of foxc1a, foxd2 and foxf1 (green) relative to dlx2a+ CNCs (red) in edn1 or jag1b mutants versus sibling controls. Numbers denote proportions of embryos with displayed patterns. Scale bars: 25 μm. (D) Summary of positive and negative inputs on Fox expression in the first two arches.
We next interrogated how these distinct expression patterns arise. As described for Foxc2, Foxd1, Foxd2, Foxf1 and Foxf2 in mouse (Jeong et al., 2004), we found that upregulation of Hh signaling, through heat shock-regulated misexpression of Shha starting at 20 hpf, increased expression of foxc1a, foxc1b, foxd1, foxd2 and foxf1 throughout most of the arches (Fig. 1B, Fig. S2). However, different effects of manipulating Bmp, Fgf, Edn1 and Jagged-Notch signaling were observed on individual Fox genes (Fig. 1B,C, Fig. S2). Misexpression of Bmp4 or Fgf3 reduced foxc1a, foxc1b, foxd1 and foxd2 yet expanded foxf1 expression. In edn1 mutants, foxc1a, foxc1b and foxd1 expression was reduced, foxf1 became diffusely expressed throughout the second arch but reduced in the first arch, and foxd2 was modestly expanded into more ventral regions. Consistent with similar expression of jag1b and foxd2 (Zuniga et al., 2010), only expression of foxd2 was lost in jag1b mutants. These results indicate combinatorial regulation of Fox expression. Shh induces all Fox genes examined; foxc1a, foxc1b and foxd1 are restricted to intermediate regions by positive Edn1 and negative Bmp and Fgf signaling; foxd2 becomes restricted to dorsal-posterior regions by positive Jagged-Notch and negative Bmp and Fgf signaling; and foxf1 is restricted to distal/ventral regions largely through positive Bmp and Fgf signaling (Fig. 1D).
Dose-dependent requirements for Fox-C genes in upper facial cartilage formation
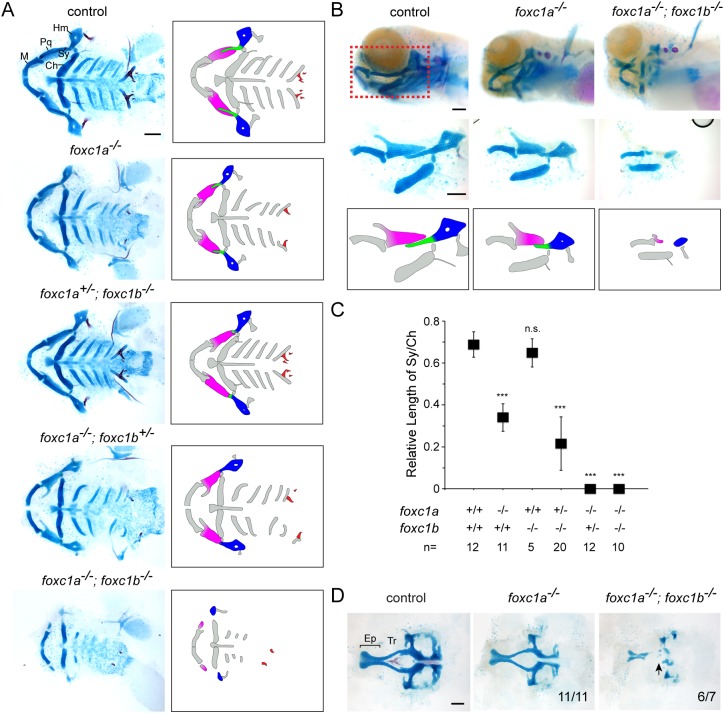
We next used TALEN or CRISPR/Cas9 mutagenesis to create frame-shift alleles for foxc1a, foxc1b, foxd1, foxd2, foxf1, foxf2a, foxf2b and foxl2, in addition to obtaining a foxl1 nonsense allele from the Sanger Center. In all cases, mutations are predicted to cause protein truncations in or before the conserved Forkhead DNA-binding domains, thus likely abrogating most, if not all, function (Fig. S3A). Homozygous mutants for foxc1b, foxd1 and foxd2 displayed no obvious larval craniofacial skeletal defects and survived past juvenile stages, with only foxd2 mutants exhibiting a growth delay by 2 months. The larval craniofacial skeleton was also unaffected in foxd1−/−; foxd2−/− double mutants (Fig. S3B). In contrast, homozygous foxc1a mutants died by 7 days post-fertilization (dpf), and displayed facial cartilage defects, as well as cardiac edema that likely accounts for the overall reduced head growth (Fig. 2). After normalizing for overall cartilage/head reduction, we found that mutants had a particularly short symplectic (Sy) cartilage, an element derived from the intermediate portion of the second arch. Sy reduction was not secondary to cardiac edema, as foxc1a+/−; foxc1b−/− animals displayed a similar Sy truncation without cardiac edema, and heterozygosity of foxc1b enhanced Sy loss in foxc1a−/− animals (Fig. 2A-C). In double homozygous foxc1a; foxc1b mutants, we observed severe reductions of upper facial cartilages, including not only the Sy but also the palatoquadrate (Pq, a dorsal-intermediate first arch cartilage) and hyomandibula (Hm, a dorsal second arch cartilage). We also observed missing or reduced trabecular cartilage of the neurocranium (Fig. 2D). However, the lower jaw Meckel's (M) and lower jaw support ceratohyal (Ch) cartilages were relatively unaffected. Photoconversion of sox10:kikGR+ arch CNCs in a similar domain to where foxc1a, foxc1b and foxd1 are co-expressed at 36 hpf resulted in labeling of the Sy, Pq and Hm cartilages by 6 dpf, i.e. those elements most affected in mutants (Fig. S4). Our results support the hypothesis that Foxc1a and Foxc1b act redundantly, and possibly cell-autonomously, in upper facial cartilage development.
Fig. 2.
Loss of upper facial cartilage in Fox-C mutants. (A) Ventral views of dissected facial skeletons, with cartilage in blue and bones and teeth in red. Schematics show affected elements in color. Ch, ceratohyal; Hm, hyomandibular; M, Meckel's; Pq, palatoquadrate; Sy, symplectic. (B) Lateral views of intact heads stained with Alcian Blue (cartilage) and Alizarin Red (bones and teeth). Shown below are unilateral dissections of skeletal elements of the first and second arches (red boxed region). Accompanying schematics show dose-dependent reductions of the Pq (magenta), Sy (green) and Hm (blue) cartilages. foxc1a−/− and foxc1a−/−; foxc1b−/− mutants also display cardiac edema and an overall smaller head. (C) Quantification of the relative length of Sy compared with Ch. ***P<0.001 versus wild-type sibling control using Student's t-test. n.s., not significant versus control (P=0.35). Error bars represent s.e.m. (D) Dissections of neurocranial cartilages show loss of the trabecular cartilages (Tr, arrow) in foxc1a−/−; foxc1b−/− mutants. Numbers denote proportions of embryos with displayed phenotypes. Ep, ethmoid plate. Scale bars: 25 μm.
Dose-dependent requirement for Fox-F genes in midline cartilage and pharyngeal tooth formation
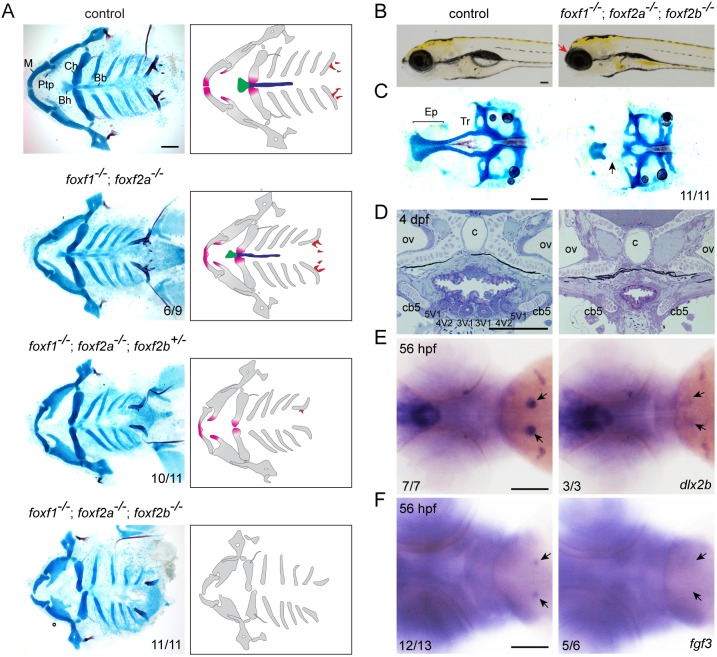
We next examined potential roles for Fox-F and Fox-L genes in facial development. Single mutants for foxf2a, foxf2b, foxl1 and foxl2, and double mutants for foxf2a; foxf2b and foxl1; foxl2, displayed no obvious craniofacial defects and survived to adulthood (Fig. S3B,C). Single foxf1 mutants also had normal larval facial skeletons yet did not survive past 10 dpf, displaying a failure of swim bladder inflation and a reduced intestine (Fig. S5), consistent with roles for Foxf1 in intestinal development in frog (Tseng et al., 2004) and mouse (Ormestad et al., 2006). In contrast, foxf1; foxf2a double mutants displayed mild midline cartilage defects, which were further enhanced by loss of fox2b alleles. In foxf1; foxf2a double mutants with at least one foxf2b allele disrupted, we observed distal truncations of the upper jaw (pterygoid process), lower jaw (M) and lower jaw support (Ch) cartilages; complete loss of midline cartilages (basihyal and basibranchials); and absence of pharyngeal teeth. We also observed absence of the trabecular cartilage of the neurocranium in foxf1; foxf2a; foxf2b triple mutants (Fig. 3A-C). Histological analysis revealed a complete absence of tooth buds in triple mutants, and in situ hybridization revealed losses of the early tooth markers dlx2b and fgf3 at 56 hpf (Fig. 3D-F). We conclude that Fox-F genes have dose-dependent requirements for the formation of distal jaw, midline and neurocranial cartilages, as well as teeth, consistent with their expression in the distal-most regions of each pharyngeal arch and the tooth-bearing seventh arch.
Fig. 3.
Loss of midline cartilage and pharyngeal teeth in Fox-F mutants. (A) Ventral views of dissected facial skeletons, with cartilage in blue and bones and teeth in red. Schematics show affected elements in color. Bb, basibranchials; Bh, basihyal; Ch, ceratohyal; M, Meckel's; Ptp, pterygoid process. (B) Lateral views of wild-type sibling control and foxf1; foxf2a; foxf2b triple mutant at 5 dpf. Mutants show anterior truncation of the head (arrow). (C) Dissections of neurocranial cartilages show loss of the trabecular cartilages (Tr, arrow) in foxf1; foxf2a; foxf2b triple mutants. Ep, ethmoid plate. (D) Sagittal sections through the tooth-forming seventh arch of a wild-type sibling control and foxf1; foxf2a; foxf2b triple mutant at 4 dpf. All teeth (3V1, 4V2 and 5V1) are missing in mutants. c, chorda; cb5, ceratobranchial 5; ov, otic vesicle. (E,F) Whole-mount in situ hybridizations show loss of dlx2b (E) and fgf3 (F) in the developing tooth regions (arrows) of foxf1; foxf2a; foxf2b triple mutants. Numbers denote proportions of embryos with displayed phenotypes or expression patterns. Scale bars: 25 μm.
Fox genes are largely dispensable for the dorsoventral patterning, proliferation and survival of arch CNCs
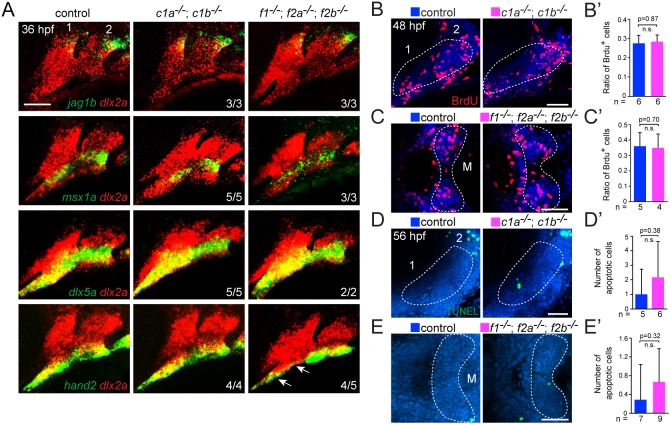
Given the regional cartilage defects of Fox-C and Fox-F mutants, we examined whether this might reflect earlier defects in the dorsoventral patterning, growth or survival of CNCs. For example, Sy defects in Fox-C mutants are similar to those seen upon loss of the dorsoventrally intermediate Dlx3-6 genes (Talbot et al., 2010), and the distal jaw truncations of Fox-F mutants are reminiscent of those seen upon loss of the ventral hand2 gene (Miller et al., 2003). However, expression of dorsal (jag1b), intermediate (dlx5a and msx1a) and ventral (hand2) genes were largely unaffected in Fox-C and Fox-F mutants, although we did observe a reduction of the hand2+ ventral first arch domain in Fox-F mutants (Fig. 4A). Examination of bromodeoxyuridine (BrdU) incorporation (48 hpf) and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining (56 hpf) also revealed no significant differences in proliferation or apoptosis, respectively, in Fox-C and Fox-F mutants (Fig. 4B-E′). These results indicate that the cartilage defects of Fox mutants are unlikely to be due to altered dorsoventral patterning, growth, or survival of CNCs, at least at the stages examined.
Fig. 4.
Dorsoventral gene expression, proliferation and apoptosis in Fox mutants. (A) Fluorescence in situ hybridizations of the first two arches (numbered) show expression of jag1b (dorsal), msx1a and dlx5a (intermediate), or hand2 (ventral) in green relative to dlx2a+ arch CNCs in red. Expression is largely unaffected in foxc1a; foxc1b double mutants (c1a−/−; c1b−/−) and foxf1; foxf2a; foxf2b triple mutants (f1−/−; f2a−/−; f2b−/−) compared with sibling controls, with the exception of reduced hand2 in the ventral first arch of foxf1; foxf2a; foxf2b triple mutants (arrows). (B-C′) Analysis of proliferation. Confocal sections show BrdU+ cells (red) compared with all nuclei (Hoechst, blue) in lateral views of the first two arches (numbered) of foxc1a−/−; foxc1b−/− embryos and ventral views of the first arch Meckel's cartilage domains (M) of foxf1−/−; foxf2a−/−; foxf2b−/− embryos, relative to sibling controls. Dashed lines represent the regions quantified in the accompanying graphs. (D-E′) Analysis of apoptosis. Maximum intensity projections show the number of TUNEL+ cells (green) in lateral views of the first two arches (numbered) of foxc1a−/−; foxc1b−/− embryos and ventral views of the first arch M cartilage domains of foxf1−/−; foxf2a−/−; foxf2b−/− embryos, relative to sibling controls. Dashed lines represent the regions quantified in the accompanying graphs. Error bars represent s.e.m. Scale bars: 50 μm. n.s., not significant.
Altered cartilage gene expression downstream of Sox9a in Fox mutants
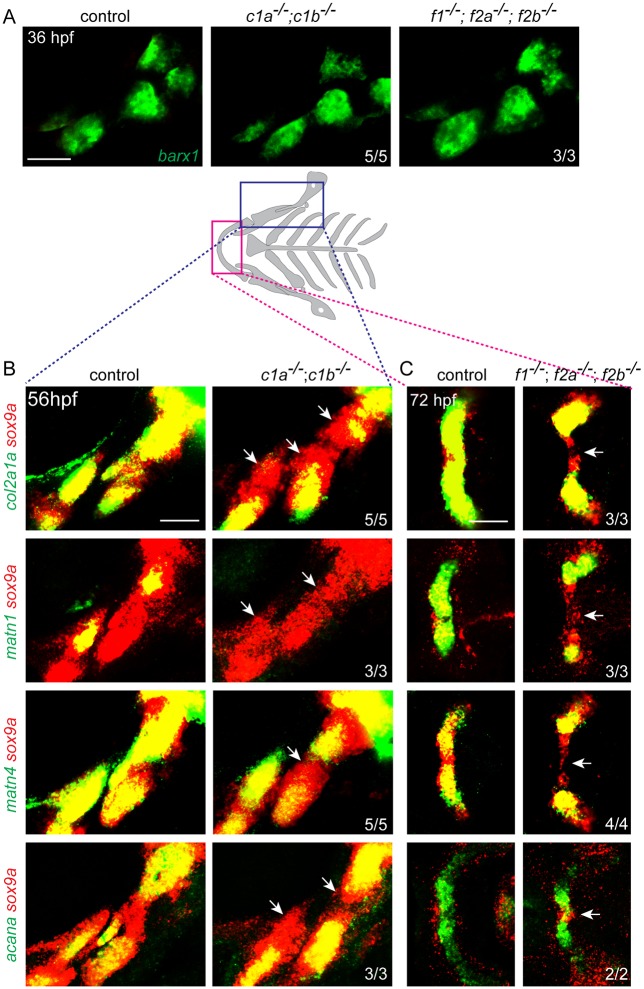
We next examined whether Fox genes might control distinct steps in cartilage differentiation. During normal zebrafish craniofacial development, barx1 and then sox9a mark CNCs in pre-cartilage condensations (Nichols et al., 2013; Barske et al., 2016). Sox9a then induces a number of cartilage differentiation genes, including those encoding for the extracellular matrix components Type II Collagen (col2a1a), Aggrecan (acana) and Matrilin (matn1 and matn4). In foxc1a; foxc1b and foxf1; foxf2a; foxf2b mutants, the initiation of barx1 and sox9a expression in the developing arches was largely unaffected (Fig. 5A-C). Reciprocally, foxc1a was expressed normally in the arches of sox9a mutants (Fig. S6A). However, expression of the Sox9 target genes col2a1a, matn1 and matn4, and to a lesser extent acana, was specifically reduced in the nascent sox9a+ upper facial cartilages affected in foxc1a; foxc1b mutants (Fig. 5B). The exact nature of col2a1a and matn4 reduction varied between affected animals. Similarly, expression of col2a1a, matn1, matn4 and acana was specifically reduced in the nascent sox9a+ ventral midline cartilages affected in foxf1; foxf2a; foxf2b mutants, as shown for the lower jaw Meckel's cartilages (Fig. 5C). We observed a similar upper face-specific loss of a col2a1:mCherry-NTR transgene in foxc1a; foxc1b mutants; in contrast, expression of a sp7:EGFP transgene that labels osteoblasts of early-forming dermal bones was unaffected (Fig. S6B). These data show that Fox-C and Fox-F factors are required for the expression of Sox9a target genes, but not sox9a itself, in distinct cartilage-forming domains of the developing face.
Fig. 5.
Fox genes are required for chondrogenic gene expression downstream of Sox9a. (A) Fluorescence in situ hybridizations show that barx1+ pre-cartilage condensations of the first two arches are unaltered in foxc1a; foxc1b double mutants (c1a−/−; c1b−/−) and foxf1; foxf2a; foxf2b triple mutants (f1−/−; f2a−/−; f2b−/−) compared with sibling controls. (B,C) Schematic of the cartilaginous facial skeleton shows regions examined by fluorescence in situ hybridization. Maximum intensity projections for foxc1a; foxc1b mutants at 56 hpf (B) and representative sections for foxf1; foxf2a; foxf2b mutants at 72 hpf (C) show expression of the chondrogenic genes col2a1a, matn1, matn4 or acana (green) relative to sox9a (red). Arrows indicate regions in which sox9a+ cells have reduced chondrogenic gene expression relative to sibling controls. Numbers denote proportions of embryos with displayed patterns. Scale bars: 50 μm.
CNC misexpression of Fox genes expands facial cartilage and reduces dermal bone
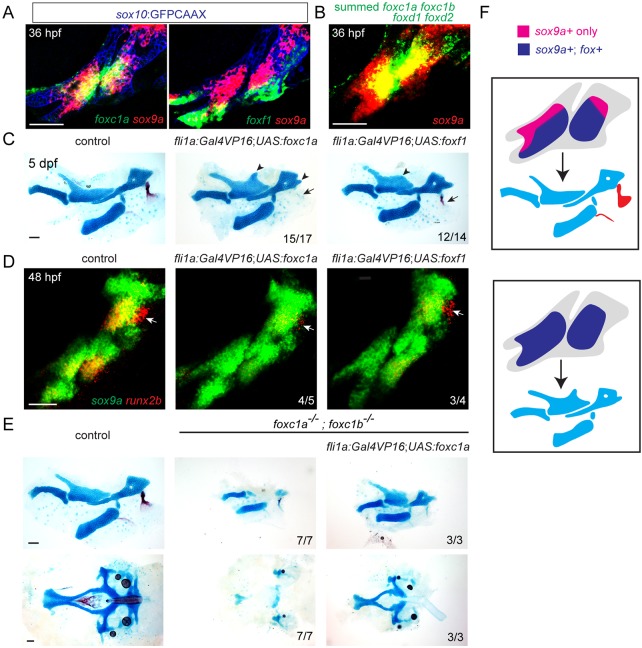
At 36 hpf, we found that sox9a is expressed in a broader domain in the first two arches than foxc1a and foxf1 (Fig. 6A). In particular, pooling of foxc1a, foxc1b, foxd1 and foxd2 probes revealed dorsal sox9a+ domains that do not appear to co-express any of the Fox genes examined (Fig. 6B,F). We therefore examined whether forced expression of Fox genes could expand cartilage formation into these sox9a-only domains. To do so, we used a fli1a:Gal4VP16 driver to misexpress UAS-driven Foxc1a or Foxf1 transgenes throughout arch CNC mesenchyme. Misexpression of either Foxc1a or Foxf1 resulted in ectopic cartilage processes dorsal to the Pq and posterior to the Hm, and loss of the opercle, an early-forming dermal bone (Fig. 6C). Consistent with sox9a expression being unaffected in Fox-C and Fox-F mutants, we did not observe an expansion of sox9a expression upon Foxc1a or Foxf1 misexpression. In addition, loss of dermal bone was reflected by earlier loss of the osteogenic gene runx2b (Fig. 6D). CNC misexpression of Foxc1a was also able to rescue the facial and neurocranial cartilage defects of foxc1a; foxc1b mutants; however, cardiac edema was not rescued, consistent with the heart being derived from mesoderm (Fig. 6E). Thus, Foxc1a function is sufficient in CNCs for facial cartilage formation. Our findings are consistent with Fox misexpression expanding the regions in which Sox9a can induce cartilage, potentially at the expense of dermal bone (Fig. 6F).
Fig. 6.
Fox CNC misexpression induces ectopic cartilage and inhibits dermal bone. (A) Confocal sections show expression of foxc1a or foxf1 (green) relative to sox9a (red) in sox10:GFPCAAX+ CNCs of the first two arches (anti-GFP, blue). (B) Maximum intensity projection shows fluorescence in situ hybridization using pooled probes against foxc1a, foxc1b, foxd1 and foxd2 (green) relative to sox9a (red). (C) Unilateral dissections of the first and second arch skeletons stained for cartilage (Alcian Blue) and bone (Alizarin Red). CNC misexpression of Foxc1a (fli1a:Gal4VP16; UAS:foxc1a) or Foxf1 (fli1a:Gal4VP16; UAS:foxf1) results in ectopic cartilage in the upper face (arrowheads), and loss of the opercle bone (arrows). (D) Double fluorescence in situ hybridizations of the first two arches show reduction of the osteoblast gene runx2b (red) in the future opercle bone (arrow) but no change in sox9a (green) upon misexpression of Foxc1a or Foxf1. (E) Dissections of the first and second arch skeletons and neurocranial cartilages. Misexpression of Foxc1a in CNCs (fli1a:Gal4VP16; UAS:foxc1a) rescues cartilage development in the upper face (top) and neurocranium (bottom). Scale bars: 50 μm. (F) Model of how arch-wide Fox misexpression expands the territory in which Sox9a induces cartilage (blue) at the expense of dermal bone (red).
DISCUSSION
The region-specific cartilage and tooth defects seen in fish lacking distinct combinations of Fox-C/F genes support the concept of a Fox code of facial skeletal patterning first proposed by Jeong et al. (2004). Whereas Edn1 and Jagged-Notch signaling interact to regulate sox9a expression along the dorsoventral arch axis (Barske et al., 2016), sox9a expression is not dependent on Fox-C/F genes. Instead, our work reveals that Fox-C/F factors are locally required for Sox9a to promote cartilage differentiation. We find that at least five signaling pathways – Hh, Fgf, Bmp, Edn1 and Jagged-Notch – interact to establish distinct expression domains of individual Fox genes. Diverse signaling pathways thus interact to position Fox and Sox expression domains independently, with the unique overlap of these two types of transcription factors positioning individual cartilage elements in the developing face.
Although we observed local reduction or loss of facial cartilage in Fox-C and Fox-F mutants, other cartilage elements (e.g. ceratohyal) were largely unaffected in both. Although Foxc1 has been implicated in cartilage development in the limb and sternum of mouse (Kume et al., 1998; Yoshida et al., 2015), we also did not observe any defects in pectoral fin cartilage in fish Fox-C or Fox-F mutants (Fig. S7). Furthermore, expression of Sox9a target genes, such as col2a1a and acana, was only partially reduced in Fox-C/F mutants. One explanation for the partial nature of defects is additional redundancy with other members of the Fox family. Indeed, we observed expression of a large number of Fox genes in the developing face, and our analysis of Fox-C and Fox-F genes revealed widespread functional redundancy. It may also be that Fox factors function to ensure maximal activation of Sox9 target genes, with Sox9 able to drive some level of cartilage gene expression even in the absence of Fox genes. Finally, particular cartilages may have a larger dependence on Fox genes for their differentiation.
The finding of upper facial cartilage defects in Fox-C zebrafish mutants, and repression of dermal bone by Foxc1a misexpression, is consistent with reported roles of Fox-C genes in mice. Foxc2 mutant mice display reductions of the pterygoquadrate cartilage and fusion of the malleus and incus cartilages (Iida et al., 1997), with these elements deriving from similar upper/dorsal regions of the mandibular arch as in fish. Foxc1 also has a role in repressing bone differentiation in mice. Foxc1 mutant mice display increased dermal bone that fuses the jaws (Hong et al., 1999; Kume et al., 2001; Inman et al., 2013) and precocious differentiation of calvarial osteoblasts, ultimately leading to reduced skull bones (Hong et al., 1999; Sun et al., 2013). However, although we observed repression of bone development by Foxc1a or Foxf1 misexpression in zebrafish, we failed to observe ectopic bone in any of the Fox mutant combinations analyzed. The neurocranial cartilage defects of Fox-C and Fox-F mutants are also potentially analogous to the cleft palate seen in Foxc2, Foxf1 and Foxf2 mouse mutants (Iida et al., 1997; Wang et al., 2003; Xu et al., 2016). In addition, tooth loss in zebrafish Fox-F mutants is consistent with expression of Foxf1 in the developing tooth mesenchyme of mice (Wang et al., 2003). It was difficult to assess whether tooth reduction in zebrafish Fox-C mutants was an indirect consequence of severe cardiac edema, but loss of FOXC1 in Axenfeld–Rieger Syndrome is associated with missing and reduced teeth (Tümer and Bach-Holm, 2009). These extensive similarities highlight conserved functions of Fox-C/F genes between fish and mammals and reveal a functional role for Fox-F genes in tooth formation.
The mechanisms by which Sox and Fox proteins interact to regulate chondrogenesis remain unclear. Sox9 is known to directly bind the enhancers of a number of cartilage genes, including Col2a1 and Acan (Lefebvre et al., 1997; Ohba et al., 2015; Yasuda et al., 2017). The observation that consensus Fox binding sites are highly enriched in the vicinity of Sox9-bound enhancers in chondrocytes (Ohba et al., 2015) suggests that Fox and Sox factors may bind to some of the same enhancers. The finding that Foxc1a and Sox9a do not regulate the expression of each other further suggests that Fox and Sox proteins act in parallel to regulate chondrocyte enhancers. One possibility is that a subset of enhancers for chondrocyte genes is co-bound by Fox-C/F and Sox9 to promote transcription. An intriguing, alternative possibility is that Fox-C/F may bind chondrocyte enhancers first, thus preparing them for later Sox9 binding and activation. Consistent with this, we find that Fox-C/F expression in the arches generally precedes sox9a expression, and then shuts off during early chondrocyte differentiation. Although it is known that FoxA factors and Foxd3 can bind enhancers in a closed chromatin conformation and displace nucleosomes to allow later gene activation (Cirillo and Zaret, 1999; Xu et al., 2009), it remains to be determined whether Fox-C/F factors might act similarly in chondrocyte precursors. Future studies that assess chromatin status in the presence or absence of Fox-C/F factors will be required to test directly how Fox and Sox factors act together to promote cartilage formation.
MATERIALS AND METHODS
Zebrafish transgenic lines and mutants
The Institutional Animal Care and Use Committee of the University of Southern California approved all animal experiments performed in this study (Protocol #10885). We staged zebrafish (Danio rerio) as described (Kimmel et al., 1995). Published lines include edn1tf216b (Miller et al., 2000), jag1bb1105 (Zuniga et al., 2011), sox9atw37 (Yan et al., 2005), Tg(UAS:bmp4;cmlc2:GFP)el49 (Zuniga et al., 2011), Tg(sp7:EGFP)b1212 (DeLaurier et al., 2010), Tg(hsp70I:Gal4)kca4 (Scheer and Campos-Ortega, 1999), Tg(sox10:kikGR)el2 and Tg(UAS:shha;cmlc2:GFP)el137 (Balczerski et al., 2012), Tg(col2a1aBAC:mCherry-NTR)el559 and Tg(sox10:GFPCAAX)el375 (Askary et al., 2015). The Gateway (Invitrogen) Tol2Kit (Kwan et al., 2007) was used to generate fli1a:Gal4VP16, UAS:fgf3, UAS:foxc1a and UAS:foxf1. fli1a:Gal4VP16 was created by combining p5E-fli1a (Das and Crump, 2012), pME-Gal4VP16, p3E-pA and pDestTol2CG2. Three independent lines were isolated, and the stable line Tg(fli1a:Gal4VP16;cmlc2:GFP)el360 was used for this study. For UAS:fgf3, UAS:foxc1a and UAS:foxf1, the coding sequences of fgf3, foxc1a and foxf1 were amplified using the primers listed in Table S3 and cloned into pDONR221 to generate pME-fgf3, pME-foxc1a and pME-foxf1, followed by LR cloning with p5E-UAS, p3E-pA, and pDestTol2CG2 or pDestTol2AB2. Two independent transgenic lines were isolated for Tg(UAS:fgf3;cmlc2:GFP), Tg(UAS:foxc1a;α-crystallin:Cerulean) and Tg(UAS:foxf1;α-crystallin:Cerulean). We used the stable lines Tg(UAS:fgf3;cmlc2:GFP)el688, Tg(UAS:foxc1a;α-crystallin:Cerulean)el700 and Tg(UAS:foxf1;α-crystallin:Cerulean)el751 for this study. foxl1sa1842 was provided by the Sanger center. TALEN mutagenesis (Sanjana et al., 2012) was used to create four alleles: foxc1ael542, foxc1ael543, foxd1el551 and foxd2el575. CRISPR/Cas9 mutagenesis (Jao et al., 2012) was used to create six alleles: foxc1bel620, foxf1el658, foxf1el660, foxf2ael616, foxf2bel621 and foxl2el615. Given that foxc1ael542 and foxf1el658; foxf2ael616; foxf2bel621 mutant embryos showed identical defects to foxc1ael543 and foxf1el660; foxf2ael616; foxf2bel621, respectively, we used foxc1ael542 and foxf1el658 for this study. See Fig. S3 for description of mutations, and Table S1 for a list of TALEN and CRISPR constructs and genotyping details. For hsp70l:Gal4 experiments, embryos were placed in a 40°C incubator from 20-24 hpf and then transferred to 28.5°C, with hsp70l:Gal4-negative siblings serving as controls. All phenotypes were scored blinded to genotype.
Skeletal staining and histology
Acid-free Alcian Blue and Alizarin Red staining of cartilage and bone was performed on 5-6 dpf larvae as described (Walker and Kimmel, 2007). Whole-mount and dissected skeletons were imaged with Leica S8APO and DM2500 microscopes, respectively. For sectioning and histology, embryos were fixed in 1.5% parafomaldehyde, 1.5% glutaraldehyde, 0.1 M sodium cacodylate (pH 7.4), 0.001% CaCl2, embedded in epon, and 1-μm-thick serial sections were obtained. Sections were stained with Toluidine Blue and observed with a Zeiss Axio Imager ZZ microscope equipped with a digital Zeiss AxioCam ICc3 camera.
In situ hybridization and immunohistochemistry
Published in situ probes include dlx2a (Akimenko et al., 1994), dlx5a (Walker et al., 2006), fgf3 (Choe and Crump, 2014), jag1b (Zuniga et al., 2010), hand2 (Angelo et al., 2000), msx1a (Akimenko et al., 1995), sox9a (Yan et al., 2002), runx2b (Paul et al., 2016), barx1 (Barske et al., 2016), col2a1a and acana (Askary et al., 2015), matn1 (Askary et al., 2016), and foxd1, foxf1 and foxf2a (Askary et al., 2017). Partial cDNAs for foxc1a, foxc1b, foxd2, foxf2b, foxl1, dlx2b and matn4 were PCR amplified with Q5 High-Fidelity DNA Polymerase (New England Biolabs) and cloned into pCR Blunt II Topo (Thermo Fisher Scientific). Sequence-verified plasmids were linearized, and antisense probes were synthesized with Sp6 or T7 RNA polymerase (Roche Life Sciences) using dioxygenin (DIG)- or dinitrophenol (DNP)-labeled nucleotides (Roche). See Table S2 for probe generation details. Fluorescence and colorimetric in situ hybridizations were performed as described (Zuniga et al., 2010). For in situ hybridization on a sox10:GFPCAAX background, after the in situ protocol we added primary antibody rabbit anti-GFP (1:200; Torrey Pines Biolabs, TP401) diluted in PBDTx (PBS with 1% bovine serum albumin, 1% DMSO, 0.1% Triton X-100), incubated overnight at 4°C, washed four times with PBDTx, added secondary antibody goat anti-rabbit (1:500; Invitrogen A11008), incubated overnight at 4°C, and washed several times with PBDTx. We used a Zeiss LSM800 confocal microscope to capture fluorescence images, with representative sections or maximum intensity projections shown as specified. Colorimetric images were obtained on a Leica S8APO microscope. Levels were modified consistently across samples in Adobe Photoshop CS6.
Proliferation and apoptosis assays
BrdU incorporation was performed as described (Laguerre et al., 2005). Briefly, live embryos were manually removed from their chorions at 46 hpf, incubated in 15 mM BrdU (Sigma-Aldrich) in 15% DMSO for 25 min at 28.5°C, harvested at 48 hpf, and fixed with 4% paraformaldehyde overnight. Fixed embryos were washed three times with PBST (PBS with 0.25% Tween-20) and twice with deionized water. Embryos were incubated in 2 N HCl for 1 h at room temperature, washed three times with PBST, and incubated in block solution (2% normal goat serum, 0.1% bovine serum albumin, 1% DMSO in PBST) for at least 2 h. Embryos were then incubated with primary antibody rat anti-BrdU (1:200; Bio-Rad, MCA2060GA) overnight at 4°C, followed by secondary antibody goat anti-rat Alexa Fluor 568 (1:500; Invitrogen, A11077) and Hoechst to visualize nuclei. The ratio of BrdU-positive to total cells in the specified regions were calculated. For apoptosis, we performed TUNEL staining using the ApopTag Fluorescein In Situ Apoptosis Detection Kit (Millipore, S7110). Briefly, fixed embryos were washed twice with PBST, transferred to Equilibration Buffer for 3 min at room temperature, and incubated in the working strength TdT enzyme for 2 h at 37°C. The reaction was stopped in working strength stop/wash buffer for 10 min at room temperature, and then embryos were incubated with anti-DIG antibody overnight at 4°C, together with Hoechst to visualize nuclei. TUNEL-positive cells were calculated in the intermediate and dorsal regions of first and second arches for foxc1a; foxc1b double mutants and the ventral midline area of the first arch for foxf1; foxf2a; foxf2b triple mutants.
Measurement of relative length of symplectic and ceratohyal cartilages
Cartilage lengths were measured using ImageJ software. The relative lengths of Sy to Ch were compared between control and the indicated mutant genotypes using a Student's t-test (two-tailed, type 3), with P<0.001 considered significant.
Analysis of cell death and proliferation
The numbers of apoptotic (TUNEL+) and proliferating (BrdU+) cells in the first two arches were compared between control and Fox-C or Fox-F mutant embryos using the Student's t-test (two-tailed, type 3), with P>0.1 considered non-significant.
Supplementary Material
Acknowledgements
We are indebted to Megan Matsutani and Jennifer DeKoeyer Crump for fish care.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: P.X., B.B., J.G.C.; Methodology: P.X., B.B., J.G.C.; Formal analysis: P.X., B.B., J.G.C.; Investigation: P.X., B.B., A.C., K.L., V.O.; Writing - original draft: P.X., J.G.C.; Writing - review & editing: P.X., J.G.C.; Supervision: A.H., J.G.C.; Project administration: J.G.C.; Funding acquisition: J.G.C.
Funding
This work was funded by the National Institute of Dental and Craniofacial Research (R01-DE018405 and R35-DE027550 to J.G.C.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.165498.supplemental
References
- Akimenko M. A., Ekker M., Wegner J., Lin W. and Westerfield M. (1994). Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 14, 3475-3486. 10.1523/JNEUROSCI.14-06-03475.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimenko M. A., Johnson S. L., Westerfield M. and Ekker M. (1995). Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development 121, 347-357. [DOI] [PubMed] [Google Scholar]
- Akiyama H., Chaboissier M. C., Martin J. F., Schedl A. and de Crombrugghe B. (2002). The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813-2828. 10.1101/gad.1017802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C., Zuniga E., Blitz I. L., Wada N., Le Pabic P., Javidan Y., Zhang T., Cho K. W., Crump J. G. and Schilling T. F. (2011). Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development 138, 5135-5146. 10.1242/dev.067801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo S., Lohr J., Lee K. H., Ticho B. S., Breitbart R. E., Hill S., Yost H. J. and Srivastava D. (2000). Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech. Dev. 95, 231-237. 10.1016/S0925-4773(00)00334-8 [DOI] [PubMed] [Google Scholar]
- Askary A., Mork L., Paul S., He X., Izuhara A. K., Gopalakrishnan S., Ichida J. K., McMahon A. P., Dabizljevic S., Dale R. et al. (2015). Iroquois proteins promote skeletal joint formation by maintaining chondrocytes in an immature state. Dev. Cell 35, 358-365. 10.1016/j.devcel.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askary A., Smeeton J., Paul S., Schindler S., Braasch I., Ellis N. A., Postlethwait J., Miller C. T. and Crump J. G. (2016). Ancient origin of lubricated joints in bony vertebrates. Elife 5, e16415 10.7554/eLife.16415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askary A., Xu P., Barske L., Bay M., Bump P., Balczerski B., Bonaguidi M. A. and Crump J. G. (2017). Genome-wide analysis of facial skeletal regionalization in zebrafish. Development 144, 2994-3005. 10.1242/dev.151712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczerski B., Matsutani M., Castillo P., Osborne N., Stainier D. Y. and Crump J. G. (2012). Analysis of sphingosine-1-phosphate signaling mutants reveals endodermal requirements for the growth but not dorsoventral patterning of jaw skeletal precursors. Dev. Biol. 362, 230-241. 10.1016/j.ydbio.2011.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barske L., Askary A., Zuniga E., Balczerski B., Bump P., Nichols J. T. and Crump J. G. (2016). Competition between jagged-notch and endothelin1 signaling selectively restricts cartilage formation in the zebrafish upper face. PLoS Genet. 12, e1005967 10.1371/journal.pgen.1005967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W., Deng J. M., Zhang Z., Behringer R. R. and de Crombrugghe B. (1999). Sox9 is required for cartilage formation. Nat. Genet. 22, 85-89. 10.1038/8792 [DOI] [PubMed] [Google Scholar]
- Choe C. P. and Crump J. G. (2014). Tbx1 controls the morphogenesis of pharyngeal pouch epithelia through mesodermal Wnt11r and Fgf8a. Development 141, 3583-3593. 10.1242/dev.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L. A. and Zaret K. S. (1999). An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4, 961-969. 10.1016/S1097-2765(00)80225-7 [DOI] [PubMed] [Google Scholar]
- Das A. and Crump J. G. (2012). Bmps and id2a act upstream of Twist1 to restrict ectomesenchyme potential of the cranial neural crest. PLoS Genet. 8, e1002710 10.1371/journal.pgen.1002710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaurier A., Eames B. F., Blanco-Sánchez B., Peng G., He X., Swartz M. E., Ullmann B., Westerfield M. and Kimmel C. B. (2010). Zebrafish sp7:EGFP: a transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis 48, 505-511. 10.1002/dvg.20639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Dagenais S. L., Erickson R. P., Arlt M. F., Glynn M. W., Gorski J. L., Seaver L. H. and Glover T. W. (2000). Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema-distichiasis syndrome. Am. J. Hum. Genet. 67, 1382-1388. 10.1086/316915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H.-K., Lass J. H. and Chakravarti A. (1999). Pleiotropic skeletal and ocular phenotypes of the mouse mutation congenital hydrocephalus (ch/Mf1) arise from a winged helix/forkhead transcriptionfactor gene. Hum. Mol. Genet. 8, 625-637. 10.1093/hmg/8.4.625 [DOI] [PubMed] [Google Scholar]
- Iida K., Koseki H., Kakinuma H., Kato N., Mizutani-Koseki Y., Ohuchi H., Yoshioka H., Noji S., Kawamura K., Kataoka Y. et al. (1997). Essential roles of the winged helix transcription factor MFH-1 in aortic arch patterning and skeletogenesis. Development 124, 4627-4638. [DOI] [PubMed] [Google Scholar]
- Inman K. E., Purcell P., Kume T. and Trainor P. A. (2013). Interaction between Foxc1 and Fgf8 during mammalian jaw patterning and in the pathogenesis of syngnathia. PLoS Genet. 9, e1003949 10.1371/journal.pgen.1003949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L.-E., Appel B. and Wente S. R. (2012). A zebrafish model of lethal congenital contracture syndrome 1 reveals Gle1 function in spinal neural precursor survival and motor axon arborization. Development 139, 1316-1326. 10.1242/dev.074344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Mao J., Tenzen T., Kottmann A. H. and McMahon A. P. (2004). Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 18, 937-951. 10.1101/gad.1190304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo A., Denduluri S., Zhang B., Wang Z., Yin L., Yan Z., Kang R., Shi L. L., Mok J., Lee M. J. et al. (2014). The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 1, 149-161. 10.1016/j.gendis.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. and Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kume T., Deng K.-Y., Winfrey V., Gould D. B., Walter M. A. and Hogan B. L. (1998). The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell 93, 985-996. 10.1016/S0092-8674(00)81204-0 [DOI] [PubMed] [Google Scholar]
- Kume T., Jiang H., Topczewska J. M. and Hogan B. L. (2001). The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 15, 2470-2482. 10.1101/gad.907301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P. and Chien C. B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Laguerre L., Soubiran F., Ghysen A., König N. and Dambly-Chaudière C. (2005). Cell proliferation in the developing lateral line system of zebrafish embryos. Dev. Dyn. 233, 466-472. 10.1002/dvdy.20343 [DOI] [PubMed] [Google Scholar]
- Lefebvre V., Huang W., Harley V. R., Goodfellow P. N. and de Crombrugghe B. (1997). SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol. Cell. Biol. 17, 2336-2346. 10.1128/MCB.17.4.2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears A. J., Jordan T., Mirzayans F., Dubois S., Kume T., Parlee M., Ritch R., Koop B., Kuo W. L., Collins C. et al. (1998). Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am. J. Hum. Genet. 63, 1316-1328. 10.1086/302109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros D. M. and Crump J. G. (2012). New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev. Biol. 371, 121-135. 10.1016/j.ydbio.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. T., Schilling T. F., Lee K., Parker J. and Kimmel C. B. (2000). sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development 127, 3815-3828. [DOI] [PubMed] [Google Scholar]
- Miller C. T., Yelon D., Stainier D. Y. and Kimmel C. B. (2003). Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development 130, 1353-1365. 10.1242/dev.00339 [DOI] [PubMed] [Google Scholar]
- Mori-Akiyama Y., Akiyama H., Rowitch D. H. and de Crombrugghe B. (2003). Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc. Natl. Acad. Sci. USA 100, 9360-9365. 10.1073/pnas.1631288100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J. T., Pan L., Moens C. B. and Kimmel C. B. (2013). barx1 represses joints and promotes cartilage in the craniofacial skeleton. Development 140, 2765-2775. 10.1242/dev.090639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba S., He X., Hojo H. and McMahon A. P. (2015). Distinct transcriptional programs underlie Sox9 regulation of the mammalian chondrocyte. Cell Rep. 12, 229-243. 10.1016/j.celrep.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormestad M., Astorga J., Landgren H., Wang T., Johansson B. R., Miura N. and Carlsson P. (2006). Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development 133, 833-843. 10.1242/dev.02252 [DOI] [PubMed] [Google Scholar]
- Paul S., Schindler S., Giovannone D., de Millo Terrazzani A., Mariani F. V. and Crump J. G. (2016). Ihha induces hybrid cartilage-bone cells during zebrafish jawbone regeneration. Development 143, 2066-2076. 10.1242/dev.131292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N. E., Cong L., Zhou Y., Cunniff M. M., Feng G. and Zhang F. (2012). A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7, 171-192. 10.1038/nprot.2011.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N. and Campos-Ortega J. A. (1999). Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech. Dev. 80, 153-158. 10.1016/S0925-4773(98)00209-3 [DOI] [PubMed] [Google Scholar]
- Sun J., Ishii M., Ting M.-C. and Maxson R. (2013). Foxc1 controls the growth of the murine frontal bone rudiment by direct regulation of a Bmp response threshold of Msx2. Development 140, 1034-1044. 10.1242/dev.085225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J. C., Johnson S. L. and Kimmel C. B. (2010). hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development 137, 2507-2517. 10.1242/dev.049700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H.-T., Shah R. and Jamrich M. (2004). Function and regulation of FoxF1 during Xenopus gut development. Development 131, 3637-3647. 10.1242/dev.01234 [DOI] [PubMed] [Google Scholar]
- Tümer Z. and Bach-Holm D. (2009). Axenfeld-Rieger syndrome and spectrum of PITX2 and FOXC1 mutations. Eur. J. Hum. Genet. 17, 1527-1539. 10.1038/ejhg.2009.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. B. and Kimmel C. B. (2007). A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82, 23-28. 10.1080/10520290701333558 [DOI] [PubMed] [Google Scholar]
- Walker M. B., Miller C. T., Coffin Talbot J., Stock D. W. and Kimmel C. B. (2006). Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev. Biol. 295, 194-205. 10.1016/j.ydbio.2006.03.028 [DOI] [PubMed] [Google Scholar]
- Wang T., Tamakoshi T., Uezato T., Shu F., Kanzaki-Kato N., Fu Y., Koseki H., Yoshida N., Sugiyama T. and Miura N. (2003). Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev. Biol. 259, 83-94. 10.1016/S0012-1606(03)00176-3 [DOI] [PubMed] [Google Scholar]
- Xu J., Watts J. A., Pope S. D., Gadue P., Kamps M., Plath K., Zaret K. S. and Smale S. T. (2009). Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 23, 2824-2838. 10.1101/gad.1861209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Liu H., Lan Y., Aronow B. J., Kalinichenko V. V. and Jiang R. (2016). A Shh-Foxf-Fgf18-Shh molecular circuit regulating palate development. PLoS Genet. 12, e1005769 10.1371/journal.pgen.1005769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H., Maeda J., Hu T., McAnally J., Conway S. J., Kume T., Meyers E. N., Yamagishi C. and Srivastava D. (2003). Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 17, 269-281. 10.1101/gad.1048903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. L., Miller C. T., Nissen R. M., Singer A., Liu D., Kirn A., Draper B., Willoughby J., Morcos P. A., Amsterdam A. et al. (2002). A zebrafish sox9 gene required for cartilage morphogenesis. Development 129, 5065-5079. [DOI] [PubMed] [Google Scholar]
- Yan Y. L., Willoughby J., Liu D., Crump J. G., Wilson C., Miller C. T., Singer A., Kimmel C., Westerfield M. and Postlethwait J. H. (2005). A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132, 1069-1083. 10.1242/dev.01674 [DOI] [PubMed] [Google Scholar]
- Yasuda H., Oh C. D., Chen D., de Crombrugghe B. and Kim J. H. (2017). A novel regulatory mechanism of type II collagen expression via a SOX9-dependent enhancer in intron 6. J. Biol. Chem. 292, 528-538. 10.1074/jbc.M116.758425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Hata K., Takashima R., Ono K., Nakamura E., Takahata Y., Murakami T., Iseki S., Takano-Yamamoto T., Nishimura R. et al. (2015). The transcription factor Foxc1 is necessary for Ihh-Gli2-regulated endochondral ossification. Nat. Commun. 6, 6653 10.1038/ncomms7653 [DOI] [PubMed] [Google Scholar]
- Zuniga E., Stellabotte F. and Crump J. G. (2010). Jagged-Notch signaling ensures dorsal skeletal identity in the vertebrate face. Development 137, 1843-1852. 10.1242/dev.049056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E., Rippen M., Alexander C., Schilling T. F. and Crump J. G. (2011). Gremlin 2 regulates distinct roles of BMP and Endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development 138, 5147-5156. 10.1242/dev.067785 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.