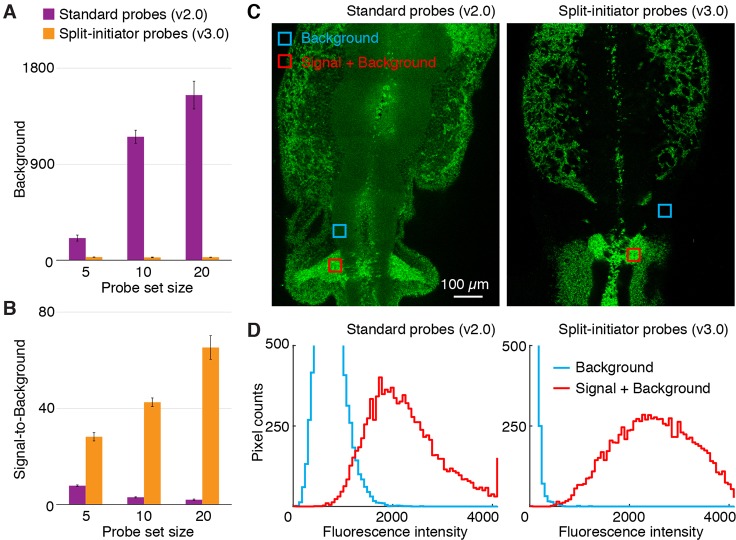
Fig. 3.
In situ validation of automatic background suppression with split-initiator probes in whole-mount chicken embryos. (A) Fluorescent background and (B) signal-to-background ratio as probe set size is increased by adding unoptimized probes: total of 5, 10 or 20 standard probes (v2.0) versus 5, 10 or 20 split-initiator probe pairs (v3.0). Any standard probes that bind non-specifically will generate amplified background, necessitating probe set optimization; split-initiator probes eliminate the potential need for probe set optimization by providing automatic background suppression. (C) Confocal micrographs in the neural crest of fixed whole-mount chicken embryos. Unoptimized probe sets: 20 standard probes (left) or 20 split-initiator probe pairs (right). See Fig. S7 (top) for the optimized standard probe set (Choi et al., 2016) with five probes. (D) Pixel intensity histograms for background and signal plus background (pixels in the depicted regions of C): overlapping distributions using unoptimized standard probes; non-overlapping distributions using unoptimized split-initiator probes. Embryos fixed at stage HH11. Target mRNA is Sox10. See Figs S5-S11 and Tables S10-S14 for additional data.