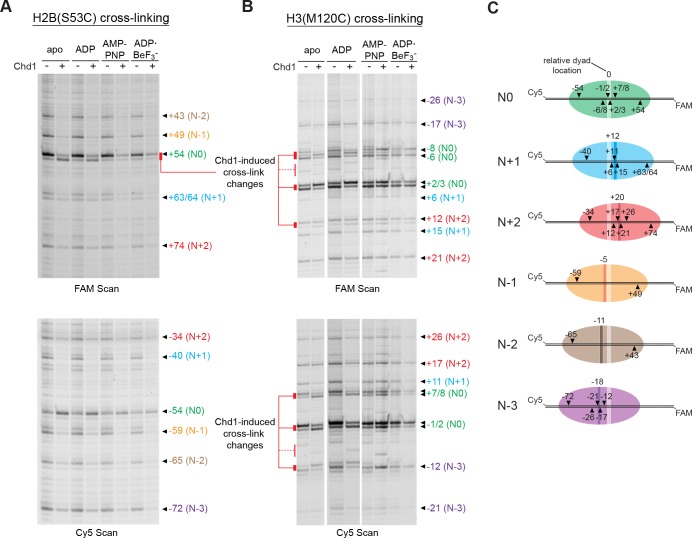
Figure 5. Chd1 binding alters histone-DNA contacts of 5S nucleosomes in a nucleotide-dependent manner.
(A) Mapping interactions of histone H2B(S53C) with 5S nucleosomal DNA. Histone cross-linking was performed in the presence or absence of 600 nM Chd1, using a sample that was enriched in centered nucleosomes. Cross-linked products were separated on urea denaturing gels and visualized by FAM or Cy5 fluorescence. Numbering of each cross-linking site is relative to the dyad of the dominant centered species, called N0 (green). Cross-links predicted to come from the same nucleosome species are given matching colors. Prominent Chd1-induced changes in histone cross-linking are highlighted by red bars. (B) Mapping interactions of histone H3(M120C) with 5S nucleosomal DNA. Cross-linking reactions were performed similarly to those described in A. (C) Schematic representations of 5S octamer positions. Cross-links observed for H2B(S53C) and H3(M120C) are indicated with black triangles pointing to the labeled DNA strand (FAM or Cy5) where they were observed. The dyad position of the enriched N0 species is indicated by a light stripe, while the dyads for the other species are shown as dark stripes, with the approximate number of nt differing from N0 given above each stripe. The 5S sequence is given in Figure 5—figure supplement 1.