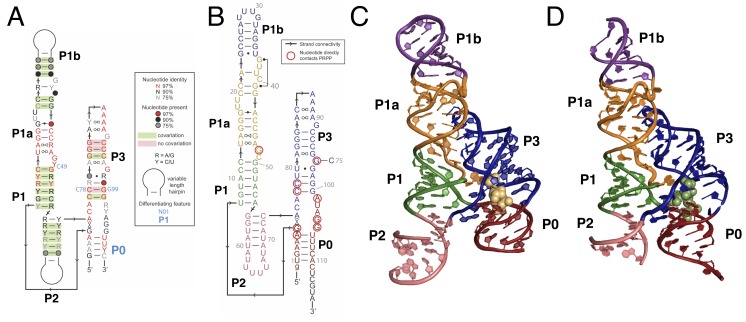
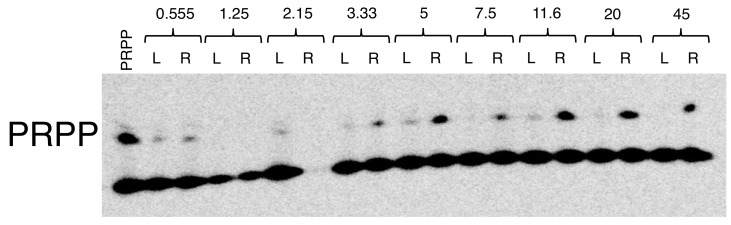
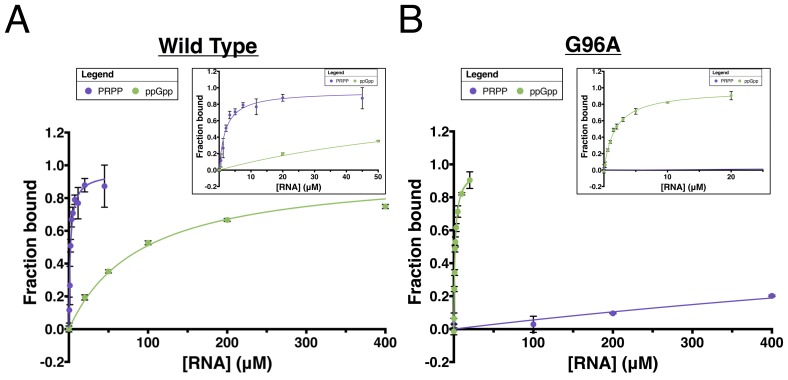
Figure 1. Overall structure of the PRPP riboswitch and its G96A mutant, which is a ppGpp aptamer.
(A) Consensus sequence of the PRPP riboswitch, adapted from Sherlock et al. (Sherlock et al., 2018b). The secondary structure has been updated to show structural information gained from the present study. The sequence is depicted as in Sherlock et al. (see key). Nucleotides noted in blue are important bioinformatic differences between PRPP riboswitches and guanidine riboswitches. Base pair notation is as published previously (Leontis and Westhof, 2001). (B) Secondary structure of the PRPP riboswitch aptamer from T. mathranii. Nucleotides are colored by paired region. Paired regions are indicated in bold. Sequence numbering is indicated in gray. Nucleotides that directly contact PRPP are circled in red, and arrows indicate strand connectivity. (C) Crystal structure of the PRPP riboswitch. Chain A is shown. The RNA is depicted as a cartoon and PRPP is depicted as yellow spheres. Nucleotides are colored by paired region as in B. (D) Crystal structure of the G96A mutant. Chain A is shown. The RNA is depicted as a cartoon and ppGpp is depicted as green spheres. Nucleotides are colored by paired region as in B.