SUMMARY
The Mis18 complex, composed of Mis16, Eic1, and Mis18 in fission yeast, selectively deposits the centromere-specific histone H3 variant, CENP-ACnp1, at centromeres. How the intact Mis18 holo-complex oligomerizes and how Mis16, a well-known ubiquitous histone H4 chaperone, plays a centromere-specific role in the Mis18 holo-complex, remain unclear. Here, we report the stoichiometry of the intact Mis18 holo-complex as (Mis16)2:(Eic1)2:(Mis18)4 using analytical ultracentrifugation. We further determine the crystal structure of Schizosaccharomyces pombe Mis16 in complex with the C-terminal portion of Eic1 (Eic1-CT). Notably, Mis16 accommodates Eic1-CT through the binding pocket normally occupied by histone H4, indicating that Eic1 and H4 compete for the same binding site, providing a mechanism for Mis16 to switch its binding partner from histone H4 to Eic1. Thus, our analyses not only determine the stoichiometry of the intact Mis18 holo-complex but also uncover the molecular mechanism by which Mis16 plays a centromere-specific role through Eic1 association.
Keywords: Kinetochore, Centromere, CENP-A, Mis16, RbAp46/48, The Mis18 holo-complex, Histone chaperone, Eic1, Analytical ultracentrifugation, Fission yeast, X-ray crystallography
E-TOC BLURB
Using structural, biochemical, and genetic approaches, An et al. unravel the oligomeric state of the fission yeast Mis18 holo-complex and the molecular mechanism by which Mis16 performs its centromere-specific function upon binding to Eic1.
INTRODUCTION
Once replicated in S phase, eukaryotic chromosomes must be evenly distributed to daughter cells in order to accurately transfer genetic material to subsequent generations. The kinetochore, a multi-protein complex that physically connects the centromere and microtubules, plays an essential role in even chromosome segregation with an error-proofing activity (Cheeseman and Desai, 2008).
Unlike the point centromere with a well-defined region of 125 base pairs of centromeric DNA as seen in budding yeast (Clarke and Carbon, 1980; Fitzgerald-Hayes et al., 1982), the regional centromere observed in most eukaryotes from fission yeast to humans, contains longer centromeric DNA ranging from several kilobases to megabases that does not possess defined consensus DNA sequences (Clarke and Baum, 1990; Murphy and Karpen, 1995; Willard, 1990). CENP-A is a centromere-specific histone H3 variant that, together with histones H4, H2A, and H2B, assembles into a centromere-specific nucleosome (Earnshaw and Rothfield, 1985; Palmer et al., 1987). In regional centromeres, it is widely appreciated that centromere identity is not determined by centromeric DNA sequence but by the presence of CENP-A (De Rop et al., 2012; Maddox et al., 2012). This indicates the epigenetic nature of CENP-A in centromere maintenance and inheritance. Ectopic localization of CENP-A to non-centromeric sites results in the formation of a neo-centromere (Amor and Choo, 2002; Scott and Sullivan, 2014; Warburton, 2001) in which all kinetochore components are recruited to the newly acquired position instead of to the original centromere (Mendiburo et al., 2011). Therefore, the mechanism by which CENP-A is specifically targeted to centromeres is an interesting and important subject to study.
In fission yeast, newly synthesized CENP-ACnp1 forms a complex with histone H4 and Scm3sp (a CENP-ACnp1-specific histone chaperone) (Pidoux et al., 2009; Williams et al., 2009). The CENP-ACnp1:H4/Scm3sp complex then localizes to centromeres with the assistance of the Mis18 holo-complex (An et al., 2015; Hayashi et al., 2014; Hayashi et al., 2004; Hirai et al., 2014; Subramanian et al., 2016; Subramanian et al., 2014). The Mis18 holo-complex, composed of Mis16, Eic1, and Mis18 in fission yeast, is a well-established CENP-ACnp1 assembly factor with two well-known biological functions (Hayashi et al., 2014; Hayashi et al., 2004; Hirai et al., 2014; Subramanian et al., 2014): (1) selective recognition of CENP-ACnp1 from dominant histone H3 through a direct interaction with Scm3sp, and (2) cell cycle-specific localization at centromeres to recruit the CENP-ACnp1-containing complex. Any genetic disturbance in components of the fission yeast Mis18 holo-complex is lethal (Hayashi et al., 2014; Hayashi et al., 2004; Hirai et al., 2014; Subramanian et al., 2014). Recently, the biological functions of each component of the Mis18 holo-complex have been described. Mis16 is responsible for specific recognition of the CENP-ACnp1:H4/Scm3sp complex through binding to the C-terminal region of Scm3sp (Scm3sp-CT) (An et al., 2015; Williams et al., 2009). Eic1 functions as an adaptor protein by connecting Mis16 and Mis18 (Hayashi et al., 2014; Hirai et al., 2014; Subramanian et al., 2014). Although the exact biological function of Mis18 has not yet been clearly delineated, the recently determined crystal structure of the Schizosaccharomyces pombe Mis18 N-terminal Yippee domain (Mis18Yippee) indicates that both the homodimer interface and the cradle-shaped binding pocket play an important role in the localization and function of the Mis18 holo-complex (Subramanian et al., 2016).
The human Mis18 holo-complex is composed of hM18BP1, hMis18α, and hMis18β (Fujita et al., 2007). Both hMis18α and hMis18β share high sequence similarity with fission yeast Mis18 (Fujita et al., 2007). Although hM18BP1 is a functional homolog of Eic1, its function and cell cycle-dependent regulation appear to be more complex (Dambacher et al., 2012; Moree et al., 2011; Pan et al., 2017; Spiller et al., 2017). The centromere-specific role of RbAp46/48, the human homolog of Mis16, has been controversial (Fujita et al., 2007; Lee et al., 2016). Unlike fission yeast Mis16, human RbAp46/48 are not constitutively associated with the rest of the Mis18 complex. Therefore, they are not considered a part of the human Mis18 holo-complex, although RNAi knockdowns of RbAp46/48 dramatically reduced CENP-A levels at centromeres (Fujita et al., 2007). Recently, significant progress has been made in characterizing the architecture of the human Mis18 holo-complex (Nardi et al., 2016; Pan et al., 2017; Spiller et al., 2017; Stellfox et al., 2016). In vitro pull-down experiments demonstrated that the N-terminal region of hM18BP1 (residues 20–130) directly recognizes the hMis18α-hMis18β complex, with the hM18BP1 dimer forming a complex with the hMis18α-hMis18β heterohexamer (Pan et al., 2017; Spiller et al., 2017). Moreover, HJURP, the human homolog of Scm3sp, was shown to interact with the oligomerized hMis18α/β C-terminal domains (Nardi et al., 2016).
Although the physiological significance of the fission yeast Mis18 holo-complex in CENP-ACnp1 recruitment and cell survival has been clearly demonstrated, the underlying molecular mechanism explaining how the Mis18 holo-complex achieves its biological function as a CENP-ACnp1 assembly factor has not been clearly defined. Particularly, Mis16 performs two seemingly non-overlapping roles: (1) as a ubiquitous protein that functions as a general histone H4 chaperone, participating in several key histone regulation processes (e.g., the HAT1, NuRD, NURF, CAF-1, and PRC2 complexes (Loyola and Almouzni, 2004; Philpott et al., 2000; Ruiz-Garcia et al., 1998), and (2) as a subunit of the Mis18 holo-complex that specifically recruits CENP-ACnp1 to centromeres via Scm3sp (An et al., 2015; Williams et al., 2009). The molecular mechanism by which Mis16 distinguishes between these two functions has not been determined.
To characterize the stoichiometry and the biochemical properties of the fission yeast Mis18 holo-complex, we purified and determined the molecular weight of the intact Mis18 holo-complex, and solved the crystal structure of Mis16 in complex with the C-terminal region of Eic1 (Eic1-CT). The molecular mass measurements indicate that the intact Mis18 holo-complex is composed of two copies of the Mis16-Eic1 heterodimer, and four copies of Mis18. Mis18 drives the oligomerization of the Mis18 holo-complex through homotetramerization. To elucidate how Mis16 selectively functions as a CENP-ACnp1 assembly factor, we solved the crystal structure of the Mis16-Eic1-CT complex at 2.3 Å resolution. The structure shows that Eic1-CT interacts with Mis16 through the same binding pocket where histone H4 binds. These results strongly suggest that when associated with Eic1, Mis16 no longer functions as a histone H4 chaperone but instead plays a centromere-restricted role. Switching its binding partner from histone H4 to Eic1 likely allows Mis16 to perform its centromere-specific function as part of the Mis18 holo-complex.
RESULTS
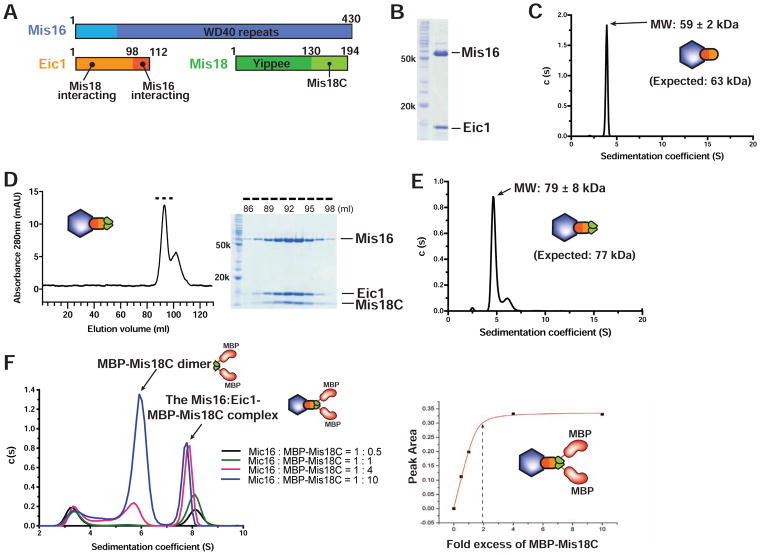
A Homodimer of the Mis18 C-terminal Domain Interacts with a Mis16-Eic1 Heterodimer
Earlier studies showed that Mis16 directly interacts with Eic13 (Hayashi et al., 2014; Hirai et al., 2014; Subramanian et al., 2014). To examine the oligomeric state of Mis16-Eic1, we first expressed and purified the Mis16-Eic1 subcomplex using the baculovirus expression system (Figure 1A and 1B). To obtain an accurate measure of the molecular mass and stoichiometry, we performed sedimentation velocity analytical ultracentrifugation (SV-AUC). SV-AUC of the purified Mis16-Eic1 subcomplex demonstrates that it sediments with a molecular weight (MW) of 59 ± 2 kDa, which corresponds to a Mis16-Eic1 heterodimer (expected MW: 63 kDa) (Figure 1C). To elucidate how Mis18 facilitates the oligomerization of the Mis18 holo-complex, we divided Mis18 into two domains as described in previous reports (Fujita et al., 2007; Nardi et al., 2016): the N-terminal Yippee-like domain (Mis18Yippee) and the C-terminal helical domain (Mis18C) (Figure 1A). We purified each domain individually and examined association with Mis16-Eic1 by size-exclusion chromatography (SEC). As shown in Figure 1D, Mis18C directly associates with Mis16-Eic1, while Mis18Yippee does not (Figure S1). The molecular mass of the Mis16-Eic1-Mis18C complex obtained with SV-AUC was 79 ± 8 kDa, suggesting a 1:1:2 molar ratio (expected MW: 77 kDa) (Figure 1E). However, the relatively low mass of Mis18C (7 kDa) makes an accurate and unambiguous estimate of oligomeric state difficult. We therefore expressed and purified Mis18C fused to maltose binding protein (MBP) (expected MW: 52 kDa). The AUC titration assay of the Mis16-Eic1-(MBP-Mis18C) complex shows that two copies of MBP-Mis18C interact with one Mis16-Eic1 heterodimer (Figure 1F).
Figure 1. Stoichiometry of the S. pombe Mis18 sub-Complexes.
(A) Domain organization of the S. pombe Mis18 holo-complex components.
(B) SDS-PAGE analysis of the purified S. pombe Mis16-Eic1 subcomplex expressed in baculovirus insect cells.
(C) SV-AUC analysis of the purified Mis16-Eic1 complex. The calculated MW corresponds to a heterodimer of Mis16 and Eic1.
(D) SEC profile of the Mis16-Eic1-Mis18C complex (left). The dashed line indicates fractions used for SDS-PAGE analysis (right).
(E) SV-AUC analysis of the Mis16-Eic1-Mis18C complex. The MW of the peak corresponds to a 1:1:2 molar ratio.
(F) SV-AUC analyses of the Mis16-Eic1 heterodimer with different concentrations of MBP-tagged Mis18C (MBP-Mis18C) (left) and its titration curve (right). See also Figure S1.
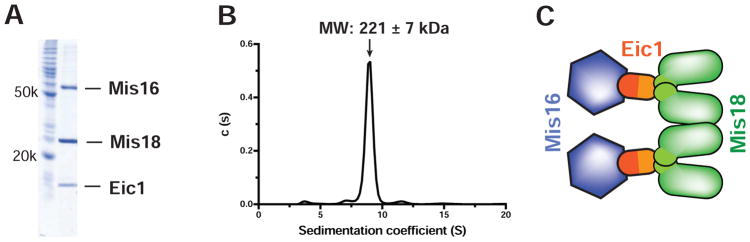
The Stoichiometry of the S. pombe Mis18 Holo-Complex is (Mis16)2:(Eic1)2:(Mis18)4
To measure the accurate molecular mass of the intact Mis18 holo-complex, we expressed and purified the S. pombe Mis18 holo-complex using baculovirus expression in insect cells (Figure 2A). Subsequent SV-AUC analysis of the Mis18 holo-complex indicates that it sediments with a molecular weight (MW) of 221 ± 7 kDa, indicating that two Mis16-Eic1 heterodimers associate with a Mis18 tetramer by forming a complex in a 2:2:4 ratio (expected MW: 218 kDa) (Figure 2B and 2C).
Figure 2. Stoichiometry of the S. pombe Mis18 Holo-Complex.
(A) SDS-PAGE analysis of the purified S. pombe Mis18 holo-complex expressed in baculovirus insect cells.
(B) SV-AUC analysis of the purified S. pombe Mis18 holo-complex. The calculated MW suggests a stoichiometry of (Mis16)2:(Eic1)2:(Mis18)4.
(C) Schematic diagram showing composition of the S. pombe Mis18 holo-complex.
See also Figure S2.
Since the stoichiometry of the Mis16-Eic1-Mis18C complex was 1:1:2 (Figure 1E and 1F), a 2:2:4 molar ratio of the Mis18 holo-complex indicates that Mis18Yippee also forms a dimer independently from the dimerization of Mis18C. A recent crystal structure of Mis18Yippee is consistent with our conclusion, showing that Mis18Yippee forms a homodimer both in solution and in the crystal structure (Subramanian et al., 2016). The same study also demonstrated that full-length Mis18 forms a homotetramer in solution (Subramanian et al., 2016), which is completely in agreement with our stoichiometry evaluation. Our SEC data additionally show that disrupting Mis18Yippee dimerization via the Mis18I31A mutation causes the mutant Mis18 holo-complex to migrate slower than wild-type (Figure S2). Thus, based on our biochemical analyses, we propose that the Mis18 holo-complex is composed of two copies of Mis16-Eic1 and four copies of Mis18, and that the oligomerization of the Mis18 holo-complex is driven by the two-fold dimerization of both Mis18Yippee and Mis18C (Figure 2C).
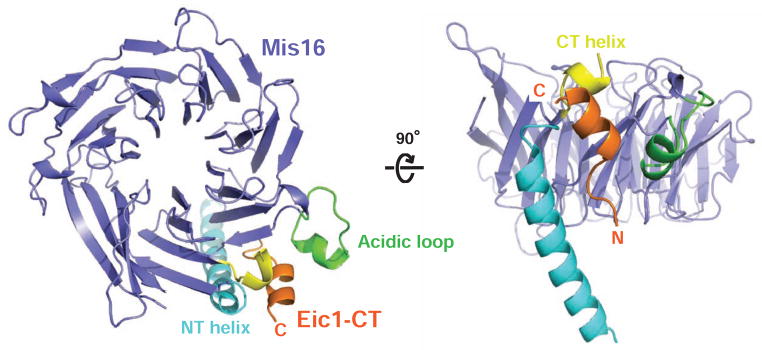
Crystal Structure of the Mis16-Eic1-CT Complex
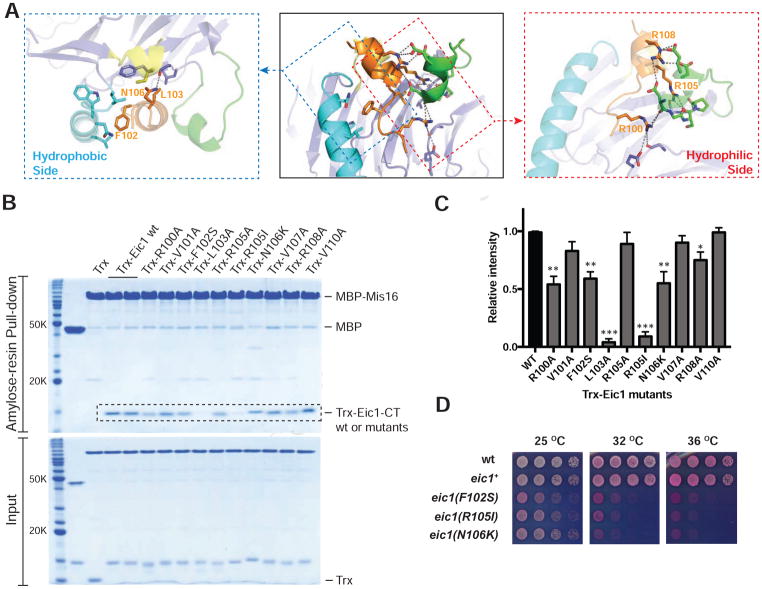
To elucidate how Eic1 recognizes Mis16 within the centromere-specific Mis18 holo-complex, we crystallized S. pombe Mis16 in complex with full-length Eic1 and recorded X-ray diffraction at 2.3 Å resolution (Figure 1B). Starting phases were obtained by molecular replacement using Schizosaccharomyces japonicus Mis16 as a search model (PDB ID: 4XYH) (An et al., 2015) (Table 1). The final refined structure has R and Rfree values of 17.2 % and 22.0 %, respectively. Although the full-length Mis16-Eic1 complex was used for crystallization, only the C-terminal region of Eic1 (Eic1-CT, residues 99–112) was clearly visible in the resultant electron density map, likely due to Eic1 degradation during crystallization (Figures 3 and S3). Eic1-CT forms an extended structure (residues 99–103) followed by a short helix (residues 104–112) and makes multiple contacts with Mis16 through hydrophobic (V101, F102, L103, V107, A109, and V110) and electrostatic/hydrogen bond (R100, R105, and R108) interactions (Figures 4A and S4A). To verify the interactions seen in the crystal structure, we expressed and purified Eic1-CT mutants fused with thioredoxin (Trx) at the N terminus and examined their direct interactions with MBP-tagged Mis16 using amylose resin pull-down assays. A series of missense mutations in Eic1-CT, which are expected to disrupt either hydrophobic (F102S and L103A) or electrostatic (R100A, R105I, and R108A) interactions, were sufficient to reduce Eic1 association with Mis16 (Figure 4B and 4C). The F102S mutation was originally identified as an Eic1 temperature-sensitive (ts) mutant (eic1-1) in vivo that substantially reduces centromeric CENP-ACnp1 levels at non-permissive temperature (36°C) (Subramanian et al., 2014). In the Mis16-Eic1-CT structure, Eic1-F102 is surrounded by Mis16 hydrophobic residues (L32, W33, and L40), so the phenylalanine to serine substitution is likely to abolish Mis16-Eic1 association at restrictive temperature (Figures 4B, 4C, and S5A). We also screened for and isolated additional S. pombe ts mutants, eic1-R105I and eic1-N106K, which display similar growth phenotypes as eic1-F102S (eic1-1) (Figure 4D). As indicated by our crystal structure, both R105I and N106K substitutions may not only disrupt hydrogen bond interactions, but also push the Eic1-CT helix out of its binding pocket and thus, significantly impair Mis16-Eic1 association (Figures 4B, 4C, and S5B). Taken together, the crystal structure of the Mis16-Eic1 complex and subsequent mutational analyses in vitro and in live cells strongly indicate that the Mis16-Eic1 interaction is essential for the physiological function of the Mis18 holo-complex at centromeres.
Table 1.
Data collection and refinement statistics.
| S. pombe Mis16 + Eic1-CT | |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.9786 |
| Space group | P43212 |
| Cell dimensions | |
| a, b, c (Å) | 154.6, 154.6, 52.7 |
| a, b, g (°) | 90, 90, 90 |
| Resolution (Å) | 50.0 – 2.3 (2.38 – 2.3)* |
| Rsym or Rmerge (%) | 9.7 (75.8) |
| I/σI | 22.4 (1.6) |
| Completeness (%) | 90.8 (59.7) |
| Redundancy | 8.0 |
| Refinement | |
| Resolution (Å) | 48.9 – 2.3 (2.36– 2.3) |
| No. reflections | 26414 (1097) |
| Rwork/Rfree | 17.2/22.0 |
| No. atoms | |
| Protein | 3172 |
| Water | 168 |
| B-factors | |
| Protein | 39.1 |
| Water | 42.0 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.931 |
| Ramachandran plot (%) | |
| Favored | 96.9 |
| Outliers | 0.26 |
| Protein Data Bank code | 5WJC |
Values in parentheses are for highest-resolution shell.
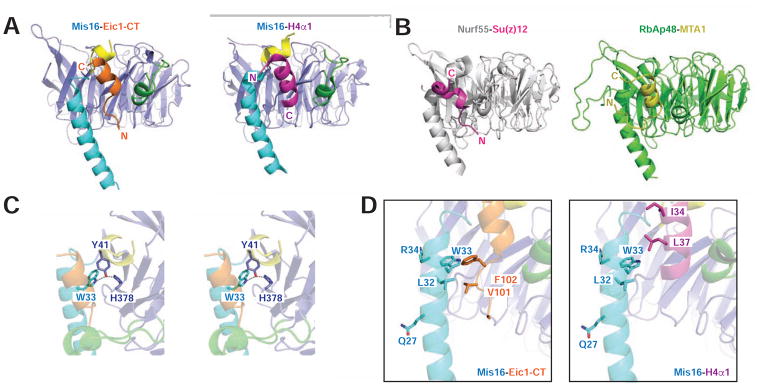
Figure 3. Crystal Structure of the S. pombe Mis16-Eic1-CT Complex.
Top- and 90°-oriented side views of the S. pombe Mis16-Eic1-CT complex. The Mis16 WD-40 repeat domain is colored slate. The binding pocket of Eic1-CT (orange) is formed by the Mis16 N-terminal helix (NT helix, cyan), acidic loop (green), and C-terminal helix (CT helix, yellow). All structures shown in the figures were generated using PYMOL (Delano Scientific, LLC).
See also Figures S3, S4, and S5.
Figure 4. In Vitro and In Vivo Mutational Analysis of Eic1-CT.
(A) Hydrophobic interactions (blue dashed boxes) and hydrophilic interactions (red dashed boxes) of Eic1-CT within the Mis16-Eic1-CT complex. Eic1 residues from the hydrophobic side mostly form hydrophobic interactions with residues in the Mis16 N-terminal helix (cyan) and WD-40 repeats (slate). The hydrophilic side residues of Eic1 form hydrogen bond and electrostatic interactions with residues from the Mis16 acidic loop (green) and WD-40 repeats.
(B) SDS-PAGE analysis of the amylose resin pull-down assay using MBP-tagged Mis16 as bait and thioredoxin (Trx)-tagged Eic1-CT (residues 91–112) (wild-type or mutant) as prey.
(C) Histogram showing normalized band intensities from pull-down experiments in (B). Band intensities for Trx-Eic1-CT mutants within the dashed box area in (B) were measured and normalized relative to that of wild-type. Results are shown as mean ± SEM of three independent measurements; *P < 0.05, ** P < 0.005, *** P < 0.0005.
(D) In vivo screen for temperature-sensitive Eic1-CT mutants identifies key residues. S. pombe cells harboring the indicated eic1 mutations were five-fold serial diluted, spotted on YES + Phloxine B media, and incubated at the indicated temperatures; dead cells stain dark pink.
See also Figures S4 and S5.
Eic1-CT Shares the Mis16 Binding Pocket with Histone H4
The Mis16 binding pocket for Eic1-CT is formed by the N-terminal helix, the acidic loop, and the C-terminal short helix of Mis16 (Figures 3 and 5A). Previously, we showed that Mis16 interacts with the first helix of histone H4 (residues 29–41, H4α1) (An et al., 2015). Notably, a structural comparison between Mis16-H4α1 and Mis16-Eic1-CT reveals that both H4α1 and Eic1-CT share the same binding pocket for interacting with Mis16, although the binding pattern is dissimilar (Figure 5A). Both Eic1-CT and histone H4α1 bind to Mis16 using a short helix, but the helix dipole of Eic1-CT is inverted relative to that of H4α1 (Figure 5A). Despite the opposite directionality of the helix, both proteins have an amphipathic helix containing both hydrophobic and positively charged residues that make multiple contacts with Mis16 (Figure S4).
Figure 5. Comparison of the Binding Patterns of Histone H4α1 and Eic1-CT to Mis16.
(A) Side views of the Mis16-Eic1-CT complex (left) and the Mis16-H4α1 complex (PDB code: 4XYI, right). The color scheme for Mis16 is the same as in Figure 3. The binding mode and positions of the N and C termini of Eic1-CT (residues 91–112, orange) and H4α1 (residues 1–48, purple) are displayed and labeled.
(B) Side views of the Nurf55-Su(z)12 complex (PDB code: 2YB8, left) (Schmitges et al., 2011) and the RbAp48-MTA1 complex (PDB code: 4PC0, right) (Alqarni et al., 2014)). Nurf55 (grey) interacts with the N-terminal fragment of Su(z)12 (residues 79–91, purple) and RbAp48 (green) interacts with the C-terminal fragment of MTA1 (residues 672–688, yellow).
(C) Stereoview of Eic1-CT binding near Mis16 Y41. Similar to the Mis16-H4α1 complex (An et al., 2015), hydrogen bonds with Mis16 Y41, W33, and H378 stabilize the binding pocket of Eic1-CT.
(D) Detailed comparison of interactions in Mis16-Eic1-CT and Mis16-H4α1 structures. In the Mis16-Eic1-CT structure, Mis16 L32 and W33 form hydrophobic interactions with Eic1 V101 and F102. However, these same Mis16 residues do not mediate any critical interactions in the Mis16-H4α1 structure.
See also Figure S6.
The observed pattern of Eic1-CT occupying the histone H4 binding pocket of Mis16, resembles that observed in RbAp48-MTA1 (NuRD complex) and Nurf55-Su(z)12 (PRC2 complex) (Figure 5B) (Alqarni et al., 2014; Schmitges et al., 2011). RbAp48 and Nurf55 are human and Drosophila homologs of Mis16 respectively, and binding of MTA1 or Su(z)12 can redirect the original histone H4 chaperone function of RbAp48 or Nurf55 to NuRD- or PRC2-specific roles respectively.
A previously described ts mutant of Mis16, mis16-53 (Y41H), fails to recruit CENP-ACnp1 to centromeres at non-permissive temperature (Hayashi et al., 2004). The crystal structure of S. japonicus Mis16 revealed that Y38 (equivalent to Y41 in S. pombe Mis16) makes hydrogen bonds with its neighboring residues W30 and H375 (W33 and H378 in S. pombe Mis16) and stabilizes the Mis16 binding pocket by maintaining the N-terminal helix intact (Figure S6) (An et al., 2015). Notably, the crystal structure of the Mis16-Eic1-CT complex demonstrates that Mis16-Eic1 interaction might be affected by the Mis16Y41H (mis16-53) mutation (Figure 5C). To examine whether this interaction is indeed influenced by Mis16Y41H and Mis16Y41A, we performed amylose resin pull-downs using MBP-Mis16Y41H and MBP-Mis16Y41A as bait and Trx-Eic1-CT or H4α1-Sumo as prey (Figure 6A and 6B). Consistent with our previous report (An et al., 2015), the Mis16-H4α1 interaction is only weakly influenced by Mis16Y41H or Mis16Y41A at permissive temperature (20°C). In contrast, the Mis16-Eic1-CT interaction is significantly reduced by these mutations even at permissive temperature (Figure 6A and 6B). This result suggests that the Mis16-Eic1-CT interaction is more sensitive to Mis16 Y41 mutations than the Mis16-H4α1 interaction.
Figure 6. Mis16 Mutants that Specifically Disrupt the Interaction with Eic1-CT, and Not with H4α1.
(A) Amylose resin pull-down assays using MBP-Mis16 wild-type or mutants as bait and either H4α1-Sumo or Trx-Eic1-CT as prey.
(B) Histograms showing normalized band intensities from pull-down experiments in (A). Band intensities for mutants within the dashed box area in (A) were measured and normalized relative to that of wild-type. Results are shown as mean ± SEM of three independent measurements; *P < 0.05 and ****P < 0.0001.
Eic1-CT Competes with Histone H4 for Mis16 Binding
Structural comparison of Mis16-H4α1 and Mis16-Eic1-CT indicates that H4α1 and Eic1-CT are mutually exclusive. To experimentally verify this hypothesis, we performed a competition assay to assess the strength of Mis16 binding to Eic1-CT in the presence of increasing amounts of H4α1. Isothermal titration calorimetry (ITC) data show that Trx-Eic1-CT binds to Mis16 with a KD of 20.8 ± 5.6 nM and H4α1-Sumo binds with a KD of 26.9 ± 8.5 nM, suggesting that the binding affinities of Eic1-CT and H4α1 for Mis16 are comparable (Figure S7B). After co-incubating equimolar quantities of MBP-Mis16 and Trx-Eic1-CT, we challenged the interaction with increasing amounts of H4α1-Sumo and measured the remaining Trx-Eic1-CT band intensity after competition in amylose resin pull-downs. As expected, the total amount of bound Trx-Eic1-CT decreased with the addition of increasing amounts of H4α1-Sumo (Figure 7A and 7B). Interestingly, challenging pre-incubated MBP-Mis16 and H4α1-Sumo with increasing amounts of Trx-Eic1-CT only weakly competed out Mis16-bound H4α1-Sumo (Figure S7A). This observation indicates that on and off rates of H4α1 association with Mis16 are lower than that of Eic1-CT. Taken together, our results suggest that Eic1 and histone H4 share the same binding pocket and compete for Mis16 binding.
Figure 7. Competition Assay of Eic1-CT and H4α1 Binding to Mis16, In Vivo Genetic Complementation Assay, and the Schematic Model of Centromere-Specific Mis16 Function Mediated by Eic1.
(A) Pre-incubated, equimolar amounts of MBP-Mis16 and Eic1-CT were challenged with either increasing amounts of Sumo or H4α1-Sumo for the competition assay, and the remaining Eic1-CT band intensity was measured following amylose resin pull-downs.
(B) Histogram showing remaining Eic1-CT band intensities (normalized) for dashed box area in (A) after the competition assay. Results are shown as mean ± SEM of three independent measurements; *P < 0.05, ** P < 0.005, ***P < 0.0005, *** P < 0.0001.
(C) Genetic complementation assay. Five-fold serial dilutions of mis16-53 (Y41H) cells expressing the indicated S. pombe Mis16 constructs integrated at the leu1 locus in the genome were spotted on complete PMG + phloxine B media supplemented with (repressed) or without (expressed) thiamine and incubated at the indicated temperatures for 3–5 days; dead cells stain dark pink. Mis16Δ1-32 only partially complements the temperature sensitivity of mis16-53 (Y41H) cells and Mis16Δ1-33 cannot complement, as observed in two independent isolates.
(D) Three missense mutations of Mis16 (E29A, L32A, and W33A) were tested for genetic complementation as described in (C). Mis16 E29, which does not make a contact with the Eic1-CT, was used as a positive control. Cells harboring a missense mutation of Mis16 (W33A) remained inviable at the restrictive temperature, while cells harboring Mis16 (E29A) or Mis16 (L32A) showed recovery of growth at 36°C when expression was induced.
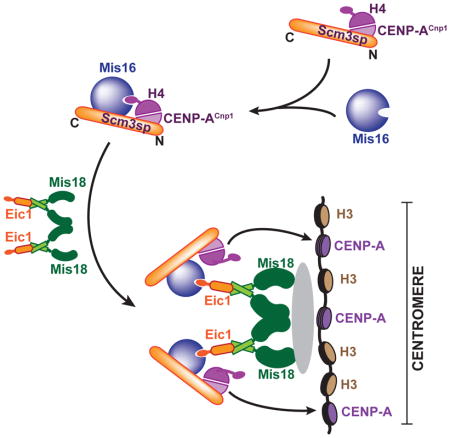
(E) Schematic diagram of the proposed mechanism by which Mis16 fulfills a centromere-specific role in fission yeast. Mis16 specifically recognizes the CENP-ACnp1:H4/Scm3sp complex through its interaction with both histone H4 and Scm3sp. Once the temporal Mis16-CENP-ACnp1:H4/Scm3sp complex approaches the (Eic1)2:(Mis18)4 complex, Eic1 replaces histone H4 in the Mis16 binding pocket by simple competition or an unknown mechanism, which results in the assembly of the Mis18 holo-complex. The Mis18 holo-complex then localizes to the centromere thereby incorporating CENP-ACnp1:H4. CENP-ACnp1:H4 might be easily dissociated from the Mis18 holo-complex and incorporated into the centromere after losing the Mis16-histone H4 interaction mediated by Eic1 occupation.
See also Figures S7 and S8.
An N-Terminally Truncated Mutant of Mis16 Specifically Disrupts the Mis16-Eic1 Interaction but not the Mis16-Histone H4 Interaction
The competitive nature of Eic1 and histone H4 binding toward Mis16 indicates that the Mis16 residues important for the Mis16-histone H4 interaction are likely to also be involved in Eic1 recognition. Structural comparison and mutational analyses confirmed that key residues required for histone H4 recognition also participate in Eic1 association (An et al., 2015; Song et al., 2008). To identify a Mis16 mutant that selectively disrupts its interaction with Eic1, we carefully inspected crystal structures of Mis16-Eic1-CT and Mis16-H4α1, and identified two N-terminal truncation mutations (Mis16Δ1-32 and Mis16Δ1-33) as candidates. Mis16 L32 and W33 participate in hydrophobic interactions with Eic1 V101 and F102, whereas both these Mis16 residues are not involved in the Mis16-H4α1 interaction (Figure 5D). Amylose resin pull-downs of Mis16Δ1-26, Mis16Δ1-32, and Mis16Δ1-33 to assess differences in binding to H4α1-Sumo and Trx-Eic1-CT demonstrated that the Mis16-Eic1 interaction is intact when both L32 and W33 (Mis16Δ1-26) are retained. However, the interaction is significantly diminished when either L32 (Mis16Δ1-32) or both L32/W33 (Mis16Δ1-33) are deleted (Figure 6A and 6B). In contrast, the Mis16-H4α1 interaction was only mildly affected in the context of the Mis16Δ1-33 truncation. These findings strongly suggest that Mis16 L32 and W33 participate in specific interactions with Eic1 but not histone H4.
To verify these observations in vivo, we performed an S. pombe genetic complementation assay by ectopically expressing Mis16Δ 1-32 and Mis16Δ1-33 in mis16-53 (Y41H) ts mutant cells. The Mis16-Eic1-CT crystal structure and pull-down assays suggest that Mis16Δ1-32 is mildly defective in interacting with Eic1 but is fully competent in recognizing histone H4. On the other hand, Mis16Δ1-33 significantly impairs the Eic1 interaction but largely retains the histone H4 interaction (Figures 5D, 6A, and 6B). Consistent with our in vitro results, the in vivo genetic complementation assay showed that Mis16Δ1-32 is able to partially rescue the temperature sensitivity of mis16-53 (Y41H) cells, while Mis16Δ1-33 completely fails to rescue the viability of mis16-53 cells at restrictive temperature (36°C) (Figure 7C). A missense mutation of Mis16 W33A also displayed complete inability to rescue the viability of mis16-53 cells at restrictive temperature (Figure 7D). Taken together, these results indicate that Mis16 W33 is specifically involved in interactions with Eic1 but not histone H4, and the Mis16-Eic1 interaction mediated by the Mis16 N terminus is crucial for centromere function and cell survival.
DISCUSSION
The fission yeast Mis18 holo-complex, composed of Mis16, Eic1, and Mis18, is an essential assembly factor for CENP-ACnp1 nucleosomes with two key biological functions: (1) centromere-specific localization in a cell-cycle dependent manner, and (2) selective recognition of the CENP-ACnp1:H4/Scm3sp complex. Any genetic disturbance in this complex results in substantial shortage of CENP-ACnp1 at centromeres, increased genome instability, and consequent loss of cell viability (Hayashi et al., 2014; Hayashi et al., 2004; Hirai et al., 2014; Subramanian et al., 2014). Recent genetic and biochemical studies of the Mis18 complex both in fission yeast and human cells have advanced our understanding of the domain organization and the biological roles of individual components (Nardi et al., 2016; Pan et al., 2017; Spiller et al., 2017; Stellfox et al., 2016; Subramanian et al., 2016). However, the molecular mechanism by which the Mis18 complex specifically recognizes the centromere and selectively recruits the CENP-ACnp1:H4/Scm3sp complex to the centromere at the atomic-level has not been elucidated.
The current study addresses two major issues regarding the fission yeast Mis18 holo-complex: (1) the stoichiometry and the oligomeric state of the intact Mis18 holo-complex, and (2) the mechanism by which Mis16, a conserved ubiquitous histone H4 chaperone, performs a centromere-specific function in the CENP-ACnp1 assembly pathway.
Comparison of Domain Organization and Oligomerization of the Fission Yeast and Human Mis18 Holo-Complexes
The human Mis18 holo-complex is composed of hMis18α, hMis18β, and hM18BP1. Recently, two groups independently reported the oligomeric state of the human Mis18 complex with the stoichiometry of hM18BP1:hMis18α:hMis18β as 2:4:2 (Pan et al., 2017; Spiller et al., 2017). The centromere-specific role of RbAp46/48 as a potential additional component of the Mis18 holo-complex, however, remains obscure (Fujita et al., 2007; Lee et al., 2016). Both human and fission yeast Mis18 holo-complexes perform the same biological function, with Mis18 and Mis16 being evolutionarily conserved (Fujita et al., 2007; Hayashi et al., 2004). However, previous studies suggest some key differences still exist. First, Mis16 in fission yeast is a constitutive subunit of the Mis18 holo-complex, whereas RbAp46/48 in humans appears to be temporarily associated with the Mis18 holo-complex (Fujita et al., 2007; Hayashi et al., 2004). However, depletion of Mis16 or RbAp46/48 displays the same phenotype, exhibiting impaired CENP-A deposition and chromosome segregation in several model organisms (Fujita et al., 2007; Hayashi et al., 2004; Lee et al., 2016). Second, the primary sequences of the adaptor proteins (Eic1 in fission yeast and hM18BP1 in humans) are not conserved, although their biological functions are. The unique function of Eic1 is to physically connect Mis16 and Mis18 (Hayashi et al., 2014; Hirai et al., 2014; Subramanian et al., 2014). In contrast, the function of hM18BP1 appears to be more complex with multiple phosphorylation sites (Dambacher et al., 2012; Fujita et al., 2007; McKinley and Cheeseman, 2014; Moree et al., 2011; Silva et al., 2012). Cell-cycle dependent phosphorylation of hM18BP1 mediated by Cdk and Plk1 regulates CENP-C-dependent centromere localization and the cell-cycle dependent assembly of the human Mis18 holo-complex (McKinley and Cheeseman, 2014; Silva et al., 2012). Third, Mis18 in fission yeast forms a homotetramer through the dimerization of both Mis18Yippee and Mis18C (Figures 1 and 2) (Subramanian et al., 2016). hMis18α and hMis18β form a heterohexamer through homodimerization of hMis18Yippee and heterotrimerization of hMis18C, although there is some disagreement as to the exact oligomeric state of hMis18α/β (Pan et al., 2017; Spiller et al., 2017), as Nardi and colleagues showed that hMis18α and hMis18β form a heterotetramer using their C-terminal helical domains (Nardi et al., 2016). Finally, in fission yeast, dimeric Mis18C associates with the N-terminal region of Eic1, and Scm3sp-CT interacts with the Mis18 holo-complex through Mis16 (Figure 1E and 1F) (An et al., 2015; Williams et al., 2009). In humans, the N-terminal Yippee domains of the hMis18α/β heterodimer recognize the hM18BP1 N-terminal domain (Pan et al., 2017; Spiller et al., 2017), and the C-terminal helical domains of hMis18α/β interact with HJURP (Nardi et al., 2016).
Although there are differences in details, the overall architecture of the human and fission yeast Mis18 holo-complexes share common features and conserved biological functions: (1) Mis18 oligomerization through both Mis18Yippee and Mis18C is the main driving force for the oligomerization of the Mis18 holo-complex, and (2) both Eic1 and hM18BP1 associate with the Mis18 dimer (Eic1 associates with the Mis18C dimer and hM18BP1 interacts with a dimer of the hMis18α Yippee domain). How the Mis18 holo-complex, whose physiological role as a CENP-A recruiting factor is evolutionarily conserved, undergoes modest reorganization and recomposition to adopt new dimensions is an interesting question that needs further investigation. Follow-up biochemical and structural studies will further advance our understanding of how similar or distinct the human and fission yeast Mis18 holo-complexes really are.
Eic1 Binding to Mis16 Redirects the Biological Function of Mis16 from a Histone H4 Chaperone to a CENP-ACnp1 Specific Assembly Factor
Mis16 and Mis18 were originally identified as upstream factors required for CENP-ACnp1 assembly in fission yeast. The temperature-sensitive mutants of Mis16 and Mis18 drastically reduce the total amount of incorporated CENP-ACnp1 in centromeric DNA at non-permissive temperatures and lead to increased frequency of uneven chromosome segregation and aneuploidy (Hayashi et al., 2004). Moreover, both Mis16 and Mis18 are considered to be essential for proper recruitment of other kinetochore components, such as Mis6, Mis15, and Mis17, to centromeres, suggesting that Mis16 and Mis18 are essential for initial kinetochore assembly (Hayashi et al., 2004). Unlike Mis18, whose role is more specific to the centromere, Mis16 and its human orthologs, RbAp46/48, are ubiquitous proteins that perform diverse physiological roles as part of histone regulating protein complexes, including the HAT1, NuRD, NURF, CAF-1, and PRC2 complexes (Loyola and Almouzni, 2004; Philpott et al., 2000; Ruiz-Garcia et al., 1998). A series of immunoprecipitation experiments showed that Mis16 participates in two distinct protein complexes in fission yeast: the Hat1-Mis16 complex and the Mis16-Eic1-Mis18 complex (Hayashi et al., 2014; Subramanian et al., 2014). Mis18 coprecipitated with Mis16, Eic1, and Eic2 (a.k.a. Mis20), but not Hat1. Hat1 only coprecipitated with Mis16. This suggests that Mis16, when already assembled in one complex, cannot simultaneously participate in another.
In this report, we present the crystal structure of the S. pombe Mis16-Eic1-CT complex (Figure 3). The structure shows that Eic1-CT inhabits the same Mis16 binding pocket that is normally occupied by histone H4α1. Our results suggest that either Eic1 or histone H4 can associate with Mis16, but not both (Figure 7A and 7B). Human and Caenorhabditis elegans homologs of Mis16, RbAp46/48 and RbAp46/48Lin53, respectively, also have centromere-specific roles similar to Mis16 (Fujita et al., 2007; Hayashi et al., 2004; Lee et al., 2016). The RNAi-mediated depletion of both RbAp46 and RbAp48 in HeLa cells diminished the centromere localization of CENP-A and led to defects in progression through interphase, raising the possibility that the physiological consequences of Mis16 depletion are conserved in humans (Hayashi et al., 2004). The same group also showed that immunoprecipitates of hMis18α contain both RbAp46 and RbAp48, although the protein levels of RbAp46/48 are not affected by hMis18α depletion (Fujita et al., 2007). RbAp46/48Lin-53 in holocentric C. elegans co-localized with CENP-AHcp3 at centromeres during metaphase regardless of centromere acetylation. In addition, depletion of RbAp46/48Lin-53 by RNAi dramatically reduced CENP-AHcp3 levels at centromeres (Lee et al., 2016). Depletion of RbAp48 in chicken DT40 cells also reduced CENP-A incorporation at centromeres to approximately 40% (Shang et al., 2016). Despite the clear centromere-specific phenotype upon depletion, RbAp46/48 have not been considered part of the hMis18 holo-complex. This is likely because (1) RbAp46/48 are not stably associated with other hMis18 components (Fujita et al., 2007), and (2) RbAp46/48 are thought to be involved in centromere acetylation as a part of the HAT1 complex and not as a part of the Mis18 holo-complex (Fujita et al., 2007). The crystal structure of the Mis16-Eic1-CT complex now suggests that Mis16 performs its centromere-specific function via Eic1 (Figure 7E). A similar mechanism has been observed in Mis16 orthologs, such as human RbAp48 (RbAp48-MTA1) and Drosophila Nurf55 (Nurf55-Su(z)12) (Figure 5B) (Alqarni et al., 2014; Schmitges et al., 2011). In both cases, a small stretch of residues (672–688 of MTA1 and 79–91 of Su(z)12) respectively occupy the histone H4 binding pocket of human RbAp48 and Drosophila Nurf55, thereby redirecting their function to NuRD- and PRC2-specific roles rather than their general histone H4 chaperone function. The crystal structure of the Mis16-Eic1-CT complex demonstrates that Mis16 adopts a similar strategy to switch its function from a general histone H4 chaperone to a centromere-specific factor by associating with Eic1. Taken together, our structure, along with previously available crystal structures (Figure 5B), suggests that Mis16 and its homologs can perform diverse biological functions by accommodating different binding partners within their binding pocket, which is normally occupied by histone H4.
The competition assay and the ITC results suggest that Eic1 has high on and off rates for Mis16 association whereas histone H4 has low on and off rates, even though they both have similar affinity for binding (Figures 7A, 7B and S7). One potential mechanism is that Mis16 may function as a chaperone that delivers Scm3sp-bound CENP-ACnp1/H4, to the Eic1-Mis18 subcomplex. We previously proposed that Mis16 specifically associates with the CENP-ACnp1:H4/Scm3sp complex by recognizing Scm3sp and histone H4 simultaneously (An et al., 2015). In particular, Scm3sp recognizes the Mis18 holo-complex mainly through an interaction mediated by the C-terminal region of Scm3sp with Mis16 (Figure S8). A possible model is that this temporal complex (Mis16-CENP-ACnp1:H4/Scm3sp complex) then docks onto the centromere-specific Eic1-Mis18 subcomplex where Eic1 replaces histone H4 to form the Mis18 holo-complex. Replacing histone H4 with Eic1 may lower the energy barrier required for centromere deposition of CENP-ACnp1 mediated by Scm3sp and Mis18. Similar mechanisms mediated by RbAp46/48 that fine-tune CENP-A deposition may exist in human cells, chicken and C. elegans3 (Fujita et al., 2007; Lee et al., 2016; Shang et al., 2016). Interestingly, in RbAp48-deficient chicken DT40 cells, HJURP, but not the Mis18 complex, fails to properly localize to centromeres, suggesting that RbAp48 likely functions as a CENP-A delivery factor in concert with HJURP in chicken (Shang et al., 2016).
M18BP1 has been shown to associate with centromeres even when CENP-C is depleted, in both Xenopus egg extracts and chicken DT40 cells (French et al., 2017; Hori et al., 2017). M18BP1 in Xenopus, chicken DT40 cells, and Arabidopsis thaliana in fact, directly binds the CENP-A nucleosome through its CENP-C-like motif. However, it has been proposed that such an interaction between M18BP1 and CENP-A is unlikely to be conserved in human and mouse cells where a CENP-C like motif appears to be absent within M18BP1 (French et al., 2017; Hori et al., 2017; Sandmann et al., 2017). In fission yeast, CENP-CCnp3 depletion is not lethal, although cells undergo a high degree of cellular stress (Tanaka et al., 2009). While we were unable to find a CENP-C-like motif within the components of the fission yeast Mis18 holo-complex, we cannot rule out the possibility that the fission yeast Mis18 holo-complex may also directly recognize CENP-ACnp1 nucleosomes.
The crystal structure of the Mis16-Eic1-CT complex reveals the molecular mechanism by which Mis16 switches its function from being a histone H4 chaperone to a centromere-specific assembly factor, by associating with Eic1. Switching its binding partner from histone H4 to Eic1 might be the key role of Mis16 in the Mis18 holo-complex, which then facilitates CENP-ACnp1 loading at centromeres.
STAR★Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents may be directed to, and will be fulfilled by the lead contact Uhn-Soo Cho (uhnsoo@med.umich.edu).
Experimental Model and Subject Details
Insect Cell culture
Sf21 cells and Hi5 cells were cultured at 27°C with 110 rpm in shaker incubator (New brunswork scientific INNOVA44R: 2 inches rotating diameter) in Insect XPRESS media (Lonza) with 100 units/mL of penicillin, 100 μg/mL of streptomycin, and 0.25 μg/mL of Gibco Amphotericin B. Cells were maintained at 0.5 – 3 × 106 cell density.
S. pombe Cell Culture
S. pombe cells were cultured in YES (Yeast Extract Supplemented) or PMG (Pombe Minimal Growth) media supplemented with or without thiamine, at 25°C, 32°C, or 36°C as indicated.
Methods Details
Protein expression and purification
Full-length S. pombe Mis16 and Eic1 were cloned into the pFastBac dual Lic expression vector, respectively, with a HisX6-maltose binding protein (MBP) tag or a HisX6-tag at the N terminus with a Tobacco Etch Virus (TEV) cleavage site to make pFastbac dual Lic MBP-Mis16-His-Eic1. S. pombe Mis18 or Mis16 WT and mutants were cloned into the pFastBac Lic vector with a HisX6-MBP tag using the same approach to generate pFastbac Lic MBP-Mis18 or pFastbac Lic MBP-Mis16. We used Top10 cells (Invitrogen) for the cloning. Baculovirus generation and protein purification were performed as described previously with minor modifications (An et al., 2015). Sf21 cell and Hi5 cells were maintained in Insect-XPRESS media (Lonza). Recombinant bacmids were produced by transformation of pFastBac plasmids into DH10Bac cells (Invitrogen). Low titer viruses were generated by transfecting 1 μg of recombinant bacmids with Cellfectin II reagents (Invitrogen) into 5× 106 cells Sf21 cells in the well of 6 well plate and amplified 2 more rounds to get high titer viruses. To express proteins, Hi5 cells were infected with high titer P3 viruses at a density of 1~2 × 106 cells per ml and further incubated for 44 hours. To purify the Mis18 holo-complex, We did same procedure with Mis16 or Mis16-Eic1 purification except HP-Q anion exchange chromatography which was excluded in Mis18 Holo complex case. The final Mis16-Eic1 complexes, in 30 mM Tris, 100 mM NaCl, 1mM TCEP, was concentrated up to 12 mg/ml using Amicon with a 30 kDa cut off (EMD Millipore) and kept at −80°C until use. Wild-type and mutants of histone H41-48 fused with Sumo at the C terminus as well as Eic199-112 (Eic1-CT) fused with thioredoxin (Trx) at the N terminus were cloned into pET3a Lic vector to generate pET3a Lic H4 1–48-Sumo or pET3a Lic Eic1 99–112-Trx and expressed in Escherichia coli Rosetta (DE3) (Novagen) with auto-inducible media (Studier, 2005). Soluble cell lysates, obtained by centrifugation after sonication, were subjected to cobalt affinity, HP-Q, and Superdex 200 size-exclusion chromatography (Prep grade 10/300, GE Healthcare) for purification. Eic1-CT proteins were kept at 4°C in 30 mM Tris-HCl (pH 8.0), 500 mM NaCl, 50% glycerol, and 1 mM TCEP, and used within 2 weeks.
Crystallization and X-ray data collection
Crystals of full-length S. pombe Mis16 in complex with Eic1 were grown using the hanging drop vapor diffusion method at 20°C by mixing in a 1:1 ratio with a reservoir solution of 100 mM Tris-HCl (pH 8.5) and 25% PEG 2000MME. Crystals obtained after 2.5 months were cryo-protected with additional 17% glycerol and then freshly frozen in liquid nitrogen. Diffraction data were collected on beamline 21ID-G at LS-CAT (Advanced Photon Source, Argonne National Laboratory, USA) at 0.9786 Å wavelength under liquid nitrogen streaming.
Data processing and structure determination
The diffraction data sets were integrated and scaled using HKL2000 (http://www.hkl-xray.com). Initial phases were obtained by molecular replacement with the program PHENIX.Phaser (McCoy et al., 2007) using the S. japonicus Mis16 structure as search model (PDB ID: 4XYH)3 (An et al., 2015). The model was further improved by repeated cycles of manual model building using COOT (Emsley and Cowtan, 2004) and refinement using PHENIX.REFINE (Adams et al., 2010). The final refinement included Translation Libration Screw (TLS) parameters and manual addition of water molecules. In total, residues 15–418 (total 430 residues) and residues 99–112 (total 112 residues) for Mis16 and Eic1 were respectively built in the electron density map. Although the full-length proteins were used for crystallization, only the C-terminal region of Eic1 (Eic1-CT, residues 99–112) was visible likely due to the protein degradation during crystallization process. The final model of the Mis16-Eic1-CT complex has R/Rfree of 0.177/0.219 and the Ramachandran statistics for favored/additional/disallowed are 96.9/2.8/0.3. The final crystal structure was validated with MolProbity (Chen et al., 2010). Crystallographic data statistics are summarized in Table 1. Figures were prepared with PyMOL (The PyMOL Molecular Graphics System, Version 1.8.6.0 Schrödinger, LLC).
Analytical ultracentrifugation
Sedimentation velocity (SV) analytical ultracentrifugation was used to determine the molecular weight as well as the stoichiometry of the Mis18 holo-complex and its sub-complexes. SV analysis was performed using a Beckman Proteome Lab XL-I analytical ultracentrifuge (Beckman Coulter, Indianapolis, IN) equipped with an AN50TI rotor. All samples were prepared in 30 mM Tris-HCl (pH 8.0) containing 100 mM NaCl and 1 mM TCEP and loaded into cells containing standard sector-shaped 2-channel Epon centerpieces with 1.2 cm path-length (Beckman Coulter, Indianapolis, IN). Samples were then allowed to equilibrate at 22°C for 1 hr in the non-spinning rotor prior to sedimentation at 40,000 rpm. Sedimentation was monitored continuously using the absorbance optics, either at 280 nm (0.33 mg/ml Mis18 holo-complex, 0.29 mg/ml Mis16-Eic1 sub-complex, and 0.32 mg/ml Mis16-Eic1-Mis18C complex) or at 292 nm (0.07 mg/ml Mis16-Eic1 sub-complex titrated with MBP-Mis18C).
The SV data was analyzed with the program SEDFIT (version 15.01b) (Schuck, 2000). Sedimentation distribution plots, shown in Figures 1C, 1D, 1F, and 2B, were generated using the continuous c(s) distribution model, with a confidence level for the ME (Maximum Entropy) regularization of 0.7. The corresponding molecular weights including their standard deviation were calculated by integration of a c(M) continuous distribution with the confidence level set to 0.5. The stoichiometry of the Mis16-Eic1-(MBP-Mis18C) complex was verified by titrating 0.07 mg/ml of Mis16-Eic1 sub-complex with a 0.5-, 1-, 4-, and 10-fold molar excess of MBP-Mis18C. The sedimentation distribution plots, c(s) as a function of s, obtained by SEDFIT were integrated and the total area of the Mis16-Eic1-MBP-Mis18C complex was plotted as a function of molar excess of MBP-Mis18C to obtain a binding isotherm. This allowed estimating the number of MBP-Mis18C molecules bound to the Mis16-Eic1 sub-complex. Buffer density as well as viscosity was calculated using SEDNTERP (http://sednterp.unh.edu/).
Amylose resin pull-down assay
MBP-tagged Mis16 and the wild-type and mutants of Trx-Eic1-CT were incubated in a 1:1 molar ratio in the presence of amylose beads (New England Biolab) pre-equilibrated with buffer A [30 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM DTT, and 0.05% NP-40 (w/v)] for 1 hr at 4°C. Amylose beads were washed three times with buffer A and the remaining proteins were subjected to denaturation for SDS-PAGE analysis. For the competition assay, MBP-tagged Mis16 was preincubated with Trx-Eic1-CT for 10 min and then the binding reactions were challenged by increasing amounts of Sumo-tagged histone H41–48 (Sumo-H4α1). The reactions were then incubated with amylose resin in the same manner. The beads were washed 3–4 times with buffer B and bead-bound proteins were applied to SDS-PAGE after denaturation. The experiments were repeated three times. To test binding between Mis18 sub-complexes and Scm3sp190-336, MBP tagged Mis18 sub-complexes and Scm3sp190-336 were incubated in a 1:3 molar ratio. The reactions were then incubated with amylose resin in the same manner. The bead-bound proteins after washing were subjected to SDS-PAGE analysis after denaturation.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) was carried out using a MicroCal VP-ITC calorimeter (Malvern Instruments) with 3 μM of Mis16 in the cell and 30 μM of Sumo, Sumo-Eic1-CT, or H41-48-Sumo as a ligand at 20°C. The final buffer of each sample was exchanged to 30 mM sodium phosphate (pH 8.0) and 100 mM NaCl using the 7K MWCO desalting column (ThermoFisher) right before ITC measurements. The dissociation constants were obtained from ITC thermograms fit to a one-site model using Origin software (OriginLab).
Screening temperature-sensitive mutants
The eic1 gene was randomly mutagenized as previously described (Subramanian et al., 2014) using the GeneMorph II random mutagenesis kit (Agilent Technologies) and fused to either a C-terminal GFP:kanR cassette or a hygR cassette alone in the 3′UTR, through a PCR-based approach. Following transformation of S. pombe cells with mutagenized eic1 constructs (GFP-tagged or untagged), kanamycin- or hygromycin-resistant temperature-sensitive colonies were screened for based on their ability to grow at permissive temperature (25°C) but not at restrictive temperature (36°C). Confirmed temperature-sensitive mutants were then verified by sequencing (F102S, R105I, N106K) and used in further analyses. The eic1 (F102S) (LS762) cells shown here are untagged (Subramanian et al., 2014), whereas eic1 (R105I) (LS1126) and eic1 (N106K) (LS1127) are both C-terminally GFP-tagged. eic1+ (LS760) served as wild-type control.
Mis16 genetic complementation assay
S. pombe Mis16 cDNA (wild-type, Δ1-32, and Δ1-33 or three missense mutations E29A, L32A and W33A) was PCR-amplified and cloned into the pDUAL-GFH41 vector, which allows for expression of C-terminally GFP-tagged Mis16 (wild-type or mutants) under the control of the medium-strength nmt41 promoter, which is induced in the absence of thiamine in the culture media (Matsuyama et al., 2004). Mis16 constructs cloned into the pDUAL-GFH41 vector were then linearized and integrated into the genome at the leu1 locus in mis16-53 (Y41H) (LS863) mutant cells. Five-fold serial dilutions of mis16-53 (Y41H) cells expressing GFP-tagged Mis16 (wild-type or mutants) from the leu1 locus (Figure 7C and 7D) were spotted onto PMG media containing phloxine B supplemented with or without thiamine and incubated at the indicated temperatures for 3–5 days.
Quantification and Statistical Analysis
Band intensities of pull down assay was quantified by imageJ program (Schneider et al., 2012) and normalized relative to that of wild-type. Results are shown as mean ± SEM of three independent measurements (n=3). Differences of binding affinity were analyzed by a Student’s t-test. (*P < 0.05, ** P < 0.005, ***P < 0.0005, **** P < 0.0001.)
Data and Software Availability
Accession codes
The coordinates and the structure factor for the reported crystal structure of the Mis16-Eic1-CT complex have been deposited in the Protein Data Bank under the ID code 5WJC.
Supplementary Material
HIGHLIGHTS.
The stoichiometry of the fission yeast Mis18 complex is (Mis16)2:(Eic1)2:(Mis18)4
The crystal structure of the Mis16-Eic1-CT complex was solved at 2.3Å resolution
Eic1 and histone H4 share the same binding pocket and are thus, mutually exclusive
Once associated with Eic1, Mis16 assumes a centromere-specific role
Acknowledgments
We thank the staff at the Advanced Photon Source LS-CAT beamlines for their advice and assistance with data collection, Mr. Hu Lee for help with protein expression and purification, Ms. Andrea Langella for technical assistance, Prof. Mitsuhiro Yanagida for the mis16-53 (Y41H) ts strain, and Dr. Marie Ary for help with editing. We thank Dr. James C. A. Bardwell for the use of his analytical ultracentrifuge and funding of his graduate student Philipp Koldewey who received support from the Howard Hughes Medical Institute of which JCAB is an investigator. We also thank Prof. Robin Allshire in whose lab initial parts of the in vivo work were performed. This work was supported by grants (1-16-JDF-017, N019154-00, AG050903 and DK111465) to U-S.C, and a QMUL start-up grant and BBSRC grant BB/R00868X/1 to L.S. J.C. was supported by the NIH Cellular and Molecular Biology Training Grant T-32-GM007315.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
Conceptualization, S.J.A. and U.S.C.; Methodology, S.J.A., P.K., J.C., L.S., and U.S.C.; Investigation, S.J.A., L.S., and U.S.C.; Writing – Original Draft, S.J.A., J.C., and U.S.C.; Writing – Review & Editing, S.J.A., P.K., J.C., L.S., and U.S.C.; Funding Acquisition, L.S., and U.S.C.; Visualization, S.J.A., J.C., and L.S.; Supervision, L.S. and U.S.C.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqarni SS, Murthy A, Zhang W, Przewloka MR, Silva AP, Watson AA, Lejon S, Pei XY, Smits AH, Kloet SL, et al. Insight into the architecture of the NuRD complex: structure of the RbAp48-MTA1 subcomplex. J Biol Chem. 2014;289:21844–21855. doi: 10.1074/jbc.M114.558940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Choo KH. Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet. 2002;71:695–714. doi: 10.1086/342730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Kim H, Cho US. Mis16 Independently Recognizes Histone H4 and the CENP-ACnp1-Specific Chaperone Scm3sp. J Mol Biol. 2015;427:3230–3240. doi: 10.1016/j.jmb.2015.08.022. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Baum MP. Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences. Mol Cell Biol. 1990;10:1863–1872. doi: 10.1128/mcb.10.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rop V, Padeganeh A, Maddox PS. CENP-A: the key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma. 2012;121:527–538. doi: 10.1007/s00412-012-0386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- French BT, Westhorpe FG, Limouse C, Straight AF. Xenopus laevis M18BP1 directly binds existing CENP-A nucleosomes to promote Ccentromeric chromatin assembly. Dev Cell. 2017;42:190–199. e110. doi: 10.1016/j.devcel.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Ebe M, Nagao K, Kokubu A, Sajiki K, Yanagida M. Schizosaccharomyces pombe centromere protein Mis19 links Mis16 and Mis18 to recruit CENP-A through interacting with NMD factors and the SWI/SNF complex. Genes Cells. 2014 doi: 10.1111/gtc.12152. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Hirai H, Arai K, Kariyazono R, Yamamoto M, Sato M. The kinetochore protein Kis1/Eic1/Mis19 ensures the integrity of mitotic spindles through maintenance of kinetochore factors Mis6/CENP-I and CENP-A. PLoS One. 2014;9:e111905. doi: 10.1371/journal.pone.0111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang WH, Hara M, Ariyoshi M, Arimura Y, Fujita R, Kurumizaka H, Fukagawa T. Association of M18BP1/KNL2 with CENP-A nucleosome is essential for centromere formation in non-mammalian vertebrates. Dev Cell. 2017;42:181–189. e183. doi: 10.1016/j.devcel.2017.06.019. [DOI] [PubMed] [Google Scholar]
- Lee BC, Lin Z, Yuen KW. RbAp46/48(LIN-53) Is Required for Holocentromere Assembly in Caenorhabditis elegans. Cell Rep. 2016;14:1819–1828. doi: 10.1016/j.celrep.2016.01.065. [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Corbett KD, Desai A. Structure, assembly and reading of centromeric chromatin. Curr Opin Genet Dev. 2012;22:139–147. doi: 10.1016/j.gde.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Shirai A, Yashiroda Y, Kamata A, Horinouchi S, Yoshida M. pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast. 2004;21:1289–1305. doi: 10.1002/yea.1181. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Localization of centromere function in a Drosophila minichromosome. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi IK, Zasadzinska E, Stellfox ME, Knippler CM, Foltz DR. Licensing of Centromeric Chromatin Assembly through the Mis18alpha-Mis18beta Heterotetramer. Mol Cell. 2016;61:774–787. doi: 10.1016/j.molcel.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Klare K, Petrovic A, Take A, Walstein K, Singh P, Rondelet A, Bird AW, Musacchio A. CDK-regulated dimerization of M18BP1 on a Mis18 hexamer is necessary for CENP-A loading. Elife. 2017;6 doi: 10.7554/eLife.23352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott A, Krude T, Laskey RA. Nuclear chaperones. Semin Cell Dev Biol. 2000;11:7–14. doi: 10.1006/scdb.1999.0346. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Garcia AB, Sendra R, Galiana M, Pamblanco M, Perez-Ortin JE, Tordera V. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J Biol Chem. 1998;273:12599–12605. doi: 10.1074/jbc.273.20.12599. [DOI] [PubMed] [Google Scholar]
- Sandmann M, Talbert P, Demidov D, Kuhlmann M, Rutten T, Conrad U, Lermontova I. Targeting of Arabidopsis KNL2 to Centromeres Depends on the Conserved CENPC-k Motif in Its C Terminus. Plant Cell. 2017;29:144–155. doi: 10.1105/tpc.16.00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KC, Sullivan BA. Neocentromeres: a place for everything and everything in its place. Trends Genet. 2014;30:66–74. doi: 10.1016/j.tig.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang WH, Hori T, Westhorpe FG, Godek KM, Toyoda A, Misu S, Monma N, Ikeo K, Carroll CW, Takami Y, et al. Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nat Commun. 2016;7:13465. doi: 10.1038/ncomms13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller F, Medina-Pritchard B, Abad MA, Wear MA, Molina O, Earnshaw WC, Jeyaprakash AA. Molecular basis for Cdk1-regulated timing of Mis18 complex assembly and CENP-A deposition. EMBO Rep. 2017;18:894–905. doi: 10.15252/embr.201643564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellfox ME, Nardi IK, Knippler CM, Foltz DR. Differential Binding Partners of the Mis18alpha/beta YIPPEE Domains Regulate Mis18 Complex Recruitment to Centromeres. Cell Rep. 2016;15:2127–2135. doi: 10.1016/j.celrep.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Subramanian L, Medina-Pritchard B, Barton R, Spiller F, Kulasegaran-Shylini R, Radaviciute G, Allshire RC, Arockia Jeyaprakash A. Centromere localization and function of Mis18 requires Yippee-like domain-mediated oligomerization. EMBO Rep. 2016;17:496–507. doi: 10.15252/embr.201541520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Toda NR, Rappsilber J, Allshire RC. Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP-A assembly. Open Biol. 2014;4:140043. doi: 10.1098/rsob.140043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Chang HL, Kagami A, Watanabe Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell. 2009;17:334–343. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Warburton PE. Epigenetic analysis of kinetochore assembly on variant human centromeres. Trends Genet. 2001;17:243–247. doi: 10.1016/s0168-9525(01)02283-1. [DOI] [PubMed] [Google Scholar]
- Willard HF. Centromeres of mammalian chromosomes. Trends Genet. 1990;6:410–416. doi: 10.1016/0168-9525(90)90302-m. [DOI] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.