Abstract
Serine proteases are one of the largest groups of enzymes, found in both eukaryotes and prokaryotes, and are responsible for many different functions. The detailed information about the hydrogen-bonds in the catalytic triad (Asp…His…Ser) of these enzymes is of importance in order to fully understand the mechanism of action. The aspartate of the triad is hydrogen bonded to the histidine but the exact nature of this bond has been under discussion for some time. It is either a common short ionic hydrogen bond (SIHB) or a delocalized low barrier hydrogen bond (LBHB) were the hydrogen bond is shorter. So far, the evidence for LBHB in proteins have not been conclusive. Here we show clear NMR evidence that LBHB does exist in NS3, a serine protease from Dengue. The one bond coupling constant between the hydrogen and nitrogen was shown to be only 52 Hz instead of the usual 90 Hz. This together with a 1H chemical shift of 19.93 ppm is evidence that the hydrogen bond distance between His and Asp is shorter than for SIHB. Our result clearly shows the existence of LBHB and will help in understanding the mechanism of the catalytic triad in the important group of serine proteases.
Introduction
The incidence of dengue virus (DENV) has grown around the world in recent decades and constitutes a major threat to human health. With increased infection rates, aided by global warming, the growth of urban areas, travel and trade, the virus is now endemic in more than 100 countries and not only restricted to tropical and subtropical regions thus making it an important subject for detailed studies.
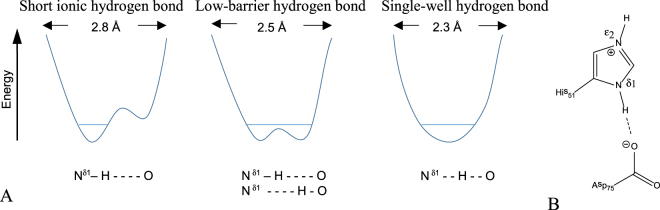
NS3 of Dengue type II (DENV2) virus is a serine protease and belongs to the large and functionally diverse group of proteolytic enzymes. Serine proteases continue to be an inspiration for mechanistic studies of enzyme catalysis and intensive efforts have been made to address questions regarding the mechanism of the serine protease catalysis1,2. One of the common features of serine proteases is the catalytic active site which consist of the triad of aspartic acid, histidine, and serine residues, Asp…His…Ser, in an interactive mode. Still, the nature of the hydrogen bond (HB) stabilization in the catalytic triad is the subject of discussion. There remains the question whether the Asp…His hydrogen bond in the catalytic triad is associated with a normal short ionic hydrogen bond (SIHB)3,4 or with a delocalized resonance effect called a low-barrier hydrogen bond (LBHB) or with a single well hydrogen bond (Fig. 1)5–7. Theories, such as the LBHB hypothesis, are inherently difficult to prove experimentally in biomolecules due to the very short-lived nature of their transition states. Furthermore, these controversies also arise from inadequate characterization of H-bonds in proteins.
Figure 1.
(A) Energy profiles for the possible hydrogen bonds for His…Asp. (B) The hydrogen bond between HNδ1 of His51 and Asp75.
The type of hydrogen bond is of importance to our understanding of the structure of the tetrahedral transition state and the possible intermediates which are necessary for a functioning enzyme. There are multiple experimental demonstrations of the transition state stabilization by HBs where a tetrahedral intermediate has been observed by a series of intermediate trapping experiments8,9. The LBHB proponents and opponents have shown several experimental observations which could be categorized into the following groups: (a) unusual low-field chemical shift (CS) of protons involved in H-bonding3,5,6,8 (b) measurement of H-bonding length by ultra-x-ray10 (c) PK studies using 15N and 13C nucleus11 (d) H/D isotope effect studies12.
Characteristic shifts in NMR chemical shielding are a well-known signature of H-bonding. The primary focus of most of the studies has been on the NMR chemical shielding effects of the catalytic His aromatic nuclei; 1H, 15N and 13C. Unusual large low- field shift of the Nδ1H protons, for single-well H-bonds (20–22 ppm) and for LBHB (17–19 ppm) has been proposed as one of the main criteria for the formation of a short, strong hydrogen bond under protonation of the catalytic His in complex with Asp in the intermediate induced complexes of serine type proteases with inhibitors5. This has been confronted in many theoretical4,13 and experimental studies3,14. It was argued that the large down field shifts that is seen for the proton between His….Asp4 is equally consistent with both the LBHB and the SIHB15. Diffraction experiments carried out using X-rays or neutrons give a direct image of the atomic positions, including that of the hydrogen atoms, however, hydrogen atoms are only visible when crystal diffraction data are at ultra-high resolution (i.e. better than 1 Å) and where the atoms are sufficiently well ordered10. The coordinate uncertainty should be much less than 0.1 Å in order for a reliable detection of geometric changes needed to distinguish between different types of H-bonds10.
Scalar through-H- bond J-coupling 1JAH, 2JAHB in A-H···B have been shown to provide an important window on H-bonding by directly exhibiting the intermolecular electronic delocalisation and ‘communication’ that underlies H-bond formation16–18. There has been shown a direct correlation between 1,2J-coupling, the low field chemical shift of proton and the length of the H-bond in organic molecules and DNA17,19,20. Unfortunately, experimental values of 1J-coupling are often not available for many H-bonded system of biological interest. This means that the currently applied experimental diagnostic of H-bonding criteria in the catalytic triad is generally incomplete and missing the direct experimental evidence of the one bond 1JNH coupling constant from NMR data. Furthermore, those few available NMR studies where the 1J-coupling in the catalytic Asp…His was observed demonstrated that all the measured values of the one bond 1JNH lay between 87 and 95 Hz and that the proton was localized on Nδ1H for at least 85%21.This fact was used as convincing evidence against the LBHB theory2 where much smaller 1JNH coupling are predicted14.
The virally encoded serine protease of Dengue virus serotype, type 2 (DENV2) lies in the N-terminal domain of NS3, with NS2B serving as a cofactor in this dimeric protease. Full length NS2B is a membrane protein, but in vitro a hydrophilic segment of 40-residues is sufficient to form an active NS2B-NS3pro complex22.
In the presence of ligand, the DENV NS2B C-terminus associates with NS3pro such that it lines the active site. This NS2B conformation is referred to as the ‘closed’ conformation, and is distinctly different from that observed for ligand free protease structures23–25. The C-terminal part of NS2B has been shown essential for proteolytic activity in both DENV26, WNV27,28 and Zika29 therefore the closed conformation is thought to be the enzymatically active structure.
The X-ray structure obtained so far of the active conformation of the DENV3 NS2B-NS3pro complexes (67% sequence identity with DENV2) with inhibitor have not had enough resolution to measure hydrogen bonds30. A low resolution modelled structure based on NMR data was recently reported, again it is not possible to say anything about the possible presence of LBHB31. The x-ray structures from related viruses are also lacking the resolution necessary to estimate the H-bond distance between His and Asp in the catalytic triad27–29. It is also not clear if the structures obtained from all these x-ray investigations shows an active conformation in which one would expect to observe LBHB. An important suggestion from the study of West Nile virus NS3 is the proposal that the active site of NS3 undergoes an induced fit involving the substrate and histidine28. This could explain the lack of LBHB in all apo forms of NS3.
Results and Discussion
Here we present experimental NMR evidence, which to the best of our knowledge, is the first time that the existence of a LBHB type H-bond for the Asp…His interaction is found in the complex of DENV2 with a substrate-analogue boronic acid mimicking the tetrahedral transition complex. Binding of the Bz-Nle-Lys-Arg-Arg-B(OH)2, to the NS3:NS2Bpro complex leads to significant changes in the NMR spectra of the complex32. In Fig. 2 1H low field spectra with or without 15N and/or 13C decoupling of the complex 15N/13C-NS3:NS2Bpro with Bz-Nle-Lys-Arg-Arg-B(OH)2 with pH varying between 5.5–8.5 are presented. There are four signals observed between 20–19 ppm and two signals around 15.57 ppm. One should note that in the absence of the ligand, no resonances are observed between 20 and 13 ppm. The pH dependent studies of the complex were performed in order to investigate any changes in pKa coming from changes in the protonation of the histidine18,33,34. A histidine not hydrogen bonded is expected to show chemical shift dependence on pH whereas histidine bound in a complex do not18,33,34. All three sets of signals (doublets in Fig. 2) in the 20–14 ppm region do not show any pH-dependence on their chemical shift and thus remain fully protonated and H-bonded over the investigated pH range.
Figure 2.
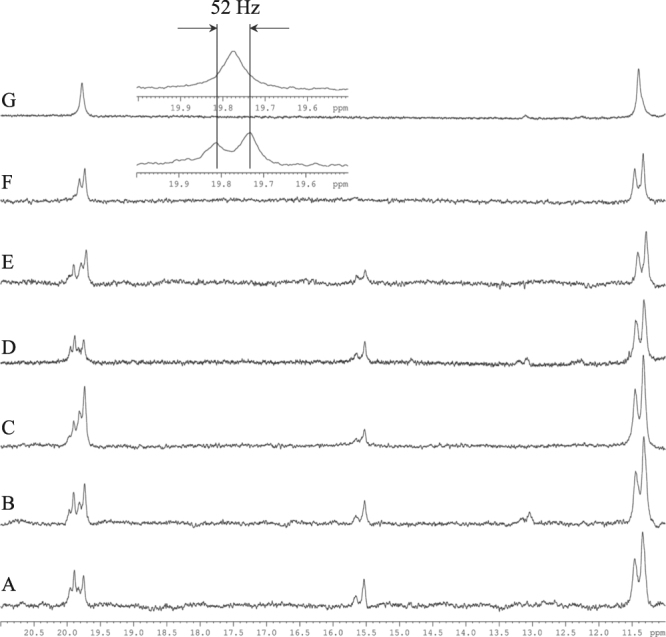
1D 1H spectra of the region 11–21ppm of the complex 15N/13C labelled NS3-NS2Bpro with, Bz-Nle-Lys-Arg-Arg-B(OH)2, without 15N/13C decoupling at different pH: (A) 5.5, (B) 6.0, (C) 6.5, (D) 7.2, (E) 8.5 all in MES buffer. In (F) pH 8.5 in Tris buffer and (G) unlabeled NS3 in the same complex at pH 8.5 in Tris buffer. The small insert shows Nδ1H of His51 at 19.772 ppm with the one bond J-coupling 1JNδ1H = 52 Hz indicated.
When decoupling of 15N and/or 13C nuclei is applied or when only the co-factor NS2B was uniformly 15N/13C labelled, the resonances are reduced to two signals at 19.933 ppm (JNH coupling = 52 Hz), 19.772 ppm (JNH coupling = 52 Hz) and one at 15.57 ppm (JNH coupling = 90 Hz) (Figs 2, S1 and S2).
This means that those signals belong to one of the aromatic NH residues of NS3pro. A spectra of the mutation of His51Asn of NS3pro in complex with NS2B and the boronic ligand does not show any extreme low field signals at 19.933, 19.772 and 15.57 ppm thus confirming the assignment of His51, see Fig. S3 in supplementary materials.
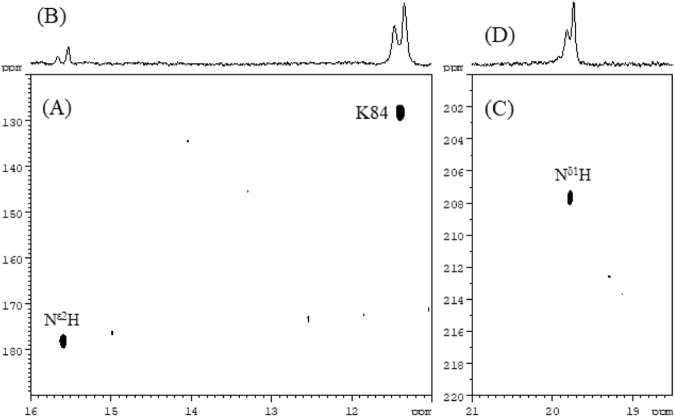
For the signal at 19.772 ppm we were able to detect a one bond 1H and 15N correlation cross peak (15N: 207.9ppm) in the TROSY type 1H 15N HSQC spectrum (Fig. 3) which allows us to unambiguously assign it to Nδ1H of His 51. For the signal at 15.57ppm 1H and 15N correlation cross peak also detected with 15N: 178.7ppm (Fig. 3) which assigned to Nε2H of His 51. The above assignment is based on the x-ray structures of serine proteases showing a possible hydrogen bonding between Nδ1 of a histidine and Asp. For the other resonance at 19.933ppm we failed to observe any 1H and 15N cross peak possibly due to different NH coupling constant (52 Hz instead of 90 Hz), line broadening and the lower concentration of the second form of the complex.
Figure 3.
Expanded 2D plots (A) and (C) of 1H-15N HSQC TROSY type spectra of the complex 5N/13C labelled NS3-NS2Bpro with Bz-Nle-Lys-Arg-Arg-B(OH)2, showing one bond 1H-15N correlation with corresponding 1D 1H spectrum presented on top (B) and (D) of the 2D spectra. In expansion (A) amide cross peak of amino acid K84 at 11.396/127.85ppm is shown as reference. Cross peak at 15.587/178.7ppm is assigned to the Nε2H of His51. In expansion (C) cross peak at 19.772/207.6ppm is assigned to the Nδ1H of His51 of the other form.
Note that there seems to be an equilibrium between two different types of complexes of the catalytic triad depending on external conditions (Fig. 2). One form, with the most intense signals at 19.772, is the most persistent in all spectra at different buffers and pH. The other one has signals at 19.933 and 15.57. This will be discussed more below.
To rule out the contribution of the 2JC-H scalar coupling to the observable splitting we have also examined 15N-NS3:NS2Bpro with Bz-Nle-Lys-Arg-Arg-B(OH)2 complex: still the coupling constant of the signals at 19.933 ppm and 19.772 ppm were 52 Hz. The coupling is about 38 Hz less than commonly reported for catalytic HN His in serine proteases which claimed to be in range 87–95Hz14. Another striking feature is that signals at 19.933 ppm and 19.772 ppm assigned by us to Nδ1H protons of the His51 in the catalytic triad are observed at almost 1.0 ppm more downfield than so far reported for either Nε2H or Nδ1H protons in protonated His induced by inhibitors9,14. However, the observed chemical shifts and one-bond coupling constants are the expected ones in LBHB if one extrapolates the results of a study of chemical shifts and coupling constants in the catalytic triad of NS3 in flaviviruses23. Only in the complex with trifluoromethyl ketone has a downfield shift at 18.9 ppm been observed and this was the main reason to claim the presence of the LBHB5. The ratio of the integrals of those three signals varies with different pH: at pH 6.5 the ratio was 0.25:0.5:0.25 correspondingly, but at pH 5.5 and 7.2 it was close to 0.3:0.3:0.3. As mentioned above, it indicates that there are an equilibrium between different forms of binding of the boronic type of inhibitor. Resonances at 19.933p pm and 15.57 ppm belong to the His of the same complex. Remarkably, the resonance at 15.57 ppm assigned to Nε2H proton and 19.933 ppm to belong to the same protonated His 51 but have different 1JNH couplings of 90 Hz and 52 Hz, respectively. Furthermore, depending on the sample preparation, one or more complex forms could be observed. As an example, in Tris buffer at pH 8.5 only one down field signal is observed, at 19.772 ppm, but the two other ones are reduced below the detection limit (Fig. 2). The difference between the two complex forms is not clear to us yet but they both show the same 1JNH coupling of Nδ1H of 52 Hz and thus LBHB.
The reason that we don’t think that the observed effects are solely linked to the presence of boron are the following: boronic type ligands are regularly used in the studies of serine proteases in order to mimic the intermediate complex. No unusual structural and functional features of the complexes have been reported in these studies35–37. The hydrogens of the His in the catalytical triad are more down field than usual in these complexes, but not to extent reported by us, and the coupling constants are either ca 90 Hz or not reported. This indicates that the presence of the boron does not in itself induce any major conformational change. Subsequently we do not think that the boronic ligand is the main reason for our down field shifts and coupling constant. Instead we believe that the reason is that we managed to trap a true mimic of the otherwise short living intermediate of the active complex due to the presence of the NS2B co-factor.
In conclusion from the above presented results it becomes clear that the small 1JNH coupling of 52 Hz of Nδ1H of His 51 indicates that the proton in the HB is located almost equidistant between Asp75 and His51 in the complex of NS3:NS2B:ligand. This result is the first evidence that serine proteases, at least for Dengue Type II, exhibit LBHB or possibly even single-well HB between the catalytic aspartate and histidine in the active site as proposed by Frey et al.5,38. Without the ligand or in free NS3, we have not observed any evidence of hydrogen bonding between His51 and Asp75. This is consistent with the induced fit proposal in the study of West Nile virus NS3 mentioned above. Our result obtained for the investigated construct indicates that we here have a very close mimic to the real transition-state of an active enzyme. Our reported findings will, together with more structural work on the complex NS3:NS2B:ligand, facilitate the development of rational structure based inhibitors that can selectively target the NS3 protease of Dengue type II (DENV2) virus. Further studies of the structure and dynamics of the complex presented here, including the two forms of His51, are ongoing.
Methods
Protein expression and purification
Reagents were from Sigma (St. Louis, MO, USA) unless otherwise stated. DENV2 NS3pro (1–185; amino acids 1476–1660 of the polyprotein) and NS2B (containing amino acids 1394–1440 of the Dengue 2 polyprotein) constructs were generated as described32. Proteins were expressed in Terrific Broth medium (MP Biomedicals) for unlabelled protein or in different isotopic labelling combinations in 1/2H, 15N, 12/13C-labelled M9 medium for labelled protein. In this study we mainly used 15N/13C labelled NS3 and unlabelled NS2B to form the complex as we wanted to focus on the catalytic triad and maximize the amount information. His51Asn mutation of the active site residue of NS3pro was introduced using the QuikChange Lightning kit (Agilent). All sequences were confirmed by Sanger sequencing.
Chemicals for isotope labelling (ammonium chloride, 15N (99%), D-glucose, 13C (99%), deuterium oxide) were purchased from Cambridge Isotope Laboratories, Inc.
Purification
NS2B and NS3pro were co-refolded by one-step dialysis overnight at 4 °C in a 2:1 molar NS2B:NS3pro ratio to maximize formation of the active complex. The refolding buffer was 25 mM Tris pH 8.5 (pH set at 4 °C), 5% glycerol, 100 mM NaCl. Thrombin (GE Healthcare) and/or TEV protease was added to a dialysis cassette (3,500 or 7,000 MWCO Slide-A-Lyzer, Thermo Fisher Scientific) to cleave off the His tag from NS2B and/or NS3pro. After refolding the solution was centrifuged at 50,000 × g to remove any precipitate or particles. Refolding yield was determined by measuring protein concentration of the two IMAC pools (NS2B: ε 5,500, MW 7.7 kDa; NS3pro: ε 36,400, MW 21.0 kDa) before refolding and comparing that to the protein concentration after refolding and centrifugation (complex: ε 41,940, MW 28.7 kDa), using a Nanodrop 1000 instrument (Thermo Scientific). The complex was then purified on an ÄKTA Explorer (GE Healthcare) by size exclusion on a HiLoad Superdex 200 column (GE Healthcare) in SEC buffer: 50 mM Tris pH 8.5 (4 °C), 5% glycerol, 50 mM NaCl.
Protease inhibitors
The NS3pro inhibitors Bz-Nle-Lys-Arg-Arg-B(OH)2 used in this study was synthesized according to the reaction schemes published in the original paper32.
Preparation of NMR samples
The NS2B-NS3pro complex was concentrated in disposable centrifugal concentrators (e.g. Amicon Ultra centifugal filter units) with a molecular weight cut-off of 10 kDa. The complex was stable during concentration and no leakage of NS2B occurred. Buffer was exchanged using gravity flow desalting columns (GE Healthcare). The MES NMR buffer contained 20 mM deuterated MES, 100 mM NaCl, 5 mM CaCl2, 0.02% NaN3, at pH 5.5, 6.0, 6.5, 7.2 or 8.5. The Tris NMR buffer contained 20 mM deuterated Tris, 100 mM NaCl, 5 mM CaCl2, 0.02% NaN3, at pH 8.5. The buffer-exchanged protein was concentrated to at least 0.3 mM.
Complexes of trifluoromethyl ketone peptidic inhibitor with other type of serine proteases have shown a downfield 1 H shift at 18.9 ppm and that has been used as the main reason to claim the presence of LBHB5 in the catalytic triad. We have observed similar behavior in Dengue NS3pro:NS2B complex with the same inhibitor (data not shown)32. To confirm that the boronic peptidic inhibitor used in this study binds in the same binding site as trifluoromethyl ketone peptidic inhibitor we used the following procedure.
The ternary NS2B-NS3pro-inhibitor complex was prepared in two steps. Firstly, the inhibitor 2,6-di-fluoro-Bz-Nle-Lys-Arg-Arg-CF3-ketone was titrated to a final concentration of 1 mM by adding 3 μL to the NS2B-NS3pro sample from a 100 mM stock solution in D2O. The formation of bound complex was monitored by the reduction of the 19F signal of unbound inhibitor and appearance of the broad signal corresponding to bound inhibitor. Secondly, the Bz-Nle-Lys-Arg-Arg-B(OH)2 inhibitor was added to the sample to replace the Bz-Nle-Lys-Arg-Arg-CF3-ketone inhibitor, which was monitored over time until the 19F signal representing the bound inhibitor had disappeared and the 19F signal of unbound inhibitor had fully reappeared.
NMR spectroscopy
NMR experiments were acquired on Bruker Avance III spectrometers operating at 14.1 and 16.4 T at a temperature of 298 K. Backbone assignment was performed as described previously32. The 1D 1H experiments were performed using the standard Bruker parameters for zgesp with a sweep width of 35 ppm. For one bond correlation 2D 1H-15N TROSY experiments (trosyetf3gpsi.2) have been used again with standard parameters and extended sweep widths: 35 ppm for 1H and 160 ppm for 15N. 1H chemical shifts are in ppm downfield from DSS. 15N chemical shifts are in ppm vs standard reference, liquid NH3.
Electronic supplementary material
Acknowledgements
The authors thank Dr, Esmeralda Woestenenk for supplying the constructs.
Author Contributions
Peter Agback and Tatiana Agback contributed equally to this work.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28441-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hedstrom L. Serine Protease Mechanism and Specificity. Chem. Rev. 2002;102:4501–4523. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 2.Polgár L. The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 2005;62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ash EL, Sudmeier JL, De Fabo EC, Bachovchin WW. A low-barrier hydrogen bond in the catalytic triad of serine proteases? Theory versus experiment. Science. 1997;278:1128–1132. doi: 10.1126/science.278.5340.1128. [DOI] [PubMed] [Google Scholar]
- 4.Schutz CN, Warshel A. The Low Barrier Hydrogen Bond (LBHB) Proposal Revisited: The Case of the Asp… His Pair in Serine Proteases. PROTEINS: Struct., Funct., Bioinf. 2004;55:711–723. doi: 10.1002/prot.20096. [DOI] [PubMed] [Google Scholar]
- 5.Frey PA, Whitt SA, Tobin JB. A Low-Barrier Hydrogen Bond in the Catalytic Triad of Serine Proteases. Science. 1994;264:1927–1930. doi: 10.1126/science.7661899. [DOI] [PubMed] [Google Scholar]
- 6.Cleland WW. Low-barrier hydrogen bonds and enzymatic catalysis. Adv. Phys. Org. Chem. 2010;44:1–17. doi: 10.1006/abbi.2000.2011. [DOI] [PubMed] [Google Scholar]
- 7.Graham JD, Buytendyk AM, Wang D, Bowen KH, Collins KD. Strong, Low-Barrier Hydrogen Bonds May Be Available to Enzymes. Biochemistry. 2014;53:344–349. doi: 10.1021/bi4014566. [DOI] [PubMed] [Google Scholar]
- 8.Bao D, Huskey WP, Kettner CA, Jordan F. Hydrogen Bonding to Active-Site Histidine in Peptidyl Boronic Acid Inhibitor Complexes of Chymotrypsin and Subtilisin: Proton Magnetic Resonance Assignments and H/D Fractionation. J. Am. Chem. Soc. 1999;121:4684–4689. doi: 10.1021/ja990180g. [DOI] [Google Scholar]
- 9.Lin J, Cassidy CS, Frey PA. Correlations of the Basicity of His 57 with Transition State Analogue Binding, Substrate Reactivity, and the Strength of the Low-Barrier Hydrogen Bond in Chymotrypsin. Biochemistry. 1998;37:11940–11948. doi: 10.1021/bi980278s. [DOI] [PubMed] [Google Scholar]
- 10.Hosur MVR, et al. Low-barrier hydrogen bonds in proteins. Crystallography Reviews. 2013;19(1):3–50. doi: 10.1080/0889311X.2013.771633. [DOI] [Google Scholar]
- 11.Everill P, Sudmeier JL, Bachovchin WW. Direct NMR Observation and pKa Determination of the Asp102 Side Chain in a Serine Protease. J. Am. Chem. Soc. 2012;134:2348–2354. doi: 10.1021/ja210091q. [DOI] [PubMed] [Google Scholar]
- 12.Westler WM, et al. Evidence for a strong hydrogen bond in the catalytic dyad of transition state analogue inhibitor complexes of chymotrypsin from proton-triton NMR isotope shifts. J. Am. Chem. Soc. 2002;124:4196–4197. doi: 10.1021/ja017860f. [DOI] [PubMed] [Google Scholar]
- 13.Lao K-U, Lankau T, Fang T-I, Zou J-W, Yu C-H. Interstitial Water and the Formation of Low Barrier Hydrogen Bonds: A Computational Model Study. International Journal of Quantum Chemistry. 2012;112:1460–1472. doi: 10.1002/qua.23140. [DOI] [Google Scholar]
- 14.Bachovchin WW. Contributions of NMR spectroscopy to the study of hydrogen bonds in serine protease active sites. Magn. Reson.Chem. 2001;39:S199–S213. doi: 10.1002/mrc.951. [DOI] [Google Scholar]
- 15.Chaudret R, Cisneros GA, Parisel O, Piquemal J-P. Unraveling Low-Barrier Hydrogen Bonds in Complex Systems with a Simple Quantum TopologicalCriterion. Chem. Eur. J. 2011;17:2833–2837. doi: 10.1002/chem.201002978. [DOI] [PubMed] [Google Scholar]
- 16.Grzesiek S, Cordier F, Jaravine V, Barfield M. Insights into biomolecular hydrogen bonds from hydrogen bond scalar coupling. Prog. Nucl. Magn. Reson. Spectrosc. 2004;45:275–300. doi: 10.1016/j.pnmrs.2004.08.001. [DOI] [Google Scholar]
- 17.Pietrzak M, et al. Symmetrization of Cationic Hydrogen Bridges of Protonated Sponges Induced by Solvent and Counteranion Interactions as Revealed by NMR Spectroscopy. Chem. Eur. J. 2010;16:1679–1690. doi: 10.1002/chem.200902259. [DOI] [PubMed] [Google Scholar]
- 18.Schubert M, et al. Probing electrostatic interactions along the reaction pathway of a glycoside hydrolase: histidine characterization by NMR spectroscopy. Biochemistry. 2007;46:7383–7395. doi: 10.1021/bi700249m. [DOI] [PubMed] [Google Scholar]
- 19.Smirnov SN, et al. Hydrogen/Deuterium isotope effects on the NMR chemical shifts and geometries of intermolecular low-barrier hydrogen-bonded complexes. J. Am. Chem. Soc. 1996;118:4094–4101. doi: 10.1021/ja953445+. [DOI] [Google Scholar]
- 20.Barfield M, Dingley AJ, Feigon J, Grzesiek S. A DFT study of the interresidue dependencies of scalar J-coupling and magnetic shielding in the hydrogen-bonding regions of a DNA triplex. J. Am. Chem. Soc. 2001;123:4014–4022. doi: 10.1021/ja003781c. [DOI] [PubMed] [Google Scholar]
- 21.Halkides CJ, Wu YQ, Murray CJ. Biochemistry. 1996;35:15941–15948. doi: 10.1021/bi961805f. [DOI] [PubMed] [Google Scholar]
- 22.Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erbel P, et al. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 24.Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandramouli S, et al. Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J. Virol. 2010;84:3059–3067. doi: 10.1128/JVI.02044-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yusof R, Clum S, Wetzel M, Krishna Murthy HM, Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage substrates with dibasic amino acids in vitro. J. Biol. Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 27.Radichev I, et al. Structure-based mutagenesis identifies important novel determinants of the NS2B cofactor of the West Nile virus two-component NS2B-NS3 proteinase. J. Gen. Virol. 2008;89:636–641. doi: 10.1099/vir.0.83359-0. [DOI] [PubMed] [Google Scholar]
- 28.Robin G, et al. Structure of West Nile virus NS3 protease: ligand stabilization of the catalytic conformation. J. Mol. Biol. 2009;385:1568–1577. doi: 10.1016/j.jmb.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, et al. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science. 2016;354:1597–1600. doi: 10.1126/science.aai9309. [DOI] [PubMed] [Google Scholar]
- 30.Noble CG, Seh CC, Chao AT, Shi PY. Ligand-bound structures of the dengue virus protease reveal the active conformation. J. Virol. 2012;86:438–446. doi: 10.1128/JVI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W-N, et al. Sensitive NMR Approach for Determining the Binding Mode of Tightly Binding Ligand Molecules to Protein Targets. J. Am. Chem. Soc. 2016;138:4539–4546. doi: 10.1021/jacs.6b00416. [DOI] [PubMed] [Google Scholar]
- 32.Woestenenk E, Agback P, Unnerståle S, Henderson I, Agback T. Co-refolding of a functional complex of Dengue NS3 protease and NS2B co-factor domain and backbone resonance assignment by solution NMR. Protein Expression and Purification. 2017;140:16–27. doi: 10.1016/j.pep.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Tsilikounas E, Rao T, Gutheil WG, Bachovchin WW. 15N and 1H NMR spectroscopy of the catalytic histidine in chloromethyl ketone-inhibited complexes of serine proteases. Biochemistry. 1996;35:2437–2444. doi: 10.1021/bi9513968. [DOI] [PubMed] [Google Scholar]
- 34.Harris TK, Turner GJ. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 35.Bone R, Shenvi AB, Kettner CA, Agard DA. Serine protease mechanism: structure of an inhibitory complex of α-lytic protease and a tightly bound peptide boronic acid. Biochemistry. 1987;26:7609–7614. doi: 10.1021/bi00398a012. [DOI] [PubMed] [Google Scholar]
- 36.Bachovchin WW, Wong WYL, Farr-Jones S, Shenvi AB, Kettner CA. Nitrogen-15 NMR spectroscopy of the catalytic-triad histidine of a serine protease in peptide boronic acid inhibitor complexes. Biochemistry. 1988;27:7689–7697. doi: 10.1021/bi00420a018. [DOI] [PubMed] [Google Scholar]
- 37.Yin Z, et al. Peptide inhibitors of dengue virus NS3 protease. Part 1: Warhead. Bioorg. Med. Chem. Lett. 2006;16:36–39. doi: 10.1016/j.bmcl.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 38.Frey PA. Strong hydrogen bonding in molecules and enzymatic complexes. Magn. Reson. Chem. 2001;39:S190–S198. doi: 10.1002/mrc.953. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.