Abstract
Recurrent parasitaemia during follow up of clinical trials of antimalarial drug efficacy results from either recrudescence of parasites surviving treatment or from parasites newly emerging from the hepatic stage of infection. Nested PCR is used to distinguish these two possibilities and the technique is difficult to standardise. There is risk of both false positive and false negative results, leading to misclassification errors. The high-resolution melt (HRM) assay was developed with pairs of conserved primers targeting blocks of merozoite surface protein 1 and 2 (msp1 and msp2) genes, and polymorphisms were compared using sequence-confirmed Plasmodium falciparum DNA samples from laboratory isolates. In this study, the HRM dissociation profiles of msp1 and msp2 amplicons were determined and validated against parasite isolates from malaria patients. The msp1 and msp2 profiles of both laboratory and clinical isolates were reproducibly differentiated by HRM. These rapid assays are performed in a closed-tube system, and so avoid cross-contamination while increasing throughput, which are two major advantages. The HRM assays offer significant gains in simplicity, speed and interpretation of results, and reduced analysis cost, for studies that require discrimination of parasite clones. Assay performance in large-scale studies utilizing DNA samples derived from filter-paper bloodspots should now be evaluated.
Introduction
Plasmodium falciparum is the most virulent of the six Plasmodium species that cause malaria in humans, being responsible for high mortality and morbidity, particularly in Africa. Efforts to control and eliminate malaria have been hampered by the emergence of drug resistant parasites1, but our ability to track and control resistance is also hampered by the high genetic diversity of P. falciparum. Further, to accurately estimate true drug efficacy in clinical trials of antimalarial drugs, recurrent infections seen after drug treatment need to be identified either as new infections arising from the liver or as recrudescent parasites persisting from the original infection. A PCR-correction method is generally deployed to distinguish between the two. In addition, ex vivo and in vitro studies to identify drug resistant parasites in complex infections also need to identify the constituent parasite clones in an infection or isolate in order to accurately capture the emergence of minor clones in the subsequent growth of parasites2,3, as is also seen in vivo4. This requires the identification of clones in paired samples, before and after treatment, and determining whether they are the same clones5.
One tool commonly used to distinguish between newly emergent and recrudescent parasites is conventional nested-PCR and gel electrophoresis detection. In this genotyping method, the genes that code the surface antigen loci of merozoite surface protein 1 and 2 (msp1,msp2) and glutamate rich protein (glurp) are amplified using sequence specific primers in a nested-PCR6. The recurrent infection is categorized by comparing the size of polymorphic repeat regions in these genes before (day 0) and after treatment (day of failure). If fragment sizes differ, the recurrent infection is categorized as a new infection, but if the post-treatment analysis produces the same fragment sizes as before treatment, these are considered recrudescent parasites, implying that treatment failure has occurred. In multi-clonal infections, minority clones constituting a small proportion of the total biomass might fall below the detection limit of the genotyping method and be missed by PCR-based detection due to competition for primers or other constituents of the reaction mix by the more abundant clones7. This was resolved by genotyping at extra time points on day 1 and day 2 post-treatment as well as at the follow-up time-point prior to the day that microscopically detected parasite recurrence occurred5,8. Using this approach, studies of treated malaria patients in Kenya and Tanzania showed the presence of additional malaria clones, differing from those at day 0 and day of failure resulting in re-classification of many recurrent infections as recrudescent infections in these extended analyses5,8,9. However, this approach requires significantly more effort, and current methods for determining different clones such as nested-PCR, microsatellite and DNA sequencing are labour-intensive, time-consuming and are prone to contamination. In addition, gel electrophoresis-based methods discriminate clones based on size differences alone and cannot detect sequence variation. In this study, a real-time quantitative PCR (qPCR) was developed with (HRM) method analysis, which is sensitive to both amplicon length and base composition, to identify distinct P. falciparum msp1 and msp2 genotypes.
Results
Primer optimization and validity of the assay
Alignments of msp1 and msp2 genes from 3D7, Dd2, 7G8, K1, FCR3, R033 and HB3 were assessed to design oligonucleotide primers that amplify the conserved regions (data not shown). To establish the specificity of the oligonucleotide primers and optimize the PCR conditions, 3D7 DNA obtained from laboratory-cultured parasites were initially used. Amplification of msp1 resulted in one minor peak and the major peak was retained in melt curve analysis but only the major one in HRM normalized graph windows. The msp2 genotyping of 3D7 DNA, however, unexpectedly produced two major melt peaks and HRM amplification curves each. It was first thought this resulted from the presence of two amplicons due to contamination. Further repeats of the msp1 and msp2 HRM analyses with a different source of DNA again gave two amplification curves for msp2, and fractionation of PCR products by gel electrophoresis confirmed that. Both msp1 and msp2 generated single band each of the predicted size. To rule out any inherent problems of the assays, we used uMELT software10 to predict the HRM curve of msp1 and msp2 gene fragment sequence of 3D7. The software correctly predicted one major and one minor curve for msp1 and two major curves for msp2 suggesting that this pattern does indicate two amplicons, but rather it is due to a bimodal melt of the double stranded DNA (dsDNA) where the base composition if not uniform throughout. The dsDNA melts in transition with regions of the amplicon that are more stable (e.g., G/C rich) melting later. These stable regions maintain their dsDNA configuration until the temperature is high enough to cause melting of the stable region, resulting in a bimodal melt profile even though only one amplicon is produced in the reaction.
To determine the limit of detection of the msp1 and msp2 HRM assays, 3D7 DNA at different parasite densities (3–0.00003%) was amplified. The assay detected as low as 1.5 parasites per µl with GCP of 77.44% and 62.44% for msp1 and msp2, respectively, relative to a 3D7 DNA sample at 1500 parasites per µl. GCP estimates increased to 92.32% and 78.41% for msp1 and msp2 genotypes respectively when 15 parasites per µl (0.0003% parasitaemia) were used. We then chose the 3D7 DNA sample with 15 parasites per µl, which is closer to limit of detection of parasites by expert or reference laboratory microscopy, to determine the inter-assay coefficient of variation (CV). The mean ± SD inter-assay CV (n = 35) for msp1 and msp2 was 93.15 ± 5.99 and 86.45 ± 9.47 respectively (Fig. 1).
Figure 1.
Normalized temperature-shifted HRM curves of msp1 and msp2 genes of P. falciparum 3D7 DNA. Each target was amplified in 35 tests with parasite DNA from a culture of 3D7 at 15 parasites per µl. The analysis software classified all samples as one genotype with (Genotype Confidence Percentage (GCP) of 71.9–99.5% for msp1 and 71.8–99.3% for msp2 genotype. Left: msp1. Right: msp2.
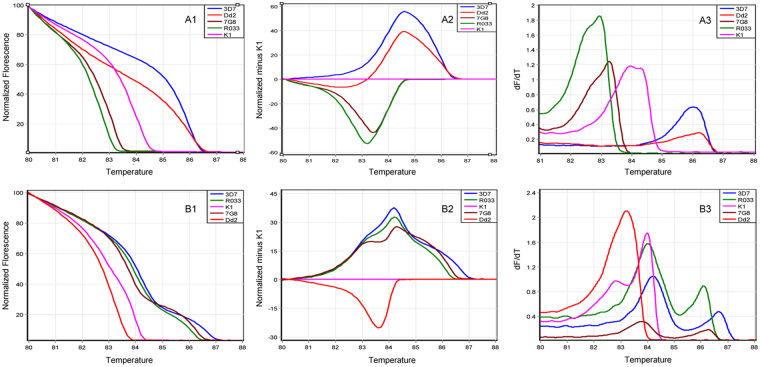
In an analysis of monoclonal parasite preparations, a distinct HRM amplification curve was produced for each of the five different clones (3D7, Dd2, 7G8, K1 and R033) using msp1 and msp2 genotypes (Fig. 2). DNA samples with 0.3%, 0.03% and 0.003% parasitaemia generated mean GCP of 76.8%, 78.2%, 99.3%, 95.5% and 88.9% for the 5 strains, respectively, when genotyped with msp1 primers and mean GCP of 78.2%, 74.5%, 86.1%, 61.5% and 87.9%, when genotyped with msp2 primers. The clones with lower melting temperature (Tm) (due to a shorter nucleotide sequence) such as 7G8, K1 and R033 in msp1 genotype were less affected by parasitaemia difference compared to the clones with higher Tm (longer nucleotide sequence) such as 3D7 and Dd2 (data not shown). The minor peak observed in 3d7 msp1 genotyping was also observed in all other laboratory isolates and was consequently excluded from HRM analysis.
Figure 2.
Normalized HRM analysis of msp1 (a1–3) and msp2 (b1–3) genotypes in 5 parasite lines. HRM amplification curves (a1 and b1), difference curves (A2 and B2) and melting curves (A3 and B3) for 5 P. falciparum laboratory strains are shown. K1 is deployed as the comparator genotype in (a2 and b2). dF/dT: negative derivative of change in florescence.
Detection of mixed laboratory clones
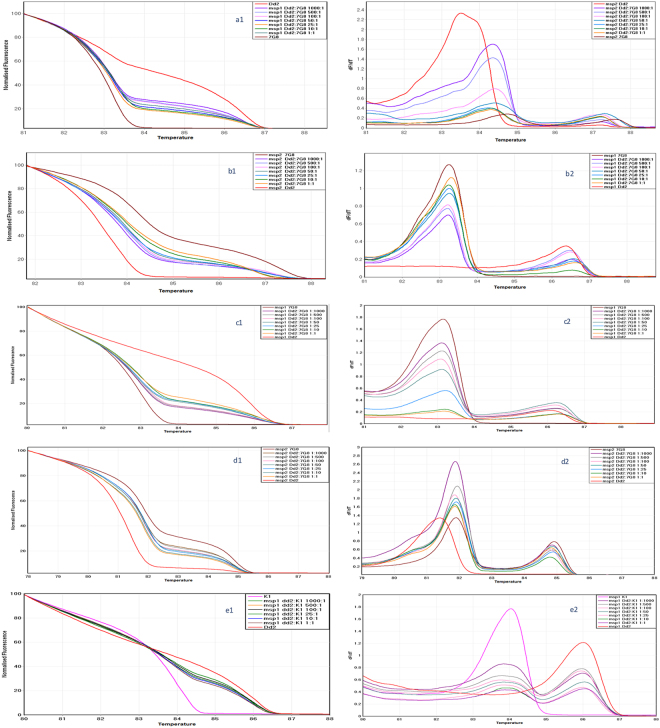
To study the sensitivity of the assay when mixed clones are present in a sample, we mixed Dd2 and 7G8, and Dd2 and K1 in different ratios. As predicted, msp1 genotype as well as the msp2 genotype for Dd2 produced two distinct HRM curves when two clones were mixed (Fig. 3). The msp2 genotype produced two HRM curves and three melt curves when two clones were present in the sample. These results have also be been confirmed in the melt curve analysis window (Table 1). If the parasitaemia of a particular laboratory strain in the mixture increases the HRM curve shifts toward the HRM curve of that particular strain. In all samples, the parasitaemia was adjusted to a maximum of 0.3% (15,000 parasites per µl) and minimum of 0.0003% (15 parasite per µl). Samples containing the same ratios of genotypes but different parasite density generally gave similar results but with smaller GCP. However, the higher the Tm value differences between mixed clones, the lower the GCP when calling the genotype similarity.
Figure 3.
msp1, msp2 HRM discrimination of artificial mixes of parasite clones at different ratios. Normalized temperature-shifted HRM amplification curves for msp1 (a1,c1 and e1) and msp2 (b1 and d1) and their corresponding melt curves (a2,b2,c2,d2 and e2) are shown for Dd2, K1 and 7G8 laboratory lines, and mixes of two strains at the ratios shown.
Table 1.
Mean melting peak and mean GCP* of single and mixed laboratory strains.
| Laboratory strain | Mix ratio | Genotype called by the software | Msp1 | Msp2 | ||
|---|---|---|---|---|---|---|
| Mean GCP * | Tm* | Mean GCP* | Tm* | |||
| 3D7 | — | 3D7 | 99.3 | 86.0 | 99.9 | 84.0 and 86.5 |
| Dd2 | — | Dd2 | 97.5 | 86.4 | 99.7 | 83.50 |
| 7G8 | — | 7G8 | 99.7 | 83.3 | 99.7 | 84.3 and 87.3 |
| K1 | — | K1 | 99.9 | 84.0 and 84.3 | 99.7 | 82.8 and 83.8 |
| R033 | — | R033 | 99.9 | 82.9 | 99.5 | 84.3 and 87.3 |
| Dd2:7G8 | 1:1 | Dd2/7G8 | 99.6 | 83.2 and 86.5 | 98.7 | 83.9 and 86.9 |
| 10:1 | Dd2/7G8 | 91.8 | 83.2 and 86.5 | 74.4 | 83.9 and 86.9 | |
| 25:1 | Dd2/7G8 | 88.3 | 83.2 and 86.5 | 68.4 | 83.9 and 86.9 | |
| 50:1 | Dd2/7G8 | 76.5 | 83.3 and 86.6 | 62.7 | 83.9 and 86.9 | |
| 100:1 | Dd2/7G8 | 73.9 | 83.2 and 86.5 | 62.4 | 83.9 and 86.9 | |
| 500:1 | Dd2/7G8 | 63.15 | 83.2 and 86.5 | 59.5 | 83.9 and 86.9 | |
| 1000:1 | Dd2/7G8 | 61.4 | 83.2 and 86.5 | 52.88 | 83.9 and 86.9 | |
| 1:1000 | Dd2/7G8 | 99.3 | 83.2 and 86.3 | 56.3 | 81.9 and 84.9 | |
| 1:500 | Dd2/7G8 | 62.8 | 83.2 and 86.3 | 61.2 | 81.9 and 84.9 | |
| 1:100 | Dd2/7G8 | 73.2 | 83.2 and 86.3 | 64.5 | 81.9 and 84.9 | |
| 1:50 | Dd2/7G8 | 78.2 | 83.3 and 86.3 | 70.2 | 81.9 and 84.9 | |
| 1:25 | Dd2/7G8 | 89.2 | 83.2 and 86.3 | 73.6 | 81.9 and 84.9 | |
| 1:10 | Dd2/7G8 | 91.2 | 83.2 and 86.3 | 78.5 | 81.9 and 84.9 | |
| 1:1 | Dd2/7G8 | 99.3 | 83.2 and 86.3 | 97.9 | 81.9 and 84.9 | |
| 1:1 | Dd2/K1 | 99.8 | 84.0 and 86.0 | — | — | |
| 10:1 | Dd2/K1 | 93.9 | 84.0 and 86.0 | — | — | |
| 25:1 | Dd2/K1 | 89.7 | 84.0 and 86.0 | — | — | |
| 50:1 | Dd2/K1 | 86.5 | 84.0 and 86.0 | — | — | |
| 100:1 | Dd2/K1 | 83.5 | 84.0 and 86.0 | — | — | |
| 500:1 | Dd2/K1 | 73.2 | 84.0 and 86.0 | — | — | |
| 1000:1 | Dd2/K1 | 71.4 | 84.0 and 86.0 | — | — | |
*For samples with a single allele, the mean GCP estimates agreement between duplicate tests while the mean GCP for the mixtures is estimated from 2 mixtures of Dd2 and 7G8 at the ratio shown, relative to the 1:1 mixture.
**Tm: melting temperature; this is estimated by the rotor-gene 6000 analysis software (version 1.7). Mixture clones of Dd2 and K1 was not done as both belong to the same allelic family.
Patient samples
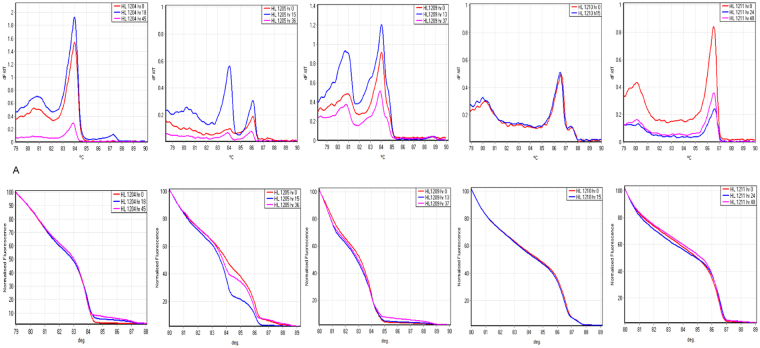
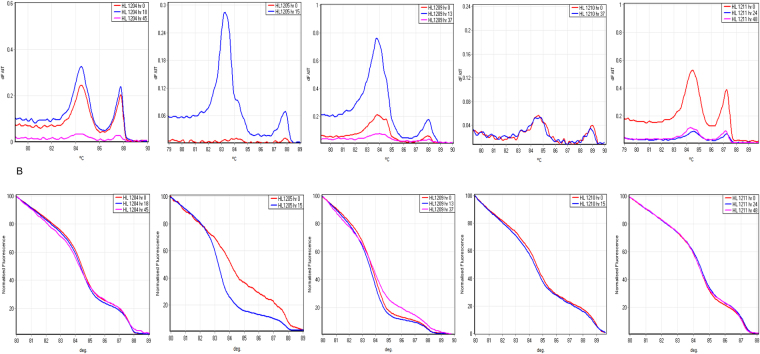
We genotyped paired pre- and post-treatment blood samples from five Hospital for Tropical Diseases (HTD) patients, and one additional unpaired pre-treatment sample. We investigated whether the assay accurately detects the number of clones in each sample and correctly classifies before and after treatment samples as the “same” or “different” infections. The HRM/melt curves generated by msp1 and msp2 genotypes for each patient are presented in Figs 4 and 5. With the exception of HL1210 and HL1211, all patient samples on post-treatment carried an extra HRM/melt curve when genotyped by msp1 compared to corresponding pre-treatment samples (Fig. 4). Generally, the additional HRM peak in the post-treatment samples displayed lower fluorescence intensity compared to other peaks, suggesting a minor clone, which was below the detection limit of the assay in the pre-treatment samples. This difference was reflected in the genotype calling of the paired samples, where HL1204 and HL1209 were classified as the “same” infection with a genotype confidence percentage (GCP) of 78.6% and 62.3%, respectively, indicating moderate similarity to the pre-treatment sample, compared to low similarity for HL1205 (<50%) (Table 2). HL1210 and HL1211 produced very similar HRM/melt curves for pre and post treatment samples, and as a result, their GCP were 90.6% and 93.1% respectively, showing much greater similarity between the two time points than the patients with an extra clone in post-treatment samples. On the other hand, msp2 genotyping produced the same HRM/melt curve in all paired patients samples with the exception of HL1205, where an extra curve was observed (Fig. 5). Consequently, the GCP was much higher (87.5–97.5%) compared to their corresponding GCP of patient samples obtained by msp1 genotyping. Paired DNA samples of HL1205 patient had GCP of less than 50% confirming the GCP obtained by msp1 genotyping.
Figure 4.
msp1 HRM analysis of paired clinical samples from the same individuals. Melting amplification curves (top graphs) and normalized temperature-shifted HRM amplification curves (bottom graphs) of msp1 genotypes in 5 patients sampled before (hr 0) and after treatment (hr 13–48). Differences in raw florescence signal between timepoints were observed, but didn’t affect HRM genotype calls after normalization. dF/dT, negative derivative of change in florescence and C°, temperature.
Figure 5.
msp2 HRM analysis of paired clinical samples from the same individuals. Melting amplification curves (top graphs) and normalized temperature-shifted HRM amplification curves (bottom graphs) of msp2 genotypes in 5 patients sampled before and after treatment.
Table 2.
Genotype calling of paired clinical samples using GCP* of msp1 and msp2.
| Patient ID | Time after treatment | Genotype called by the software compared to hr 0 | Msp1 Confidence % relative to hr 0 | Msp2 Confidence % relative to hr 0 |
|---|---|---|---|---|
| HL 1204 | hr 0 | HL1204 | 99.53 | 99.41 |
| hr 18 | HL1204 | 82.17 | 82.95 | |
| hr 45 | HL1204 | 59.52 | 55.65 | |
| HL 1205 | hr 0 | HL1205 | 98.59 | 90.75 |
| hr 15 | Variation | <50 | <50 | |
| hr 36 | HL1205 | 70.27 | Negative | |
| HL 1209 | hr 0 | HL1209 | 99.77 | 98.72 |
| hr 13 | HL1209 | 56.35 | 69.63 | |
| hr 37 | HL1209 | 57.80 | <50 | |
| HL 1210 | hr 0 | HL1210 | 94.75 | 98.52 |
| hr 15 | HL1210 | 73.23 | 74.35 | |
| hr 40 | Negative | NA | NA | |
| HL 1211 | hr 0 | HL1211 | 99.83 | 95.89 |
| hr 24 | HL1211 | 65.63 | 88.57 | |
| hr 48 | HL1211 | 77.51 | 94.24 |
All clinical samples after treatment had the same genotype compared to their corresponding samples before treatment (hr 0) samples with a GCP of more than 50%. Exceptions are HL1205 and HL1209, where both had a post-treatment genotype different from the pre-treatment in one or more loci. A second time-point after treatment was included for comparison purposes and in order to further evaluate the performance of msp1 and msp2 HRM genotyping for low parasitaemia samples. *GCP, Genotype Confidence Percentage, percentage of similarity of post-treatment sample to its corresponding pre-treatment sample.
The allelic family of the DBS field samples was determined relative to the HRM melt curve of the positive controls with known allelic family (RO33 for msp1 or Dd2 for msp2 and K1 and 3D7 for both), which represent the three different HRM melt curve observed in the initial validation experiment. Any sample within 0.2 °C degree of the sample with known HRM melt curve belongs to the same clone (Supplementary Fig. S1A–C).
All DBS field samples collected on day 0 showed the same HRM melt curve (the same Tm value) using both msp1 and msp2 assays compared to the samples collected 7 days later (Fig. S1A–C). On the other hand, the day 14 sample of one child showed one less clone compared to day 0 and day 7 while the day 21 sample of another child had one different clone compared to day 0 and 7 (Supplementary Table S1). The msp1 and msp2 HRM melt curve results were similar to clones generated by gel-based electrophoresis. One sample (K011) on day 28 was negative by msp1 but showed similar HRM melt curve on msp2 though slightly different Tm was observed (84.2 on d0 and 83.8 on day 28). However, the normalized HRM curve classified the two samples as “different”. This is consistent with the gel-based PCR, which generated different clones on d0 (K1 and RO33) and d28 (K1 and MAD20) suggesting a possible new infection (Supplementary Fig. S2).
To confirm the above HRM melt curve analysis, a GCP analysis was performed on all field samples. Samples collected before treatment and 7 days after treatment were classified as the “same” with a mean GCP of 85% (64–99%) for msp1 and 82% (61–99%) for msp2 in all samples. Samples collected after day 7 (day 14, 21 and 28) were classified as “different” with a GCP of less than 50% relative to day 0 sample. This was in complete agreement with the gel-based PCR results (Supplementary Fig. S2).
GCP cut-off value for distinguishing between recrudescence and new infection
When compared to DNA prepared from a 3D7 culture at 3% parasitaemia, DNA from the lowest parasitaemia 3D7 culture (0.0003%) produced GCP estimates of 77.44% and 62.44% for msp1 and msp2 respectively. When Dd2 and 7G8 were artificially mixed, DNA from a 10:1 to 1000:1 mixtures at 0.3% parasitaemia generated a GCP estimate for msp1 and msp2 of >50% in comparison with a 1:1 mixture at the same parasite density (Table 1). This GCP estimate is a better reflection of actual field isolates, and fits with the WHO definition of a new infection, which requires that all alleles, in one or all loci tested, should be different in both time-points. Therefore, we opted to use GCP of 50% as a cut off value to distinguish recrudescence from new infections. This cut-off value was applied for the paired pre- and post-treatment patient samples and accurately classified four of the five as the “same” infections with a satisfactory GCP (≥62.3%). This was expected as the paired samples were obtained before and 13–48 hrs after antimalarial treatment and new infections are not expected at this time.
Genetic diversity study
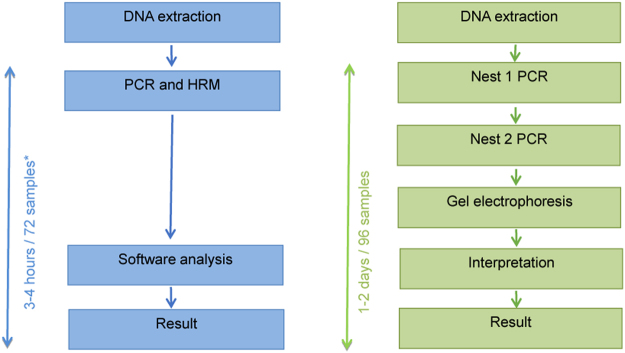
To assess the general relevance of the assay for applications requiring analysis of genetic diversity, the difference graph was used to cluster clones based on their similarities. Using the standard gel-based msp1 and msp2 genotyping, parasite isolates are broadly classified into K1, MAD20 and R033, and FC and IC/3D7 sub family clones respectively. Since the Tm value for the K1 clone was found to lie in the middle of the range of values across all laboratory isolates tested, this clone was used as a calibrator and the software calculated the florescence signal of each clone relative to the signal of K1 and to generate a difference graph (Fig. 2). The approach correctly clustered the msp2 clones into two groups, cone comprising 3D7, 7G8 and R033, and the second represented by Dd2. These groups correspond to IC/3D7 and FC sub family alleles originally described by Snounou and colleagues respectively6,7. The msp1 difference graph clustered the clones into three groups containing 3D7, Dd2 (MAD20), 7G8 (R033). In gel-based discrimination, 3D7 and K1 are grouped together but in the HRM assay, they cluster into different groups, reflecting difference in the DNA sequence and length. Interestingly, the patient DNA samples HL1204 and HL1209, HL1205 and HL1212, and HL1210 and HL1211 clustered into 7G8/R033, Dd2 (MAD20) and 3D7 allele types respectively with GCP ranging from 42.6% to 78.2% when genotyped with msp1. When genotyped with msp2 HL1204, HL1210 and HL1211 clustered into IC/3d7 sub family clones while HL1205, HL1209 and HL1212 clustered in both IC/3D7 and FC type clones suggesting that both alleles were present in the samples. This has been previously reported for these isolates using gel electrophoresis, where the samples contained more than 3 or more clones of FC type and more than 1 or more clones of IC/3D7 type3. Using HRM, the classification of parasite clones into clusters of sub-families takes 3–4 hrs compared to 1–2 days using PCR and gel-electrophoresis methods (Fig. 6).
Figure 6.
Benefits of msp1 and msp2 genotyping by real-time PCR and HRM. Summary of significant gains in simplicity, speed and interpretation of results from utilizing the qPCR-HRM genotyping approach compared to the conventional msp1 and msp2 genotyping using gel electrophoresis. *3–4 hours per run (72 samples for rotor-gene and 96 samples for other platforms).
Discussion
Genotyping of the msp1 and msp2 loci has been deployed to assess multiplicity of infection, to evaluate strain-specific selection in vaccine efficacy studies, to estimate the genetic diversity of P. falciparum in population genetic studies, and to distinguish between recrudescent and novel genotypes in recurrent infections occurring in participants treated for malaria in the course of antimalarial drug efficacy trials. These studies have most commonly used PCR and gel electrophoresis, for discrimination among different msp1 and msp2 alleles6,11. The specificity of PCR- identification of recrudescent infections can be improved by deploying multiple post-treatment time points5,9. However, if using the nested-PCR and gel-electrophoresis method, this approach is laborious, resource intensive and can cause difficulties in interpretation. The current study reports the development of real-time PCR based HRM technology for msp1 and msp2 genotyping a new tool for in vivo and in vitro drug and vaccine efficacy studies. HRM analysis is a closed tube approach for fast, accurate, high throughput genotyping and provides a simple method for PCR-correction. This eliminates the limitations of gel electrophoresis and subjective characterisation of the different alleles. Unlike electrophoresis, HRM can distinguish alleles based on differences in base composition, sequence and amplicon size, delivers quantitative estimate of allele similarity (GCP), and has been successfully evaluated for detection of drug resistance malaria parasites12.
The msp1 and msp2 HRM assays correctly discriminated alleles from five different single-clone laboratory isolates, those of polyclonal DNA samples derived from whole blood and DBS. The assays detected densities as low as 1.5 parasites per µl accurately, suggesting that the method is as sensitive as the nested-PCR electrophoresis method. However, the mixed clones evaluated were artificially prepared and the ability of HRM assays to precisely estimate the probability of missed alleles in natural field isolates remains to be determined. This was clear when paired patient samples were genotyped and analysed using HRM, where at least one allele was missed in two patient samples compared to the nested-PCR and gel electrophoresis method3. It is not clear whether this is due to an intrinsic sensitivity difference in detecting minor clones or the detection of false positive alleles by nested-PCR and gel electrophoresis, possibly due to amplicon contamination. HRM assays avoid contamination and risk of PCR product carry-over as the method is non-nested and uses a closed-tube readout.
Both msp1 and msp2 HRM genotyping identified one or more extra alleles in post-treatment samples from some of the patients. This was also observed previously when the same samples were genotyped by PCR and gel electrophoresis, suggesting that this is a general observation regardless of the PCR-based genotyping methods used3,13, best explained by changes in the relative abundance of the constituent circulating clones in vivo before and after treatment5,8. In polyclonal infections, these changes in relative abundance mean certain genotypes may remain undetected at one time point, but recur later leading to false classification. These minority clones are missed by PCR-based detection methods due to competition for primers or other constituents of the reaction mix by the more abundant clones7. In a study in Papua New Guinea, detection of minor clones decreased with an increase in multiplicity of infection, suggesting the dominance of major clones among PCR amplicons detected14. A similar study carried out in Uganda reported a higher probability of mixed infections in high transmission areas, and this affected classification of treatment outcomes15. A slow-clearing minority variant present but not detected at admission, could cause recrudescence but would be identified in the recurrent sample as a new infection, rather than recrudescence of parasite genotypes present at the time of treatment. This has recently been demonstrated in a multiplicity of infection study to detect and quantify the markers of antimalarial drug resistance16. Therefore, the failure to detect minor alleles at day 0 is likely to lead to misclassification of some infections17. The data in this study and previous reports3,5,8 demonstrate that inclusion of the additional time points day 1, day 2 and day before failure, in addition to pre-treatment and day of failure, improve specificity when determining the complexity and origins of recurrent infections.
One other strength of the HRM assay is that the clones are discriminated based not only on length but also sequence differences and this feature allows the HRM assay to overcome the resolution limitation observed in the gel electrophoresis-based methods6. This is particularly important in high transmission settings, where two distinct clones may generate PCR products with the same size but which differ in sequence. These will be indistinguishable by gel electrophoresis. Others have attempted to resolve this by classifying any pair of samples that contain matched and unmatched alleles as new infections18, but this can clearly lead to underestimation of recrudescence infections. WHO/MMV guideline states that, in treated malaria patients, “ a ‘re-infection’ [i.e. new infections] is a subsequent occurring parasitaemia in which all the alleles in parasites from the post-treatment sample are different from those in the admission sample, for one or more loci tested”19. This definition seeks to minimize misclassification error and maximize accuracy of classification.
One of the shortcomings of the HRM assays is that some of the msp1 and msp2 alleles generate more than one melting peak, due to the inherent two-phased nature their DNA melting profile. For distinguishing recrudescence from new infections, this shortcoming can be overcome as the HRM amplification curves of the samples before and after treatment are compared with each other, and any additional peak due to the inherent nature of DNA melting will be reflected in both samples. The interpretation of those melting peaks will be more challenging if the genotyping purpose is to study parasite diversity and complexity of infection, although we have shown that this can be overcome by including known comparators for both msp1 and msp2 genotyping. Measurement of the amplification signal of each clone relative to these comparators thus permits classification of allelic variants into clusters.
In addition to drug and vaccine efficacy studies, HRM genotyping can be applied in studies of parasite biology – for example, to assess parasite cloning experiments, and in monitoring the identity, integrity and clonality of propagated parasite lines, if necessary direct from cryo-preserved material. The HRM method can also be deployed as tool for in vitro drug sensitivity studies, particularly for polyclonal parasite isolate in which a subset of genotypes preferentially survive in vitro cultures, as this often reflect survival in in vivo therapeutic efficacy studies20. Our data warrant the large-scale evaluation of the performance of the real-time PCR and HRM approach on DNA samples derived from filter paper bloodspots collected in field studies.
Materials and Methods
Plasmodium falciparum DNA strains and field samples
Parasite DNA from malaria cases treated at the Hospital for Tropical Diseases (HTD), London, UK was utilized for this study. Clinical isolates were obtained from six patients: pre-treatment and post-treatment samples were available for patients HL1204, HL1205, HL1209, HL1210 and HL1211 and only pre-treatment for patient HL1212. Details of the patient history, parasitaemia and treatment have previously been published3. Eighteen paired dried blood spot (DBS) samples were available to validate the HRM on DBS field samples. For 15 children, day 0 (before treatment) and day 7 (7 days after treatment) were available. For two children, day 0 and day 14 or day 28 samples were available and for one child, day 0, 7 and 21 samples were available. The samples were collected between January and July 2014 from asymptomatic children aged 5 to 12 years attending schools near Mbita, Kenya, as part of a study on mosquito behaviour and odor profile of malaria-infected individuals. Study site, sample collection and other details of the study has already been published21,22. Culture adapted laboratory isolates 3D7, Dd2, 7G8, K1, R033 and D10 were obtained from the Malaria Research and Reference Reagent Repository (http://MR4.org). Parasite cultures were tightly synchronized as ring-stage trophozoites to stimulate infected peripheral blood in vivo.
PCR
New primer sets were designed to amplify block 1 of msp1 and block 3 of msp2 genes. Pairs of oligonucleotide primers for msp1 (Msp1HRM_F: TAGAAGATGCAGTATTGACAGGT and Msp1HRM_R: CAGCGTAAGATTTAGCATCTGAATC) and msp2 (Msp2HRM_F: AGCAACACATTCATAAACAATGCT) and Msp2HRM_R: TCCATGTTGTCCTGTACCTTTATTC) flanking the target regions were designed with the PCR amplicon expected to yield 346 bp and 518 bp respectively. The msp1 primers are conserved in the commonly used isolates K1, MAD20 and Ro33, and the msp2 primers in FC27 and 3D7/IC respectively. Amplification of DNA was performed in a 25 µl reaction volume on a Rotor-gene 6000 thermal cycler (QIAGEN, Germany). The reaction mixture contained 5 µl of extracted genomic DNA, 200 nM of each primer, 3 mM of MgCl2, 300 nM of each dNTP, 5 µM SYTO 9 green florescence nucleic acid stain (Invitrogene), 1X NH4 reaction buffer (Bioline, UK) and 1 U Taq polymerase (Biolione, UK). PCR conditions were one cycle of 94 °C for 2 min, 40 cycles of 94 °C for 15 sec, 54 °C for 20 sec and 72 °C for 40 sec. To verify the specificity of the primer sets, PCR products were detected by gel electrophoresis of 10 µl from each reaction on 2% agarose gels. Gel-based PCR assays targeting msp1 and msp2 genes were amplified using previously reported methods6 and clone similarity between time points was carried out using previously published approaches5,18. Gels were made and run in TBE buffer (sigma-Aldrich, UK) and 5ul of loading buffer (Bioline, UK) were added to each sample prior to electrophoresis.
Optimization and validation
The specificity of the msp1 and msp2 primers was established by running gel-electrophoresis of the PCR products. The sensitivity and limit of detection of the assay was estimated using a five-fold serial dilution of 3D7 in blood starting at 3% parasitaemia. The sensitivity to detect clones in mixed infection was estimated using a mixture of Dd2 and 7G8, and Dd2 and K1 Plasmodium falciparum in culture medium at 5% haematocrite in a ratio of 1:1, 10:1, 25:1, 50:1, 100:1, 500:1 and 1000:1, and Dd2:7G8 in a ratio of 1:1, 10:1, 25:1, 50:1, 100:1, 500:1 and 1000:1 with a minimum parasitaemia of 15 parasites per µl. The reproducibility of the msp1 and msp2 HRM assays was determined by carrying out 35 replicate tests of 15 parasites per µl of 3D7 strain.
HRM curve acquisition
QPCR HRM analysis was performed in a Rotor-gene 6000 thermal cycler (Corbett Life Science). In order to determine the optimal melting temperature for differentiation of P. falciparum clones, the PCR products were subjected to a ramping of 0.1 °C s−1 between 75 °C and 90 °C. All specimens were performed in duplicate and their melting profiles were analysed using Rotor-gene 6000 software (version 1.7.87) and the HRM algorithm provided. Visual examination of the melt curves data from a panel of laboratory isolates across a gradient of different temperature ramp increments was performed to select the optimum temperature in which most laboratory isolates show distinct conventional melt curves.
Genotyping analysis
For genotype analysis, temperature-shifted and normalized amplification curves were used in in the HRM analysis. Normalization regions of 82.0–82.5 and 88.0–88.5 were used as a standard but were modified for some clinical samples depending on the melting temperature peaks observed. The threshold in the melting curve analysis was adjusted to ensure that all positive samples generate interpretable dF/dT profiles and melt peak estimates. A fixed threshold was not deployed as the florescence signal amplitude varied across samples. The Genotype Confidence Percentage (GCP) is a value attributed to each genotype/allele compared with the genotype of a calibrator, with a value of 100% indicating an exact match. For clinical samples, the pre-treatment sample (hr 0) was a calibrator.
Clonal and cluster analysis
Melt curve analysis was used to verify the presence of multiple clones in each parasite isolate or culture. Clone similarity was signified by assigning names of known genotypes (strains) to samples with similar melting temperature to the known genotypes. PCR amplicons from five different P. falciparum laboratory isolates were subjected to HRM curve analysis. The difference-curve graph was used to classify the different msp1 and msp2 clones into clusters based on florescence signal difference relative to K1 (reference strain).
Analysis methods
Validation of each result was assessed using CT value, end-point florescence level and amplification efficiency. The real-time data were analysed using different modules available in the Rotor-gene software. A sample was re-analysed if its CT value was ≥30, or if it had lower amplification end point compared to the majority plots, or if amplification efficiency differ from other reactions or fell below an amplification value of 1.4 (2 = 100% amplification efficiency). The reproducibility of the msp1 and msp2 assays was measured by calculating the coefficient of variation (mean ± standard deviation) of GCP of replicate tests.
Availability of data and material
All culture-adapted parasite lines described have been deposited in the European Malaria Reagent Repository (http://www.malariaresearch.eu/) and are freely available to researchers. All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Ethics approval and consent to participate
All research was performed in accordance with LHSTM relevant guidelines. Voluntary informed written consent and travel history was obtained from patients presenting with malaria to the HTD, or the Accident and Emergency Department of University College London Hospitals (UCLH). Approval for the study was obtained from the Research Ethics Committee of the University College London Hospitals (Application number: 07/Q0505/60), and include in their manuscript a statement confirming that informed consent was obtained from all participants and/or their legal guardians
Electronic supplementary material
Acknowledgements
The authors thank Prof Abdoulaye A. Djimde and Dr. Don van Schalkwyk for their valuable comments. We also thank Annette Obukosia Busula, Jetske Gudrun de Boer, Geoffrey Omondi Olweru, David John Odoyo and Ailie Robinson for providing us with the Kenyan dried bloodspot field samples.
Author Contributions
C.J.S. and K.B.B. contributed to study design. N.D. and K.B.B. performed laboratory work. C.J.S. and K.B.B. analysed the data. C.J.S. and K.B.B. wrote the manuscript. All authors have contributed to and approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28179-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ashley EA, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gosi P, et al. Evaluation of parasite subpopulations and genetic diversity of the msp1, msp2 and glurp genes during and following artesunate monotherapy treatment of Plasmodium falciparum malaria in Western Cambodia. Malar J. 2013;12:403. doi: 10.1186/1475-2875-12-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Schalkwyk DA, et al. Culture-adapted Plasmodium falciparum isolates from UK travellers: in vitro drug sensitivity, clonality and drug resistance markers. Malar J. 2013;12:320. doi: 10.1186/1475-2875-12-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henriques G, et al. Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J Infect Dis. 2014;210:2001–2008. doi: 10.1093/infdis/jiu358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beshir KB, et al. Residual Plasmodium falciparum parasitemia in Kenyan children after artemisinin-combination therapy is associated with increased transmission to mosquitoes and parasite recurrence. J Infect Dis. 2013;208:2017–2024. doi: 10.1093/infdis/jit431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snounou, G. & Beck, H. P. The use of PCR genotyping in the assessment of recrudescence or reinfection after antimalarial drug treatment. Parasitol Today14, 462–467, doi:S0169-4758(98)01340-4 (1998). [DOI] [PubMed]
- 7.Juliano JJ, Gadalla N, Sutherland CJ, Meshnick SR. The perils of PCR: can we accurately ‘correct’ antimalarial trials? Trends Parasitol. 2010;26:119–124. doi: 10.1016/j.pt.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martensson A, et al. Influence of consecutive-day blood sampling on polymerase chain reaction-adjusted parasitological cure rates in an antimalarial-drug trial conducted in Tanzania. J Infect Dis. 2007;195:597–601. doi: 10.1086/510910. [DOI] [PubMed] [Google Scholar]
- 9.Sawa P, et al. Malaria transmission after artemether-lumefantrine and dihydroartemisinin-piperaquine: a randomized trial. J Infect Dis. 2013;207:1637–1645. doi: 10.1093/infdis/jit077. [DOI] [PubMed] [Google Scholar]
- 10.Dwight Z, Palais R, Wittwer CT. uMELT: prediction of high-resolution melting curves and dynamic melting profiles of PCR products in a rich web application. Bioinformatics. 2011;27:1019–1020. doi: 10.1093/bioinformatics/btr065. [DOI] [PubMed] [Google Scholar]
- 11.Felger I, et al. Plasmodium falciparum: extensive polymorphism in merozoite surface antigen 2 alleles in an area with endemic malaria in Papua New Guinea. Exp Parasitol. 1994;79:106–116. doi: 10.1006/expr.1994.1070. [DOI] [PubMed] [Google Scholar]
- 12.Daniels R, et al. Rapid, field-deployable method for genotyping and discovery of single-nucleotide polymorphisms associated with drug resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2012;56:2976–2986. doi: 10.1128/AAC.05737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenhouse B, et al. Validation of microsatellite markers for use in genotyping polyclonal Plasmodium falciparum infections. Am J Trop Med Hyg. 2006;75:836–842. [PMC free article] [PubMed] [Google Scholar]
- 14.Koepfli C, et al. How much remains undetected? Probability of molecular detection of human Plasmodia in the field. Plos One. 2011;6:e19010. doi: 10.1371/journal.pone.0019010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slater M, et al. Distinguishing recrudescences from new infections in antimalarial clinical trials: major impact of interpretation of genotyping results on estimates of drug efficacy. Am J Trop Med Hyg. 2005;73:256–262. [PubMed] [Google Scholar]
- 16.Hastings IM, Nsanzabana C, Smith TA. A comparison of methods to detect and quantify the markers of antimalarial drug resistance. Am J Trop Med Hyg. 2010;83:489–495. doi: 10.4269/ajtmh.2010.10-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juliano JJ, et al. Misclassification of drug failure in Plasmodium falciparum clinical trials in southeast Asia. J Infect Dis. 2009;200:624–628. doi: 10.1086/600892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cattamanchi A, Kyabayinze D, Hubbard A, Rosenthal PJ, Dorsey G. Distinguishing recrudescence from reinfection in a longitudinal antimalarial drug efficacy study: comparison of results based on genotyping of msp-1, msp-2, and glurp. Am J Trop Med Hyg. 2003;68:133–139. [PubMed] [Google Scholar]
- 19.WHO/MMV. Methods and Techniques for Clinical Trials on Antimalarial Drug Efficacy: Genotyping to Identify Parasite Populations. Geneva: Wolrd Health Organization. http://apps.who.int/malaria/docs/drugresistance/MalariaGenotyping.pdf, accessed on 04 March 2018 (2008).
- 20.Nsobya SL, Kiggundu M, Joloba M, Dorsey G, Rosenthal PJ. Complexity of Plasmodium falciparum clinical samples from Uganda during short-term culture. J Infect Dis. 2008;198:1554–1557. doi: 10.1086/592506. [DOI] [PubMed] [Google Scholar]
- 21.Beshir KB, et al. Plasmodium falciparum parasites with histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in two endemic regions of Kenya. Scientific reports. 2017;7:14718. doi: 10.1038/s41598-017-15031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson A, et al. Plasmodium-associated changes in human odor attract mosquitoes. Proc Natl Acad Sci USA. 2018;115:E4209–E4218. doi: 10.1073/pnas.1707856115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.