Abstract
Polyploidization is a widespread phenomenon, especially in flowering plants that have all undergone at least one event of whole genome duplication during their evolutionary history. Consequently, a large range of plants, including many of the world’s crops, combines more than two sets of chromosomes originating from the same (autopolyploids) or related species (allopolyploids). Depending on the polyploid formation pathway, different patterns of recombination will be promoted, conditioning the level of heterozygosity. A polyploid population harboring a high level of heterozygosity will produce more genetically diverse progenies. Some of these individuals may show a better adaptability to different ecological niches, increasing their chance for successful establishment through natural selection. Another condition for young polyploids to survive corresponds to the formation of well-balanced gametes, assuring a sufficient level of fertility. In this review, we discuss the consequences of polyploid formation pathways, meiotic behavior and recombination regulation on the speciation success and maintenance of polyploid species.
Keywords: polyploidy, genome evolution, diploidization, meiosis, unreduced gametes, recombination, crossover
Introduction
Meiosis is the fundamental process by which are formed the gametes in all sexual organisms. Largely investigated in the last decades (for review see Mercier et al., 2015; Zickler and Kleckner, 2015), this process consists in a single phase of DNA replication followed by two divisions, where first, pairs of parental chromosomes (i.e., homologs) and then, sister chromatids separate into four cells of a tetrad. During the first division, occurrence of meiotic recombination is determinant for ensuring both genome stability and generation of diversity, through one of its products: the crossovers. Indeed, in addition to maintain pairs of homologs physically linked at the end of metaphase I (i.e., bivalents), crossovers result in reciprocal exchanges of DNA between non-sister chromatids. At least one crossover is required per bivalent to obtain well-balanced gametes and avoid the formation of aneuploid progenies. However, as a result of the so-called phenomenon of interference, rarely more than three crossovers are formed per bivalent in a meiosis, typically widely spaced from one another and primarily located on chromosome extremities.
In polyploids, which are widespread in plants even in major crops (e.g., cotton, rapeseed, and wheat), the situation is delicate as they combine two genomes or more deriving from the same (autopolyploidy) or related (allopolyploidy) species (Stebbins, 1947). While all Angiosperms have experienced at least one event of whole genome duplication in their evolutionary history (Jiao et al., 2011), polyploidization remains an active and ongoing process recognized as a major driving force for plants speciation (Comai, 2005; Alix et al., 2017). Indeed, polyploids may occupy new ecological niches (Stebbins, 1985; Blaine Marchant et al., 2016) and often display higher adaptability than their progenitors, as evidenced by their better tolerance to abiotic stresses (McIntyre, 2012; Allario et al., 2013). However, the reasons of such a speciation success are not well-understood given that polyploidization initially results in a depletion of variability, due to the limited number of parental genotypes, and frequently confers instant reproductive isolation (Husband and Schemske, 2000; Husband and Sabara, 2004). In this review, we aim to highlight how meiotic recombination may favor this success by (1) conditioning the genetic variability of newly formed polyploids, (2) expanding the allelic combinatorial possibilities in the following generations, and (3) ensuring the proper segregation of multiple homologs and/or related chromosomes (i.e., homoeologs) in established auto- and allopolyploids, respectively.
The Routes Leading to Polyploidy Combined With the Occurrence of Meiotic Recombination Condition the Initial Allelic Variability
A novel polyploid individual may form via several routes (Ramsey and Schemske, 1998; Tayalé and Parisod, 2013) (Figure 1). Depending on the formation pathway, the genetic status (i.e., level of heterozygosity vs. homozygosity) of newly formed polyploids will highly differ and may impact their performance and speciation success, as evidenced by gains recorded in highly heterozygous plants for growth, fertility, and yield (Bingham, 1980; Stebbins, 1980; Werner and Peloquin, 1991). While immediate consequences of polyploidization were mostly investigated by inducing the somatic doubling of chromosomes through chemical treatment (Tamayo-Ordóñez et al., 2016), this path does not fully mimic what happened in nature in terms of occurrence and variability. Indeed, although possible when mitotic non-disjunction of sister-chromatids arises either in meristem tissue of sporophytes, zygote or young embryo, this route remains rarely observed spontaneously and restricts the number of alleles fixed per locus in auto- and allotetraploids (Ramsey and Schemske, 1998).
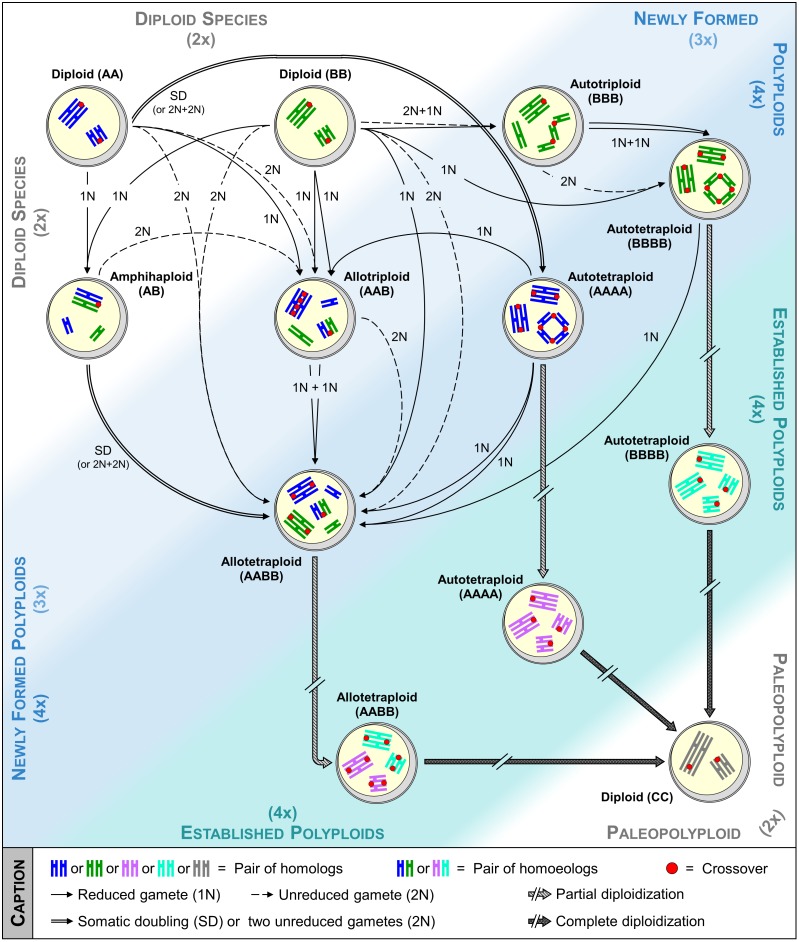
FIGURE 1.
From the formation toward the complete diploidization of polyploids: insights of meiotic pairing and recombination. Auto- and allotetraploids can be formed via many routes, which may involve intermediates (e.g., triploids and amphihaploids). While pairing and recombination are initially perturbed, as represented by multiple/illegitimate associations and by increasing number of crossovers, a diploid-like meiosis is setting up through a strict regulation of meiotic recombination, a process referred as partial diploidization. Following millions of years, diploidization becomes complete through a return at a diploid stage resulting in a chromosome number reduction. On this figure, insights of pairing and recombination mostly based on research performed in Arabidopsis, Brassica, Lolium, and Phlox genera are presented. Within each pollen mother cell drawn, the chromosomes of the same size and same color derive from the same species, while those of different colors derive from two different species.
Nowadays, it is accepted that polyploids predominantly arise sexually, through the generation of gametes having the somatic (2n) rather than the haploid (n) number of chromosomes; a phenomenon referred as ‘gametic non-reduction’ (Harlan and de Wet, 1975). Indeed, production of unreduced gametes has been observed across widely disparate phyla (Veilleux, 1983; Bretagnolle and Thompson, 1995), at frequencies typically averaging from 0.1 to 2.0% in natural populations (Kreiner et al., 2017). Moreover, polyploidy induction may have been facilitated throughout plants evolutionary history via greatly enhanced frequencies of unreduced gametes. For instance, abiotic stresses such as temperature fluctuation often favor the production of unreduced gametes (Mason et al., 2011; Pécrix et al., 2011; De Storme et al., 2012), which is striking in regard to the coincidence of ancient WGD events with adverse climatic events (Vanneste et al., 2014; Van de Peer et al., 2017). On the other hand, mutation of certain genes may also have facilitated polyploidization, especially when promoting unreduced gametes in both male and female meiosis, as observed for OSD1 and TAM in Arabidopsis thaliana (d’Erfurth et al., 2009, 2010; Wang et al., 2010). Although a plethora of cytologic mechanisms has been described (De Storme and Geelen, 2013), unreduced gametes commonly arise in plants through First (FDR) or Second Division Restitution (SDR), corresponding to the defect of meiosis I or II, respectively. Thus, depending on their origin, unreduced gametes will display different genetic makeups (Bretagnolle and Thompson, 1995; Brownfield and Köhler, 2011). In the strict sense, the non-disjunction of homologs in FDR is combined with the abolishment of recombination, yielding unreduced gametes retaining the full heterozygosity of the initial individual (Figure 2); note however that in some instance a partial loss of variability happens due to the occurrence of recombination, a mechanism referred as FDR-like (Ramanna and Jacobsen, 2003). In contrast, SDR, consisting in the exclusive separation of recombined homologs, always results in partially homozygous unreduced gametes, from the crossover location toward the end of a chromosome arm (Figure 2).
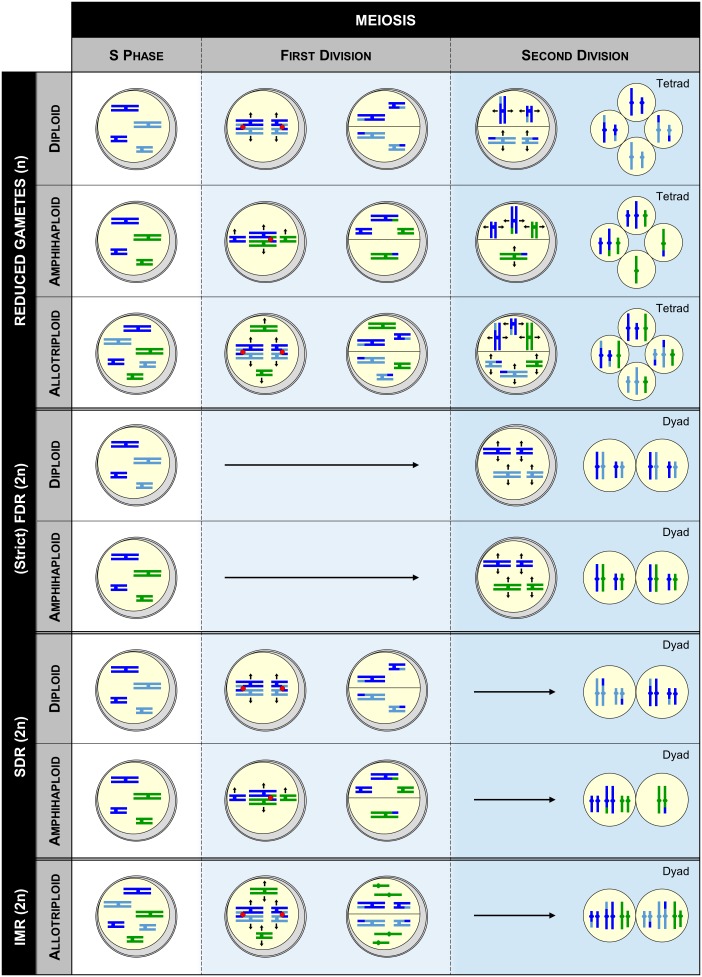
FIGURE 2.
Genotypic outcome of gametes deriving from diploid, amphihaploid and allotriploid heterozygous individuals when meiosis either occurs normally (WT) or undergoes a meiotic restitution (FDR, SDR, or IMR). FDR, SDR, and IMR correspond to first, second, and intermediate meiotic restitution, respectively. Within each pollen mother cell drawn, the chromosomes of the same size colored in light and dark blue derive from the same species (differentiation showing the heterozygosity), while those colored in blue and green derive from two different species. Note that this figure is not exhaustive, as other mechanisms such as FDR-like may occur (see the text).
Two major routes may lead to the formation of both auto- and allotetraploids, either directly or indirectly via a triploid-bridge (Figure 1). Theoretically, the highest level of variability is obtained when tetraploids arise directly from the merger of two unreduced gametes provided by different diploid individuals, especially when they belong to the (strict) FDR-type. Indeed, increasing proportions of tri- and tetra-alleles per locus are expected in autotetraploids, while allotetraploids systematically benefit from a full heterozygosity between homologs and homoeologs (Watanabe et al., 1991). Nevertheless, a partial loss of variability may occur between homologs when tetraploids originate from SDR (or FDR-like) unreduced gametes. Except in case of spontaneous mutation preventing the separation of homologs or chromatids in male and female meiosis, two unreduced gametes have relatively low chance to merge in a single step (Ramsey and Schemske, 1998). Therefore, even though a lower variability is expected by this route, it is suggested that tetraploid species are most presumably formed in two steps via a triploid bridge (Husband, 2004). Indeed, triploids resulting through the merger of reduced and unreduced gametes are fertile in a large range of plant lineages (Ramanna and Jacobsen, 2003) and may form a tetraploid either by self-fertilization or intercross with one diploid progenitor. From the first path (self-fertilization), two reduced gametes of the triploid individual are required. However, the resulting plants are often aneuploids, decreasing the likelihood of successful speciation (Figure 2; Leflon et al., 2006). From the second path (intercross), the triploid must provide an unreduced gamete, likely arising from an indeterminate meiotic restitution (IMR), where FDR and SDR occur simultaneously for unpaired and paired chromosomes, respectively (Figure 2; De Storme and Geelen, 2013).
Allotetraploids may also arise from amphihaploid or autotetraploid intermediates, most presumably in a single step, even though both can first promote an allotriploid bridge (Figure 1). However, these paths may have a low natural occurrence. Indeed, while amphihaploids are generally poorly fertile (Mason and Pires, 2015), the success of allotetraploid formation through an autotetraploid closely relates to the ability of this latter to generate well-balanced reduced gametes (see the following sections). The path by which the autotetraploids arose conditions the variability per subgenome of the resulting allotetraploid. On the other hand, the amphihaploid path will provide a lower variability than all routes previously described as only one chromatid of each chromosome is assigned per dyad through (strict) FDR-type, maintaining exclusively the heterozygosity between homoeologs (Figure 2). Even more harmful, SDR-type (and FDR-like) may result in unbalanced dyads due to the occurrence of crossovers between homoeologs (Cifuentes et al., 2010).
Managing the Variability in Newly Formed Polyploids Through Disturbed Meiotic Recombination
Although newly formed polyploids may sometimes successfully establish via the main use of vegetative reproduction, as in the young allodecaploid Spartina anglica (Ainouche et al., 2004), most favor sexual reproduction (Comai, 2005; Nasrallah, 2017). In addition to harboring a higher variability (Figure 1), populations deriving from this latter route will benefit from different patterns of meiotic recombination, increasing their chance of adaptation in new ecological niches and their speciation success through natural selection.
While the generation of diversity is usually hampered, polyploid plants are often able to overcome the limits arising from the tight regulation of meiotic recombination (Mercier et al., 2015). Indeed, as reported in Arabidopsis, Brassica, Gossypium, Phlox, and Zea genera, newly formed auto- and allotetraploids exhibit higher crossover frequencies between their homologous chromosomes than their diploid progenitors (Bingham et al., 1968; Raghuvanshi and Pathak, 1975; Desai et al., 2006; Leflon et al., 2010; Pecinka et al., 2011). For instance, in allotetraploids Brassica napus (AACC, 2n = 4x = 38), resulting from the hybridization of Brassica rapa (AA, 2n = 2x = 20) and Brassica oleracea (CC, 2n = 2x = 18) (Nagaharu, 1935), about twice as many crossovers were detected between A homologs than in diploid AA plants; whilst both displayed identical A genotypes (Leflon et al., 2010). Similarly, substantial increase of crossover frequencies was found in resynthesized triploids, typically lower than in tetraploids (Gymer and Whittington, 1975; Raghuvanshi and Pathak, 1975), but astonishingly more elevated in Brassica AAC allotriploids (Leflon et al., 2010). Moreover, Pelé et al. (2017) have recently pointed out that this crossovers boost occurring within the A genome was strikingly associated with reduced interference and dramatic changes in the shape of recombination landscapes. While the molecular mechanisms remain unknown, observations made in Brassica aneuploids suggest that this phenomenon is genetically controlled. Indeed, Suay et al. (2014) demonstrated that the boost of crossovers arising in AAC allotriploids relates to specific additional C chromosomes; the single C09 explaining 50% of the overall variation. Although exceptions have been reported in Clitoria ternatea and Secale cereale (Hazarika and Rees, 1967; Gandhi and Patil, 1997), enhanced recombination frequencies may have huge repercussions on the speciation success. The wider diversity of resulting gametes may indeed accelerate the elimination of deleterious alleles and facilitate in the long run adaptation of neopolyploids to adverse environmental situations.
In newly formed allopolyploids, meiotic recombination may also occur between the homoeologous chromosomes, as reported in diverse species including Brassica napus, Coffea arabica, Nicotiana tabacum, and Tragopogon miscellus (Song et al., 1995; Lim et al., 2004; Gaeta et al., 2007; Gaeta and Chris Pires, 2010; Xiong et al., 2011; Chester et al., 2012; Lashermes et al., 2014). Detected as early as the first meiosis of resynthesized allotetraploids (Szadkowski et al., 2010), homoeologous recombination frequency often correlates with the existing collinearity between homoeologs and varies according to the route of polyploid formation (Szadkowski et al., 2011; Rousseau-Gueutin et al., 2016). For instance, while such events are almost inexistent in the previously described Brassica AAC allotriploids (Leflon et al., 2006; Pelé et al., 2017), they commonly occur in ACC allotriploids and AC amphihaploids (Cifuentes et al., 2010; Yang et al., 2017). Moreover, the resulting homoeologous exchanges are smaller and more frequent when arising from unreduced gametes of amphihaploids rather than by somatic doubling (Szadkowski et al., 2011). These homoeologous exchanges deeply impact the variability and gene content of newly formed polyploids. For instance, the young allotetraploid Coffea arabica showed about 5% of homoeologous gene loss since its formation (Lashermes et al., 2014). Even more astonishing, up to 10% of genes are impacted after only three generations following the resynthesis of Brassica napus (Rousseau-Gueutin et al., 2016), highlighting that homoeologous exchanges are a major cause of gene copy number variation in Brassica napus varieties (Hurgobin et al., 2018). In some instances, these structural changes are at the origin of phenotypic variations, such as flowering time divergence, seed quality or disease resistance (Pires et al., 2004; Zhao et al., 2006; Stein et al., 2017), which may have contributed in the ability of allopolyploid species to exploit a wider range of environmental conditions.
Ensuring a Diploid-Like Meiosis to Get Fully Established Through Overall or Targeted Depletion of Meiotic Crossovers
The presence of more than one possible partner to pair and recombine with may however lead to the generation of unbalanced gametes and reduced fertility, whenever illegitimate or multiple associations arise between chromosomes at Metaphase I of meiosis (Ramsey and Schemske, 2002). Nevertheless, while commonly (but not systematically) observed in resynthesized polyploids, such associations unfrequently occur in the established ones (Figure 1 and Supplementary Table 1). Considering the contrasted examples summarized in Supplementary Table 1, it seems that the mechanisms leading to a diploid-like meiosis (referred as ‘partial diploidization’) may either already exist in the parental diploids or are set up after the polyploid formation. Indeed, some species show a global genome stasis since their first meiosis (see Gossypium hirsutum), while others revealed increasing proportion of bivalents in the following generations (see Arabidopsis thaliana, Pennisetum typhoides). Although, this may be species-specific, the partial diploidization requires a particular regulation of meiotic recombination that differs according to the polyploids type.
In autopolyploids, multiple copies of every chromosome are true homologs thereby sharing the same chance to pair and recombine with each other. Consequently, when all homologs align in parallel during the Prophase I of meiosis, multiple associations may occur (Lloyd and Bomblies, 2016). However, while these associations are dissolved prior to Metaphase I in established autopolyploids, which primarily form bivalents through a random assortment of homologs into pair (i.e., polysomic inheritance), they are frequently maintained in those resynthesized that exhibit trivalents and/or tetravalents (Supplementary Table 1). Theoretically, a sharp reduction in the overall number of crossovers can overcome this fate, especially by ensuring a single crossover per chromosome (Lloyd and Bomblies, 2016). Although exceptions were reported, this theory has gained concrete support in the autotetraploid Arabidopsis arenosa. Indeed, while multivalents and increased crossover rates are observed following polyploidy induction, natural accessions exhibit predominantly bivalents with on average 1.09 crossover (Carvalho et al., 2010; Pecinka et al., 2011; Yant et al., 2013). The molecular basis of the overall crossover number reduction in established autopolyploids remains unknown but it is suggested to result from elevated interference given that the obligatory crossover is maintained per homolog pair (Bomblies et al., 2016). Additionally, genomic comparison of Arabidopsis arenosa and its related diploids evidenced the selection of a few meiotic genes involved in the process of crossover formation, thereby providing a list of candidates to test (Yant et al., 2013).
In allopolyploids the situation is even more challenging because of their hybrid origin. Indeed, generation of balanced gametes requires that chromosomes form pairs, instead of multivalents, and that pairs are restricted to homologs (i.e., disomic inheritance). Targeted rather than overall reduction in the number of crossovers is therefore more relevant for dissolving illegitimate associations occurring when homoeologs align in parallel during the Prophase I (Lloyd and Bomblies, 2016). Consistently, allopolyploids seem to maintain elevated crossover rates between their homologs throughout their evolution. Indeed, like resynthesized allotetraploids, cultivars of Brassica napus show twice more crossovers than related diploids (Wang et al., 2012; Chalhoub et al., 2014; Cai et al., 2017). Although efficient, homoeologs recognition is not completely error proof as small homoeologous exchanges may be detected in modern allopolyploids (Lloyd et al., 2018), but to a lesser extent than in the resynthesized allopolyploids (Supplementary Table 1). Previously thought to result from the increased divergence between homoeologous genomes (Feldman et al., 1997), it is now considered that this process is more likely genetically controlled (Jenczewski and Alix, 2004). So far, only the Pairing homoeologous 1 (Ph1) locus acting in the hexaploid bread wheat (Triticum aestivum, AABBDD, 2n = 6x = 42) has been molecularly characterized (Sears, 1976; Griffiths et al., 2006). Briefly, this latter corresponds to a cluster of defective cyclin dependent kinases-like (CDKs) and methyl-transferase genes, where is inserted a paralog of the major crossover gene ZIP4 that is responsible for the Ph1 phenotype (Knight et al., 2010; Greer et al., 2012; Martín et al., 2014, 2017). Indeed, this latter ZIP4 copy was recently shown to promote homologous recombination while inhibiting the maturation of crossovers between homoeologs (Rey et al., 2017). Moreover, although the underlying gene remains unknown, a Ph2 locus acting on the synapsis progression has been identified in wheat, likely promoting the Ph1 efficiency rather than directly suppressing homoeologs crossovers (Martinez et al., 2001; Sutton et al., 2003). Finally, two further genomic regions limiting homoeologous recombination have been mapped in Arabidopsis suecica (BYS) and Brassica napus (PrBn) (Liu et al., 2006; Henry et al., 2014). However, while BYS explains less than 10% of the variability, the efficiency of PrBn in the allotetraploid Brassica napus remains unclear as it was detected through a segregating population of amphihaploids and may thereby act exclusively in a single dose (Nicolas et al., 2009; Grandont et al., 2014).
Conclusion and Perspective
In this review, we showed that a particular regulation of meiotic recombination may have huge repercussions on the level of genetic diversity and genome stability of polyploids, and thereafter on their speciation success through natural selection. While the molecular basis of meiotic recombination has been strongly investigated in diploid species (for review see Mercier et al., 2015; Zickler and Kleckner, 2015), with recent discoveries of genes and factors (i.e., genomic and epigenetic) controlling formation and frequency of crossovers (Fernandes et al., 2017; Ziolkowski et al., 2017; Serra et al., 2018; Underwood et al., 2018), far less is known in polyploids. However, it has been shown that following polyploidization, duplicated copies of genes regulating meiosis and recombination process are preferentially lost (De Smet et al., 2013; Lloyd et al., 2014). Therefore, with a special attention on meiotic dosage-sensitive genes, and by taking advantage of the increasing number of sequenced polyploid plant genomes as well as of the major advances in NGS and genome editing (Crispr-Cas9) technologies, it will be possible to better understand the molecular mechanisms governing regulation of meiotic recombination in polyploids, from their formation toward their establishment. This increased knowledge on meiotic recombination will thereafter facilitate the growth of genetic diversity or introgression of gene of interest in polyploid crops.
Author Contributions
AP organized and prepared the major part of the manuscript. A-MC and MR-G contributed to writing and reviewing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Julie Ferreira de Carvalho (UMR IGEPP, France) and Dr. Julia Zinsmeister (Enza Zaden B.V., Netherlands) for their critical review of the manuscript.
Footnotes
Funding. This work was partly supported by BAP INRA Department and ANR CROC: Project ANR-14-CE19-0004. AP was supported by a fellowship from BAP INRA and Conseil Régional de Bretagne.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00907/full#supplementary-material
References
- Ainouche M. L., Baumel A., Salmon A. (2004). Spartina anglica CE Hubbard: a natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biol. J. Linn. Soc. 82 475–484. 10.1111/j.1095-8312.2004.00334.x [DOI] [Google Scholar]
- Alix K., Gérard P. R., Schwarzacher T., Heslop-Harrison J. S. (2017). Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants. Ann. Bot. 120 183–194. 10.1093/aob/mcx079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allario T., Brumos J., Colmenero-Flores J. M., Iglesias D. J., Pina J. A., Navarro L., et al. (2013). Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 36 856–868. 10.1111/pce.12021 [DOI] [PubMed] [Google Scholar]
- Bingham E. T. (1980). “Maximizing heterozygosity in autopolyploids,” in Polyploidy, ed. Lewis W. H. (Boston, MA: Springer; ), 471–489. 10.1007/978-1-4613-3069-1_24 [DOI] [PubMed] [Google Scholar]
- Bingham E. T., Burnham C. R., Gates C. E. (1968). Double and single backcross linkage estimates in autotetraploid maize. Genetics 59 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaine Marchant D., Soltis D. E., Soltis P. S. (2016). Patterns of abiotic niche shifts in allopolyploids relative to their progenitors. New Phytol. 212 708–718. 10.1111/nph.14069 [DOI] [PubMed] [Google Scholar]
- Bomblies K., Jones G., Franklin C., Zickler D., Kleckner N. (2016). The challenge of evolving stable polyploidy: could an increase in “crossover interference distance” play a central role? Chromosoma 125 287–300. 10.1007/s00412-015-0571-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretagnolle F., Thompson J. D. (1995). Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 129 1–22. 10.1111/j.1469-8137.1995.tb03005.x [DOI] [PubMed] [Google Scholar]
- Brownfield L., Köhler C. (2011). Unreduced gamete formation in plants: mechanisms and prospects. J. Exp. Bot. 62 1659–1668. 10.1093/jxb/erq371 [DOI] [PubMed] [Google Scholar]
- Cai C., Wang X., Liu B., Wu J., Liang J., Cui Y., et al. (2017). Brassica rapa genome 2.0: a reference upgrade through sequence re-assembly and gene re-annotation. Mol. Plant 10 649–651. 10.1016/j.molp.2016.11.008 [DOI] [PubMed] [Google Scholar]
- Carvalho A., Delgado M., Barão A., Frescatada M., Ribeiro E., Pikaard C. S., et al. (2010). Chromosome and DNA methylation dynamics during meiosis in the autotetraploid Arabidopsis arenosa. Sex. Plant Reprod. 23 29–37. 10.1007/s00497-009-0115-2 [DOI] [PubMed] [Google Scholar]
- Chalhoub B., Denoeud F., Liu S., Parkin I. A., Tang H., Wang X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345 950–953. 10.1126/science.1253435 [DOI] [PubMed] [Google Scholar]
- Chester M., Gallagher J. P., Symonds V. V., da Silva A. V. C., Mavrodiev E. V., Leitch A. R., et al. (2012). Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. U.S.A. 109 1176–1181. 10.1073/pnas.1112041109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes M., Eber F., Lucas M. O., Lodé M., Chèvre A.-M., Jenczewski E. (2010). Repeated polyploidy drove different levels of crossover suppression between homoeologous chromosomes in Brassica napus allohaploids. Plant Cell 22 2265–2276. 10.1105/tpc.109.072991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6 836–846. 10.1038/nrg1711 [DOI] [PubMed] [Google Scholar]
- De Smet R., Adams K. L., Vandepoele K., Van Montagu M. C., Maere S., Van de Peer Y. (2013). Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc. Natl. Acad. Sci. U.S.A. 110 2898–2903. 10.1073/pnas.1300127110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N., Copenhaver G. P., Geelen D. (2012). Production of diploid male gametes in Arabidopsis by cold-induced destabilization of postmeiotic radial microtubule arrays. Plant Physiol. 160 1808–1826. 10.1104/pp.112.208611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N., Geelen D. (2013). Sexual polyploidization in plants – cytological mechanisms and molecular regulation. New Phytol. 198 670–684. 10.1111/nph.12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I., Cromer L., Jolivet S., Girard C., Horlow C., Sun Y., et al. (2010). The cyclin-A CYCA1; 2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet. 6:e1000989. 10.1371/journal.pgen.1000989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I., Jolivet S., Froger N., Catrice O., Novatchkova M., Mercier R. (2009). Turning meiosis into mitosis. PLoS Biol. 7:e1000124. 10.1371/journal.pbio.1000124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Chee P. W., Rong J., May O. L., Paterson A. H. (2006). Chromosome structural changes in diploid and tetraploid A genomes of Gossypium. Genome 49 336–345. 10.1139/g05-116 [DOI] [PubMed] [Google Scholar]
- Feldman M., Liu B., Segal G., Abbo S., Levy A. A., Vega J. M. (1997). Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J. B., Seguéla-Arnaud M., Larchevêque C., Lloyd A. H., Mercier R. (2017). Unleashing meiotic crossovers in hybrid plants. Proc. Natl. Acad. Sci. U.S.A. 115 2431–2436. 10.1073/pnas.1713078114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta R. T., Chris Pires J. (2010). Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol. 186 18–28. 10.1111/j.1469-8137.2009.03089.x [DOI] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., Iniguez-Luy F., Leon E., Osborn T. C. (2007). Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19 3403–3417. 10.1105/tpc.107.054346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S., Patil V. P. (1997). Colchicine-induced autotetraploidy in Clitoria ternatea L. Cytologia 62 13–18. 10.1508/cytologia.62.13 [DOI] [Google Scholar]
- Grandont L., Cuñado N., Coriton O., Huteau V., Eber F., Chèvre A. M., et al. (2014). Homoeologous chromosome sorting and progression of meiotic recombination in Brassica napus: ploidy does matter! Plant Cell 26 1448–1463. 10.1105/tpc.114.122788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E., Martín A. C., Pendle A., Colas I., Jones A. M., Moore G., et al. (2012). The Ph1 locus suppresses Cdk2-type activity during premeiosis and meiosis in wheat. Plant Cell 24 152–162. 10.1105/tpc.111.094771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S., Sharp R., Foote T. N., Bertin I., Wanous M., Reader S., et al. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439 749–752. 10.1038/nature04434 [DOI] [PubMed] [Google Scholar]
- Gymer P. T., Whittington W. J. (1975). Hybrids between Lolium perenne and Festuca pratensis. New Phytol. 74 295–306. 10.1111/j.1469-8137.1975.tb02618.x [DOI] [Google Scholar]
- Harlan J. R., de Wet J. M. J. (1975). On Ö. Winge and a prayer: the origins of polyploidy. Bot. Rev. 41 361–390. 10.1007/BF02860830 [DOI] [Google Scholar]
- Hazarika M. H., Rees H. (1967). Genotypic control of chromosome behaviour in rye X. Chromosome pairing and fertility in autotetraploids. Heredity 22 317–332. 10.1038/hdy.1967.44 [DOI] [Google Scholar]
- Henry I. M., Dilkes B. P., Tyagi A., Gao J., Christensen B., Comai L. (2014). The BOY NAMED SUE quantitative trait locus confers increased meiotic stability to an adapted natural allopolyploid of Arabidopsis. Plant Cell 26 181–194. 10.1105/tpc.113.120626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurgobin B., Golicz A. A., Bayer P. E., Chan C.-K. K., Tirnaz S., Dolatabadian A., et al. (2018). Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol. J. 16 1265–1274. 10.1111/pbi.12867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband B. C. (2004). The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biol. J. Linn. Soc. 82 537–546. 10.1111/j.1095-8312.2004.00339.x [DOI] [Google Scholar]
- Husband B. C., Sabara H. A. (2004). Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae). New Phytol. 161 703–713. 10.1046/j.1469-8137.2004.00998.x [DOI] [PubMed] [Google Scholar]
- Husband B. C., Schemske D. W. (2000). Ecological mechanisms of reproductive isolation between diploid and tetraploid Chamerion angustifolium. J. Ecol. 88 689–701. 10.1046/j.1365-2745.2000.00481.x [DOI] [Google Scholar]
- Jenczewski E., Alix K. (2004). From diploids to allopolyploids: the emergence of efficient pairing control genes in plants. Crit. Rev. Plant Sci. 23 21–45. 10.1080/07352680490273239 [DOI] [Google Scholar]
- Jiao Y., Wickett N. J., Ayyampalayam S., Chanderbali A. S., Landherr L., Ralph P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473 97–100. 10.1038/nature09916 [DOI] [PubMed] [Google Scholar]
- Knight E., Greer E., Draeger T., Thole V., Reader S., Shaw P., et al. (2010). Inducing chromosome pairing through premature condensation: analysis of wheat interspecific hybrids. Funct. Integr. Genomics 10 603–608. 10.1007/s10142-010-0185-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiner J. M., Kron P., Husband B. C. (2017). Evolutionary dynamics of unreduced gametes. Trends Genet. 33 583–593. 10.1016/j.tig.2017.06.009 [DOI] [PubMed] [Google Scholar]
- Lashermes P., Combes M. C., Hueber Y., Severac D., Dereeper A. (2014). Genome rearrangements derived from homoeologous recombination following allopolyploidy speciation in coffee. Plant J. 78 674–685. 10.1111/tpj.12505 [DOI] [PubMed] [Google Scholar]
- Leflon M., Eber F., Letanneur J. C., Chelysheva L., Coriton O., Huteau V., et al. (2006). Pairing and recombination at meiosis of Brassica rapa (AA) ×Brassica napus (AACC) hybrids. Theor. Appl. Genet. 113 1467–1480. 10.1007/s00122-006-0393-0 [DOI] [PubMed] [Google Scholar]
- Leflon M., Grandont L., Eber F., Huteau V., Coriton O., Chelysheva L., et al. (2010). Crossovers get a boost in Brassica allotriploid and allotetraploid hybrids. Plant Cell 22 2253–2264. 10.1105/tpc.110.075986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. Y., Matyasek R., Kovarik A., Leitch A. R. (2004). Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 82 599–606. 10.1111/j.1095-8312.2004.00344.x [DOI] [Google Scholar]
- Liu Z., Adamczyk K., Manzanares-Dauleux M., Eber F., Lucas M. O., Delourme R., et al. (2006). Mapping PrBn and other quantitative trait loci responsible for the control of homeologous chromosome pairing in oilseed rape (Brassica napus L.) haploids. Genetics 174 1583–1596. 10.1534/genetics.106.064071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A., Blary A., Charif D., Charpentier C., Tran J., Balzergue S., et al. (2018). Homoeologous exchanges cause extensive dosage-dependent gene expression changes in an allopolyploid crop. New Phytol. 217 367–377. 10.1111/nph.14836 [DOI] [PubMed] [Google Scholar]
- Lloyd A., Bomblies K. (2016). Meiosis in autopolyploid and allopolyploid Arabidopsis. Curr. Opin. Plant Biol. 30 116–122. 10.1016/j.pbi.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Lloyd A. H., Ranoux M., Vautrin S., Glover N., Fourment J., Charif D., et al. (2014). Meiotic gene evolution: can you teach a new dog new tricks? Mol. Biol. Evol. 31 1724–1727. 10.1093/molbev/msu119 [DOI] [PubMed] [Google Scholar]
- Martín A. C., Rey M. D., Shaw P., Moore G. (2017). Dual effect of the wheat Ph1 locus on chromosome synapsis and crossover. Chromosoma 126 669–680. 10.1007/s00412-017-0630-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín A. C., Shaw P., Phillips D., Reader S., Moore G. (2014). Licensing MLH1 sites for crossover during meiosis. Nat. Commun. 5:4580. 10.1038/ncomms5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M., Cuñado N., Carcelén N., Romero C. (2001). The Ph1 and Ph2 loci play different roles in the synaptic behaviour of hexaploid wheat Triticum aestivum. Theor. Appl. Genet. 103 398–405. 10.1007/s00122-001-0543-3 [DOI] [Google Scholar]
- Mason A. S., Nelson M. N., Yan G., Cowling W. A. (2011). Production of viable male unreduced gametes in Brassica interspecific hybrids is genotype specific and stimulated by cold temperatures. BMC Plant Biol. 11:103. 10.1186/1471-2229-11-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason A. S., Pires J. C. (2015). Unreduced gametes: meiotic mishap or evolutionary mechanism? Trends Genet. 31 5–10. 10.1016/j.tig.2014.09.011 [DOI] [PubMed] [Google Scholar]
- McIntyre P. J. (2012). Polyploidy associated with altered and broader ecological niches in the Claytonia perfoliata (Portulacaceae) species complex. Am. J. Bot. 99 655–662. 10.3732/ajb.1100466 [DOI] [PubMed] [Google Scholar]
- Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M. (2015). The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66 297–327. 10.1146/annurev-arplant-050213-035923 [DOI] [PubMed] [Google Scholar]
- Nagaharu U. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7 389–452. [Google Scholar]
- Nasrallah J. B. (2017). Plant mating systems: self-incompatibility and evolutionary transitions to self-fertility in the mustard family. Curr. Opin. Genet. Dev. 47 54–60. 10.1016/j.gde.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Nicolas S. D., Leflon M., Monod H., Eber F., Coriton O., Huteau V., et al. (2009). Genetic regulation of meiotic cross-overs between related genomes in Brassica napus haploids and hybrids. Plant Cell 21 373–385. 10.1105/tpc.108.062273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A., Fang W., Rehmsmeier M., Levy A. A., Scheid O. M. (2011). Polyploidization increases meiotic recombination frequency in Arabidopsis. BMC Biol. 9:24. 10.1186/1741-7007-9-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pécrix Y., Rallo G., Folzer H., Cigna M., Gudin S., Le Bris M. (2011). Polyploidization mechanisms: temperature environment can induce diploid gamete formation in Rosa sp. J. Exp. Bot. 62 3587–3597. 10.1093/jxb/err052 [DOI] [PubMed] [Google Scholar]
- Pelé A., Falque M., Trotoux G., Eber F., Nègre S., Gilet M., et al. (2017). Amplifying recombination genome-wide and reshaping crossover landscapes in Brassicas. PLoS Genet. 13:e1006794. 10.1371/journal.pgen.1006794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires J. C., Zhao J., Schranz M. E., Leon E. J., Quijada P. A., Lukens L. N., et al. (2004). Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. 82 675–688. 10.1111/j.1095-8312.2004.00350.x [DOI] [Google Scholar]
- Raghuvanshi S. S., Pathak C. S. (1975). Polyploid breeding and possibility of raising double varieties in Phlox drummondii, Hook. Cytologia 40 355–363. 10.1508/cytologia.40.355 [DOI] [Google Scholar]
- Ramanna M. S., Jacobsen E. (2003). Relevance of sexual polyploidization for crop improvement–A review. Euphytica 133 3–8. 10.1023/A:1025600824483 [DOI] [Google Scholar]
- Ramsey J., Schemske D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29 467–501. 10.1146/annurev.ecolsys.29.1.467 28034953 [DOI] [Google Scholar]
- Ramsey J., Schemske D. W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33 589–639. 10.1146/annurev.ecolsys.33.010802.150437 [DOI] [Google Scholar]
- Rey M. D., Martín A. C., Higgins J., Swarbreck D., Uauy C., Shaw P., et al. (2017). Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol. Breed. 37:95. 10.1007/s11032-017-0700-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin M., Morice J., Coriton O., Huteau V., Trotoux G., Nègre S., et al. (2016). The impact of open pollination on the structural evolutionary dynamics, meiotic behavior and fertility of resynthesized allotetraploid Brassica napus L. G3 7 705–717. 10.1534/g3.116.036517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears E. R. (1976). Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 10 31–51. 10.1146/annurev.ge.10.120176.000335 [DOI] [PubMed] [Google Scholar]
- Serra H., Lambing C., Griffin C. H., Topp S. D., Nageswaran D. C., Underwood C. J., et al. (2018). Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc. Natl. Acad. Sci. U.S.A. 115 2437–2442. 10.1073/pnas.1713071115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Lu P., Tang K., Osborn T. C. (1995). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. U.S.A. 92 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G. L. (1947). Types of polyploids: their classification and significance. Adv. Genet. 1 403–429. 10.1016/S0065-2660(08)60490-3 [DOI] [PubMed] [Google Scholar]
- Stebbins G. L. (1980). “Polyploidy in plants: unsolved problems and prospects,” in Polyploidy, ed. Lewis W. H. (Boston, MA: Springer; ), 495–520. 10.1007/978-1-4613-3069-1_26 [DOI] [PubMed] [Google Scholar]
- Stebbins G. L. (1985). Polyploidy, hybridization, and the invasion of new habitats. Ann. Mo. Bot. Gard. 72 824–832. 10.2307/2399224 [DOI] [Google Scholar]
- Stein A., Coriton O., Rousseau-Gueutin M., Samans B., Schiessl S. V., Obermeier C., et al. (2017). Mapping of homoeologous chromosome exchanges influencing quantitative trait variation in Brassica napus. Plant Biotechnol. J. 15 1478–1489. 10.1111/pbi.12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suay L., Zhang D., Eber F., Jouy H., Lodé M., Huteau V., et al. (2014). Crossover rate between homologous chromosomes and interference are regulated by the addition of specific unpaired chromosomes in Brassica. New Phytol. 201 645–656. 10.1111/nph.12534 [DOI] [PubMed] [Google Scholar]
- Sutton T., Whitford R., Baumann U., Dong C., Able J. A., Langridge P. (2003). The Ph2 pairing homoeologous locus of wheat (Triticum aestivum): identification of candidate meiotic genes using a comparative genetics approach. Plant J. 36 443–456. 10.1046/j.1365-313X.2003.01891.x [DOI] [PubMed] [Google Scholar]
- Szadkowski E., Eber F., Huteau V., Lodé M., Coriton O., Jenczewski E., et al. (2011). Polyploid formation pathways have an impact on genetic rearrangements in resynthesized Brassica napus. New Phytol. 191 884–894. 10.1111/j.1469-8137.2011.03729.x [DOI] [PubMed] [Google Scholar]
- Szadkowski E., Eber F., Huteau V., Lodé M., Huneau C., Belcram H., et al. (2010). The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186 102–112. 10.1111/j.1469-8137.2010.03182.x [DOI] [PubMed] [Google Scholar]
- Tamayo-Ordóñez M. C., Espinosa-Barrera L. A., Tamayo-Ordóñez Y. J., Ayil-Gutiérrez B., Sánchez-Teyer L. F. (2016). Advances and perspectives in the generation of polyploid plant species. Euphytica 209 1–22. 10.1007/s10681-016-1646-x [DOI] [Google Scholar]
- Tayalé A., Parisod C. (2013). Natural pathways to polyploidy in plants and consequences for genome reorganization. Cytogenet. Genome Res. 140 79–96. 10.1159/000351318 [DOI] [PubMed] [Google Scholar]
- Underwood C. J., Choi K., Lambing C., Zhao X., Serra H., Borges F., et al. (2018). Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 28 519–531. 10.1101/gr.227116.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., Mizrachi E., Marchal K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18 411–424. 10.1038/nrg.2017.26 [DOI] [PubMed] [Google Scholar]
- Vanneste K., Baele G., Maere S., Van de Peer Y. (2014). Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary. Genome Res. 24 1334–1347. 10.1101/gr.168997.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veilleux R. (1983). Diploid and polyploid gametes in crop plants: mechanisms of formation and utilization in plant breeding. Plant Breed. Rev. 3 253–288. 10.1002/9781118061008.ch6 [DOI] [Google Scholar]
- Wang W., Huang S., Liu Y., Fang Z., Yang L., Hua W., et al. (2012). Construction and analysis of a high-density genetic linkage map in cabbage (Brassica oleracea L. var. capitata). BMC Genomics 13:523. 10.1186/1471-2164-13-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jha A. K., Chen R., Doonan J. H., Yang M. (2010). Polyploidy-associated genomic instability in Arabidopsis thaliana. Genesis 48 254–263. 10.1002/dvg.20610 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Peloquin S. J., Endo M. (1991). Genetic significance of mode of polyploidization: somatic doubling or 2n gametes? Genome 34 28–34. 10.1139/g91-005 [DOI] [Google Scholar]
- Werner J. E., Peloquin S. J. (1991). Significance of allelic diversity and 2n gametes for approaching maximum heterozygosity in 4 x potatoes. Euphytica 58 21–29. 10.1007/BF00035336 [DOI] [Google Scholar]
- Xiong Z., Gaeta R. T., Pires J. C. (2011). Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. 108 7908–7913. 10.1073/pnas.1014138108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wei X., Shi G., Wei F., Braynen J., Zhang J., et al. (2017). Molecular and cytological analyses of A and C genomes at meiosis in synthetic allotriploid Brassica hybrids (ACC) between B. napus (AACC) and B. oleracea (CC). J. Plant Biol. 60 181–188. 10.1007/s12374-016-0221-2 [DOI] [Google Scholar]
- Yant L., Hollister J. D., Wright K. M., Arnold B. J., Higgins J. D., Franklin F. C. H., et al. (2013). Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 23 2151–2156. 10.1016/j.cub.2013.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Udall J. A., Quijada P. A., Grau C. R., Meng J., Osborn T. C. (2006). Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor. Appl. Genet. 112 509–516. 10.1007/s00122-005-0154-5 [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (2015). Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7:a016626. 10.1101/cshperspect.a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski P. A., Underwood C. J., Lambing C., Martinez-Garcia M., Lawrence E. J., Ziolkowska L., et al. (2017). Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 31 306–317. 10.1101/gad.295501.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.