Graphical abstract
Method name: Phenol-chloroform RNA extraction
Keywords: RNA purification, RNA contamination, RNA extraction, RNA purity, Phenol-chloroform extraction, RNA quantification, NRVMs, NRVM, RT-qPCR
Abstract
Accurate and reliable analysis of gene expression depends on the extraction of pure and high-quality RNA. However, while the conventional phenol-chloroform RNA extraction is preferable over silica-based columns, particularly when cost is a concern or higher RNA yield is desired, it can result in significant RNA contamination. Contaminants including excess phenol, chloroform, or salts, can have significant impacts on downstream applications, including RNA quantification and reverse transcription, that can skew data collection and interpretation. To overcome the issue of RNA contamination in the conventional phenol-chloroform based RNA extraction method, we have optimized the protocol by adding one chloroform extraction step, and several RNA washing steps. Importantly, RNA quality and purity and accuracy in the quantification of RNA concentration were significantly improved with the modified protocol, resulting in reliable data collection and interpretation in downstream gene expression analysis.
-
•
Our protocol is customized by the addition of a second chloroform extraction step. Chloroform is carefully pipetted so as to not disturb the interphase layer. Any contaminants accidentally removed from interphase will be present in subsequent steps and can result in RNA contaminated with protein or phenol. The additional chloroform step increases RNA purity.
-
•
Additionally, the addition of 2 additional ethanol washes, initially intended to remove any residual salts from the isopropanol RNA precipitation step, also removed residual phenol contamination, enhancing RNA purity.
-
•
In summary, these modifications serve to enhance not only the purity of the RNA but, also increase the accuracy and reliability of RNA quantification.
Specifications Table [please fill in right-hand column of the table below]
| Subject area | Biochemistry, Genetics and Molecular Biology |
|---|---|
| More specific subject area |
|
| Method name | Phenol-Chloroform RNA Extraction |
| Name and reference of original method | Our method is an enhanced version of the Qiazol Total RNA extraction protocol: https://www.qiagen.com/us/resources/download.aspx?id=61c3ddbd-69c1-4b68-ab89-a428f14a9245&lang=en, which is the commercially available form of the chloroform-phenol/guanidine-based RNA extraction protocol originally developed and validated by Piotr Chomczynski and Nicoletta Sacchi in their publication [Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction, Chomczynski P, Sacchi N., Anal Biochem. 1987, Apr;162(1):156-9]. The publication was later updated, [The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on,’ Chomczynski P, Sacchi N., Nat Protoc. 2006;1(2):581-5]. All 3 methodologies are based on the use of guanidinium-phenol-chloroform to promote phase separation of biological mixtures and subsequent selective isolation of molecules of interest. |
| Resource availability | n/a |
Method details
Background
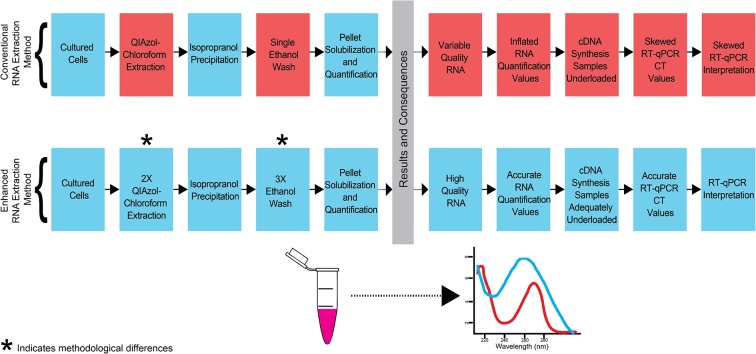
Ribonucleic acid (RNA) is a major macromolecule essential in various biological processes, from serving as a template for protein synthesis to catalyzing biological reactions. As such, isolated RNA is widely used in a number of molecular biology assays, including gene expression analysis via reverse-transcription quantitative polymerase chain reaction (RT-qPCR) arrays, and next generation sequencing. However, accurate and reliable gene expression data rely on the proper extraction of purified and high-quality RNA. Typically, RNA extraction is performed using one of two methods: phenol-chloroform based extraction or extraction using commercially available silica spin column kits. Phenol-chloroform based RNA extraction relies on the use of acid guanidinium thiocyanate-phenol-chloroform to promote phase separation of biological mixtures and subsequent selective isolation of molecules of interest [1,2]. Commercially available column-based kits on the other hand are often more straightforward, but can be relatively expensive compared to phenol-chloroform extraction methods, particularly when extracting RNA from a large number of samples. Additionally, phenol-chloroform based RNA extraction is advantageous when extracting RNA from small quantities of cells or tissues because it yields 2.4–93 times more RNA than silica column based protocols [[3], [4], [5]]. However, while phenol-chloroform extraction using proprietary phenol-based reagents (e.g., TRIzol; Thermo Fisher Scientific, Waltham, MA and QIAzol; Qiagen, Hilden Germany) is generally more economical, significant contaminants including phenol, guanidine, chloroform, and salt can remain in samples. The presence of these contaminants can affect both the quantitation of RNA on spectrophotometers as well as impact downstream assays. To overcome the issue of RNA contamination in the conventional phenol-chloroform based RNA extraction method (old), we have optimized the protocol by adding an additional chloroform step and several subsequent RNA washing steps using 75% ethanol (new). Fig. 1 demonstrates the differences between the old and new protocols. Importantly, we demonstrate that RNA quality and purity were significantly improved with the enhanced protocol, as was accuracy in the quantification of RNA concentration. Moreover, relative to samples with low levels of contaminants, cycle threshold (Ct) values after reverse-transcription and RT-qPCR were lower and less variable when using the new method, translating to a greater ability to detect low abundance transcripts and to detect differences between groups more reliably. Importantly, although the total time required to complete the enhanced protocol will vary by experience, lab equipment, and sample number, investigators should expect the enhanced protocol to lengthen the amount of time required to perform RNA extraction by a minimum of 30 min relative to the conventional protocol.
Fig. 1.
Comparison of the Enhanced RNA Extraction Method to the Conventional Extraction Method.
Table of defined steps in RNA extraction using the conventional (old) method versus our enhanced protocol (new). Notably, steps not included in the old protocol include: an additional chloroform step and several subsequent RNA washing steps using 75% ethanol.
Required reagents and equipment
-
•
Optional: RNase AWAY (Thermo Fisher Scientific, Waltham, MA)
-
•
QIAzol (or other proprietary phenol-based reagent)
-
•
Chloroform
-
•
Isopropanol
-
•
Ethanol (EtOH)
-
•
Nuclease-free water
-
•
1.5 mL capped tubes
-
•
1 mL pipette, 0.2 mL pipette and tips
-
•
Optional: 0.2 mL gel loading pipette tips
-
•
Refrigerated centrifuge or centrifuge chilled to 4 ℃
-
•
65 ℃ heat block
Procedure
Preparation
-
1
Prepare a clean area, using RNase AWAY spray or other chemical decontaminant.
Homogenization and RNA extraction
-
2
Aspirate media and add 500 μL of QIAzol to each well of cells.
Note: 500 μL of QIAzol is sufficient for 100,000–800,000 cells (plated in various well/plate sizes). If more QIAzol is necessary, volumes of chloroform and isopropanol in subsequent steps should be adjusted accordingly.
-
3
Let sit at room temperature (RT) for 3 min.
-
4
Scrape adherent cells using cell scraper and transfer to 1.5 mL tube.
-
5
Add 100 μL RNase-free chloroform to each tube containing 500 μL QIAzol.
-
6
Shake vigorously by hand for 15 s.
-
7
Let sit at RT for 3 min.
-
8
Centrifuge at 4 ℃ ≤12,000×g for 15 min.
-
9
Add 100 μL chloroform to a new 1.5 mL tube.
-
10
Transfer RNA-containing upper aqueous phase (clear supernatant) into chloroform.
-
11Repeat chloroform extraction 1 time:
-
aShake vigorously by hand for 15 s.
-
bLet set at RT for 3 min.
-
cCentrifuge at 4 ℃ ≤ 12,000×g for 15 min.
-
a
RNA precipitation
-
12
Add 250 μL RNase-free isopropanol to a new 1.5 mL tube.
-
13
Transfer RNA-containing upper aqueous phase (clear supernatant) into isopropanol.
-
14
Invert by hand 10–20 times to mix.
-
15
Let sit at RT for 10 min.
-
16
Centrifuge at 4 ℃ ≤12,000×g for 10 min to precipitate RNA.
Note: There should be a visible small white pellet following precipitation, however if RNA concentration is very low, a pellet may not be visible.
-
17
Remove supernatant and discard.
Note: Supernatant can be removed at this step by carefully tilting the tube, and pouring the supernatant out, without disturbing the pellet. Some remaining supernatant is fine.
RNA wash
-
18
Add 1 mL of 75% EtOH in nuclease-free water to pellet.
-
19
Centrifuge at 4 ℃ 7500×g for 5 min.
-
20
Remove supernatant and discard.
Note: Supernatant can be removed at this step by carefully tilting the tube, and pouring the supernatant out, without disturbing the pellet. Some remaining supernatant is fine.
-
21Repeat EtOH wash 2 times:
-
aAdd 1 mL of 75% EtOH in nuclease-free water to pellet.
-
bCentrifuge at 4 ℃ 7500×g for 5 min.
-
cRemove supernatant and discard.
-
dAdd 1 mL of 75% EtOH in nuclease-free water to pellet.
-
eCentrifuge at 4 ℃ 7500×g for 5 min.
-
fRemove supernatant and discard.
-
a
-
22
Pulse spin samples at RT.
-
23
Carefully remove remaining supernatant with pipette without disturbing the RNA pellet (gel loading tips work best).
-
24
Leave tubes open at RT for 3–5 min to evaporate EtOH.
Note: Alternatively, excess EtOH can be removed by carefully tipping tubes upside down onto a Kimwipe (Kimberly-Clark, Irving, TX) or other wicking paper.
-
25
Heat tubes open at 65 ℃ for 2–5 min to evaporate any remaining EtOH.
RNA solubilization
-
26
Add 20 μL of nuclease-free water to the RNA pellet.
Note: Volume of water to be added can be optimized for desired RNA concentration. Heat tubes at 65 ℃ for 2–5 min to solubilize RNA.
-
27
Vortex tubes 5–10 s, pulse spin, and place solubilized RNA on ice immediately.
-
28
Quantify RNA concentration and purity.
-
29
Use RNA for downstream applications or freeze at -80 ℃ immediately.
* Indicates methodological deviations from the conventional phenol-based RNA extraction protocol.
Method validation
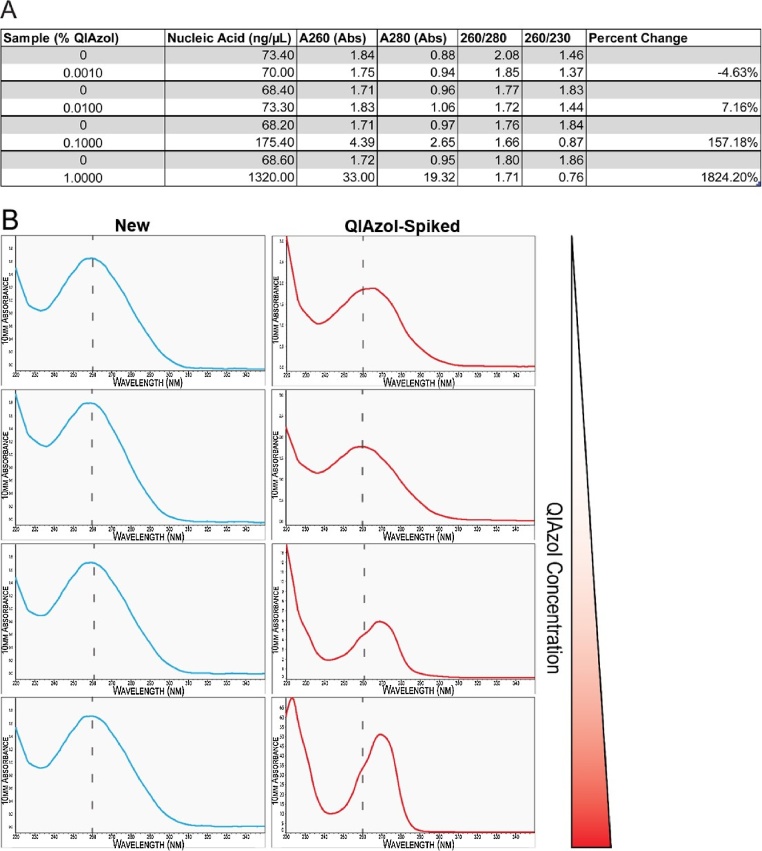
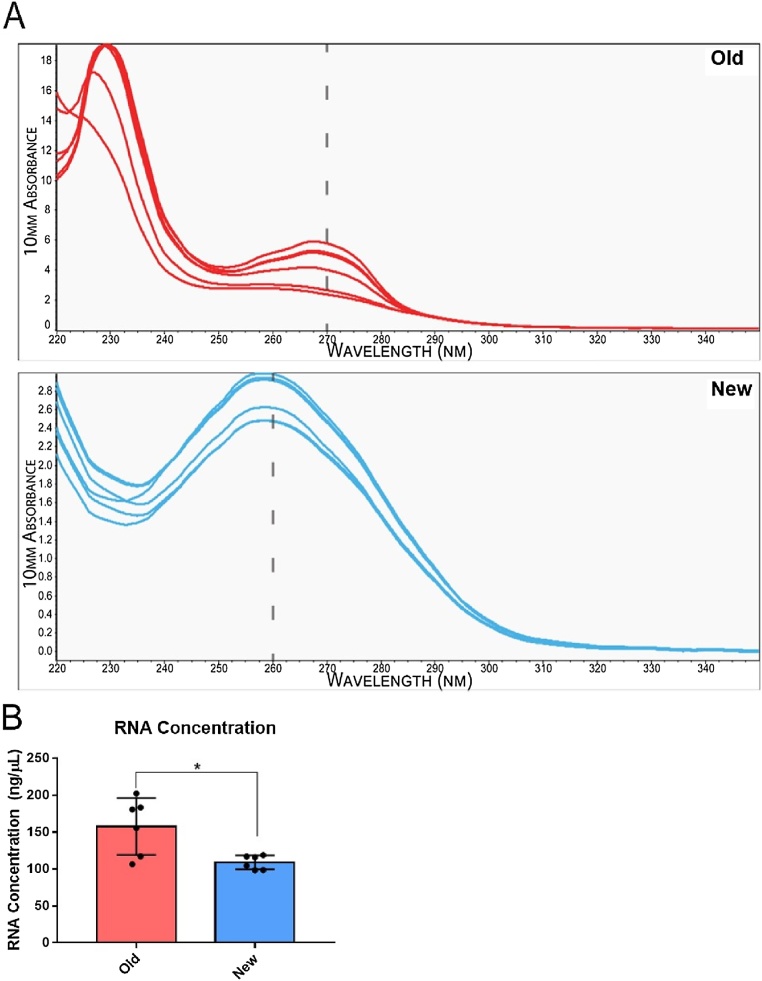
To quantify the impact of proprietary phenol-based reagents in purified RNA samples, QIAzol reagent was added to baseline concentrations of purified RNA from primary neonatal rat ventricular myocytes (NRVM). Baseline RNA concentrations using the new method and absorbance spectra were first recorded using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA). Then, to mimic typical phenol-based reagent contamination that is often observed with the standard (old) RNA extraction protocol, QIAzol was deliberately added to final concentrations of 0.001%, 0.010%, 0.10% and 1.0% (v/v %). Following QIAzol addition, NanoDrop readings were repeated to measure the impact of QIAzol contamination on both RNA concentration and purity based on the absorption spectrum (Fig. 2A and B). Our results indicate a linear relationship between the concentration of QIAzol contamination and an errant value of RNA concentration (ng/μL). The lowest concentration of QIAzol for example, 0.0010%, resulted in a change from baseline of -4.63%, while the highest concentration of QIAzol, 1.0%, resulted in a 1824.20% change from baseline (Fig. 2A and B). The intermediate amounts of QIAzol contamination, 0.1% and 0.01%, resulted in a 57.8% and 7.6% change respectively (Fig. 2A and B). Moreover, as the concentration of QIAzol increased, a distinct shift in the absorption spectrum towards higher wavelengths was observed, with a typical peak absorption at 270 nm, rather than 260 nm (Fig. 2B). In contrast, the absorption spectrum from RNA samples extracted with the enhanced (new) protocol have a distinct peak at 260 nm, which is the maximum wavelength at which DNA and RNA absorb UV (Fig. 3A), while using the standard (conventional) RNA extraction protocol frequently results in absorption spectra and RNA concentrations that resemble QIAzol-spiked samples (Fig. 3A). Notably, a second distinct and substantial peak appeared at approximately 230 nm indicating further non-nucleic acid residual contamination (Fig. 3A). Additionally, the quantification of RNA containing phenol-based contaminants was typically significantly inflated (p < 0.05), as compared to RNA purified using the new protocol (Fig. 3B).
Fig. 2.
Low Amounts of Phenol-Based Reagent Contamination Impacts RNA Quantitation.
RNA extraction utilizing our enhanced protocol was performed on 4 independent NRVM samples. (A) Depicted in the table are 4 representative baseline RNA concentrations measured using a NanoDrop 2000. QIAzol was added to purified RNA to achieve final concentrations of 0.0010%, 0.010%, 0.10% and 1.0%. RNA concentrations were re-measured and results recorded to determine the effect of QIAzol on concentration and absorption spectra of relevant wavelengths. (B) NanoDrop absorption spectra from 4 representative RNA samples purified with the enhanced RNA extraction protocol pre- and post-QIAzol spike-ins. Peak UV absorbance occurs at 260 nm in purified RNA (blue) and at 270 nm following addition of QIAzol (red). Dashed line corresponds to 260 nm on X-axis. Gradient on right hand side increases in red corresponding with increasing QIAzol concentration in QIAzol spiked samples (red) ranging from the lowest QIAzol concentration 0.001% to the highest concentration 1.00%.
Fig. 3.
Representative Spectra Comparing the Enhanced RNA Extraction Method to the Conventional Extraction Method.
RNA was extracted from six independent NRVM samples using either the conventional RNA extraction protocol (old-red) or the enhanced protocol (new-blue). (A) The spectra were recorded with a NanoDrop 2000 and overlayed for comparison within and between groups. Dashed line appears over 270 nm in the old protocol (red) and 260 nm in the new protocol (blue). (B) Box and whisker plots (min to max) of RNA concentrations based on the absorption spectra in RNA samples extracted using the old protocol versus the new protocol. Asterisk (*) denotes a significant difference between groups, unpaired T-test (*p < 0.05), n = 6 old, n = 6 new.
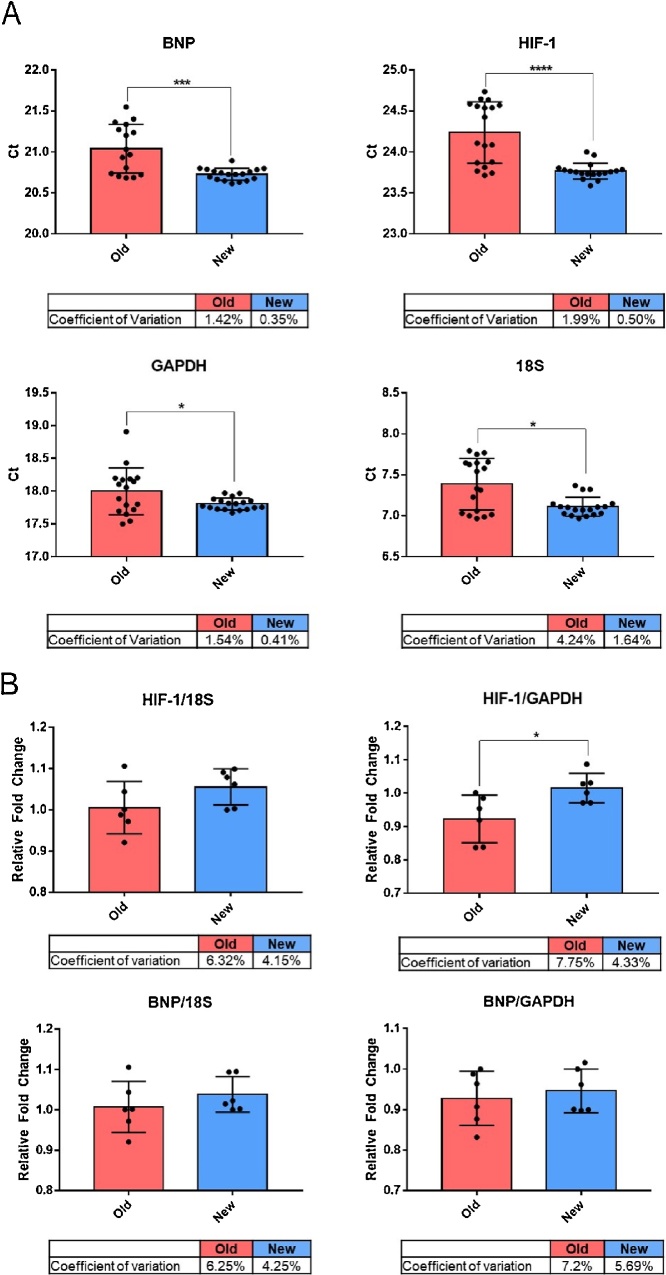
To quantify the effect of proprietary phenol-based reagents on RNA used in downstream applications, we performed RT-qPCR on contaminated and high-purity samples (old vs. new) of purified RNA from NRVM. Gene targets were chosen such that our data represent genes (in NRVM) with relatively high basal expression (natriuretic peptide type-B, BNP), low basal expression (hypoxia inducible factor, HIF-1), as well as two common housekeeping genes, ribosomal 18S RNA and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Our RT-qPCR results indicate that RNA purified using the old method resulted in significantly increased (p < 0.05) Ct values, as well as more variable Ct values (increased standard deviation, standard error, and higher coefficient of variation percentage) from samples of the same group relative to samples extracted using the newly adapted method (Fig. 4A). Moreover, even after normalization of each target to appropriate housekeeping genes (e.g., 18S or GAPDH) variability between samples remained higher in samples extracted using the old methods versus the modified extraction method (Fig. 4B).
Fig. 4.
RT-qPCR Analysis Comparing the Enhanced RNA Extraction Method to the Conventional Extraction Method.
RT-qPCR was performed on reverse-transcribed RNA that was extracted from six independent NRVM samples using either the conventional RNA extraction protocol (old) or the enhanced protocol (new). (A) Bar graphs of individual Ct values for genes BNP, HIF-1, GAPDH and 18S. For all groups, asterisk (*) denotes a significant difference between groups, unpaired T-test (*p < 0.05, ***p<0.005, ****p<0.001). Below each graph is the coefficient of variation for all Ct values. For all targets, n = 6 old, n = 6 new, all run in triplicate. (B) Bar graphs of average triplicate values for targets HIF-1 and BNP normalized to either housekeeping gene 18S or GAPDH. For all groups, asterisk (*) denotes a significant difference between groups, unpaired T-test (*p < 0.05). Below each graph is the coefficient of variation for all Ct values. For all targets, n = 6 old, n = 6 new.
NRVM were isolated from the ventricles of 1- to 2-day-old Sprague Dawley rats (Charles River, Wilmington, MA) by enzymatic digestions as described [6]. All animal protocols are in accordance with PHS Animal Welfare Assurance, ID A3269-01, and approved by the University of Colorado, Denver - Animal Care and Use Committee.
Conclusions
Contamination of RNA with proprietary phenol-based reagents such as QIAazol negatively impact RNA quantification, purity and results of downstream experiments. We demonstrated that RNA quality and purity were significantly improved with the modified protocol, as was accuracy in the quantification of RNA concentration based on the absorption spectra. Moreover, relative to samples with low levels of contaminants, Ct values after reverse-transcription and RT-qPCR were lower and less variable with the new method, translating to a greater ability to detect low abundance transcripts and to detect differences between groups more easily. While column-based RNA purification methods yield high-quality RNA, reagent cost becomes prohibitive especially when the number of samples is high. Additionally, RNA yield using column-based extraction methods is typically lower than non column-based purification methods. Our adaptable enhanced RNA extraction protocol effectively removes residual amounts of phenol-based contaminants without adding exorbitant demands on time, training or cost, and without decreasing overall RNA yield. Similar results were seen using the proprietary phenol-based reagent TRIzol (unpublished data). As such, we anticipate that these findings will be of interest to investigators who routinely quantify mRNA expression, particularly when extracting RNA from a large number of samples or from small quantities of cells or tissues.
Sources of funding
This work was supported by National Institutes of Health [HL126928 to S.M., HL107715 to B.S., HL119533 to C.S.], the Addison Scott Memorial Fund, the Boedecker Foundation, the Nair Family and the Jack Cooper Millisor Chair in Pediatric Heart Disease. LST was supported by 5T32HL007822-18. AMG was supported by NIH R01 HL126928-03S1.
Disclosures
CCS: Equity in miRagen, Inc., BLS: Research support from Forest Laboratories, Inc. CCS, BLS, SDM are founders and scientific advisors for CoramiR Biomedical, LLC.
Acknowledgements
We would like to acknowledge the Pediatric Cardiovascular Research Laboratory team at the University of Colorado, Denver, particularly those members involved in the preparation of neonatal rat ventricular myocytes.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.mex.2018.05.011.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. http://www.ncbi.nlm.nih.gov/pubmed/2440339 Internet, cited 2018 May 3, Available from: [DOI] [PubMed] [Google Scholar]
- 2.Chomczynski P., Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat. Protoc. 2006;1:581–585. doi: 10.1038/nprot.2006.83. http://www.ncbi.nlm.nih.gov/pubmed/17406285 Internet, cited 2018 May 3, Available from: [DOI] [PubMed] [Google Scholar]
- 3.Deng M.Y., Wang H., Ward G.B., Beckham T.R., McKenna T.S. Comparison of six RNA extraction methods for the detection of classical swine fever virus by real-time and conventional reverse transcription–PCR. J. Vet. Diagn. Invest. 2005;17:574–578. doi: 10.1177/104063870501700609. http://www.ncbi.nlm.nih.gov/pubmed/16475517 Internet, cited 2018 Feb 23, Available from: [DOI] [PubMed] [Google Scholar]
- 4.Santiago-Vazquez L.Z., Ranzer L.K., Kerr R.G. Comparison of two total RNA extraction protocols using the marine gorgonian coral pseudopterogorgia elisabethae and its symbiont Symbiodinium sp. Electron. J. Biotechnol. 2006;9 http://www.ejbiotechnology.info/content/vol9/issue5/full/15/index.html Internet, cited 2018 Feb 23, Available from: [Google Scholar]
- 5.Xiang X., Qiu D., Hegele R.D., Tan W.C. Comparison of different methods of total RNA extraction for viral detection in sputum. J. Virol. Methods. 2001;94:129–135. doi: 10.1016/s0166-0934(01)00284-1. http://www.ncbi.nlm.nih.gov/pubmed/11337047 Internet, cited 2018 Feb 23, Available from: [DOI] [PubMed] [Google Scholar]
- 6.Sucharov C.C., Dockstader K., Nunley K., McKinsey T.A., Bristow M. β-Adrenergic receptor stimulation and activation of protein kinase A protect against α1-adrenergic-mediated phosphorylation of protein kinase D and histone deacetylase 5. J. Card Fail. 2011;17:592–600. doi: 10.1016/j.cardfail.2011.03.006. http://www.ncbi.nlm.nih.gov/pubmed/21703532 Internet, cited 2016 Apr 5, Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.