Abstract
Background:
Circulating miRNAs (miRs) in the biofluids such as serum and plasma act as potential biomarkers for early diagnosis, treatment and prognosis. In the present study, an attempt made to see the expression of miR-21 in serum of 20 cases of Oral sub-mucous fibrosis (OSMF), 20 cases of Oral squamous cell carcinoma and 40 healthy volunteers. The expression of miR-21 was evaluated in relation to different demographical and clinicopathological features such as sex, tobacco, pan-masala, alcohol, smoking and clinical staging respectively with an aim to identify correlation with oral pre-cancer and cancer stages.
Materials and Methods:
The relative expression level of miR-21 was determined by quantitative real-time RT-PCR (qRT-PCR) in the sera of 20 OSCC, 20 OSMF patients and 40 healthy subjects as a control. Association between expression of miR-21 and OSCC clinical stages and demographical parameters such as sex, pan-masala, tobacco, smoking, alcohol have also been analyzed in detail.
Results:
The results obtained by t-test revealed significant increase in the expression level of miR-21 in OSCC as compared to OSMF. The study also revealed the positive correlation between higher miR-21 expression and pan-masala chewers as shown by t-test. The statistical test, ANOVA has also indicated a positive correlation between up-regulation of miR-21 in the clinical stages of the OSCC.
Conclusion:
The results of present study indicated up-regulation of circulating miR-21 in serum of OSCC as compared to OSMF (p=0.001), this study also elucidated the positive correlation between miR-21 expression in OSCC/OSMF patients, only one demographical parameter (Pan-masala) and negative correlation for other parameters such as sex, tobacco, smoking, alcohol etc. Other findings suggested a significant increase (p=0.000) in the expression of miR-21 in clinical staging (I-IV) of oral cancer. More studies are needed to validate it as potential diagnostic and prognostic biomarker for OSMF and OSCC for better management.
Keywords: Circulating miRNA, non-invassive biomarker, miR-21
Introduction
Oral sub-mucous fibrosis (OSMF) is a chronic, progressive, pre-malignant condition related with consumption of betal nut, tobacco, pan-masala chewing, alcohol and smoking. It shows pre-cancerous lesion in the oral cavity and is seen mainly in the subcontinent of India and South East Asia (Ranganathan et al., 2004). The prevalence of OSMF has increased over the last four decades from 0.03% to 6.42% in India (Pindborg et al., 1968; Hazarey et al., 2007). An estimate of 5 million OSMF patients in India has been reported for the previous published data (Aziz SR, 2010). Areca nut with tobacco increases the frequency of OSMF and leads to significant morbidity by transformation into oral squamous cell carcinoma (OSCC) (Haider et al., 2000).
Among all cancers, oral cancer is the top three cancers in India (Elango et al., 2006). Common sites of OSCC are the floor of the mouth, gingiva, palate, lip or in the tongue (Elango et al., 2006). The most common risk factors for oral cancer are betal nut chewing, chewing tobacco, cigarette smoking and severe alcoholism (Chaturvedi et al., 2013; Jornet et al., 2015). Poor diet and poor dental care are also the reason for oral cancer (Lin et al., 2011). Highest incidence of oral cancer is found in India, South and South Asian countries. The percentage of oral cancer is 90-95% squamous cell carcinoma in India (Sharma et al., 2015). According to the statistics of 2012, In India the incidence of oral cancer patients in males and females was 53,842 and 23,161 respectively (Gupta et al., 2014). Oral cancer is the eighth most common cancer worldwide (Duarte et al., 2007). Over past several decades in-spite of screening examination facilities, incidence of oral cancer and associated mortality has increased. OSCC can be cured if detected at early stage because prognosis of OSCC is stage specific, patients with initial stage (stage I) have 80-90% of overall survival rate while this is confined to only 30% with higher stage tumors of larger size (stage IV). For the detection of oral cancer, several earlier techniques such as cytological screening and biopsy procedures have been employed but they had low sensitivity. Hence, a non-invasive sensitive and specific procedure have been widely accepted to treat the patients of OSCC at an early stage (Beckwith et al., 2003; Brindle et al., 2003; Carraro et al., 2007; Duarte et al., 2007; Makinen et al., 2006; Odunsi et al., 2005; Stubbs et al 2002; Tsang et al., 2002; Whitehead et al., 2005).
Molecular studies have evolved novel diagnostic, therapeutic and prognostic biomarkers to improve survival rate of patients through diagnosis of malignancies at an early stage (Siegel et al., 2015; Kanaan et al., 2012; Mudduluru 2011). Many studies have been identified about miRs in serum/plasma/urine in different type of cancer including OSCC. Hence circulating miRs may play a vital role as diagnostic and prognostic biomarkers in human cancer. The miRs are non-coding RNA molecules that alter many cellular processes such as differentiation of cell, cell cycle progression and apoptosis. They are also involved in progression of cancer processes via playing the role as oncogenes and tumor suppressor genes (Mudduluru et al., 2011; Mitchell et al., 2008; Schwarzenbach et al., 2011). The miR-21 is an oncogene that targets many genes such as phosphate and tensin homologue, programmed cell death 4 and tropomycin 1(alpha) (Cheng et al., 2012). Up-regulation of miR-21 have been reported in many epithelial cell derived tumors head and neck, esophageal, lung, gastric, colon, pancreatic, prostate cancer and breast cancer. It is also over-expressed in some hematological malignancies such as leukemia, lymphoma and multiple myeloma (Liu et al., 2010).
In the present study, miR-21 expression has been analyzed in serum of OSMF, OSCC as well as healthy control individuals to identify the higher expression of non invasive novel circulating miR-21 in OSCC as compared to OSMF and healthy controls. It can be used as diagnostic tool for effective detection of OSCC at an early stage after validation.
Materials and Methods
Subjects and Sample Collection
In the present study, 20 OSMF patients, 20 OSCC patients and 40 healthy controls were registered (Table 1). The blood samples were collected from OSCC patients attending Surgical Oncology Department from King George’s Medical University (K.G.M.U.), Lucknow, while the blood samples of OSMF and healthy controls were collected from the Department of Dentistry, Era’s Lucknow Medical College and Hospital (ELMC and H), Lucknow between February 2015-February 2017. All participants gave their written informed consent and the study was approved by Ethics committee of ELMC and H, Lucknow. All the patients were enrolled in the present study according to inclusion and exclusion criteria as mentioned below:
Table 1.
miRNA-21 Expression in Demographical Parameters of OSMF and OSCC Cases
| Demographical | Frequency | Percentage | Mean fold change | Standard deviation | p-value |
|---|---|---|---|---|---|
| Parameter | |||||
| Tobacco Chewers | |||||
| Yes | 38 | 95 | 1.904 | 1.874 | |
| No | 2 | 5 | 2.185 | 0.8697 | 0.503 |
| Smoking | |||||
| Yes | 13 | 32.5 | 2.532 | 1.421 | |
| No | 27 | 67.5 | 1.66 | 1.66 | 0.166 |
| Pan Masala | |||||
| Yes | 26 | 65 | 1.193 | 0.9015 | |
| No | 14 | 35 | 3.304 | 2.31 | 0 |
| Alcohol | |||||
| Yes | 8 | 20 | 1.1386 | 0.8177 | |
| No | 32 | 80 | 2.1291 | 1.961 | 0.201 |
Inclusion Criteria
All the fresh diagnosed cases of OSCC, OSMF and healthy controls who consented to participate in the study have been included in the study.
Exclusion Criteria
Cases of SCC that either has or had in past, any other malignancy.
Patients who have AIDS or any other known Immunodeficiency disorder.
Any patient who is in terminal stage of a disease and therefore not operable.
Collection and storage of serum sample and RNA isolation
Before surgical tumor resection, blood samples were collected in clot activator tube from each participant. The basic demographic and clinical information details were taken from all participants. Depending upon histological type and grade of tumor and cancer stage, staging of tumors have been done according to American Joint Committee on Cancer (AJCC) (Mitchell et al., 2008; Schwarzenbach et al., 2011). For serum separation, blood was kept for 45 minutes to allow clotting and thereafter it was processed according to Qiagen Kit (Germany) protocol. The Blood was centrifuged at 40C for 3,000 rpm for 10 minutes (REMI). To remove other contaminants like erythrocytes the supernatant fluid was re-centrifuged at 4°C at 13,500 rpm for 10 mins to yield better utility of miR for further processing of total RNA isolation. The Serum was stored at -80°C until processing for total RNA isolation. According to manufacturers protocol, total RNA were extracted from serum samples using Qiazol reagent (Qiagen, Germany). For this, the miRNeasy mini Kits (Qiagen, Germany) and miRNeasy serum/plasma (Qiagen, Germany) were used to extract miRs from serum samples. In addition, miRNeasy procedures reduce the chances of contamination with salt or phenols, that interfere with further processing. According to manufacturer’s protocol, 200 µl should be taken for RNA extraction.
cDNA synthesis (Reverse Transcription)
For the cDNA synthesis, total RNA sample should be 1.5 µl HiSpec buffer, 4 µl Nucleic Mix, 2 µl miScript master mix 2 µl and RNase free water10.5 µl. The total volume of final mixture should be 20 µl. The reverse transcription PCR reaction condition was 95°C for 5 mins and 37°C for 60 mins and then held at 4°C.
Real-Time PCR
After cDNA synthesis, for the miR based qRT-PCR assays, according to manufacturer’s instruction, each PCR reaction was performed in triplicates using SYBR Green Master Mix (Qiagen, Germany). Each reaction was carried out in a volume of 25 µl, containing 1 µl cDNA, 2.5 µl universal primer, 2.5 µl PCR primers along with 12.5 µl 2x QuantiTect SYBR Green PCR Master Mix were mixed with RNase free water. The PCR amplification reaction included de-naturation at 94°C for 15 seconds followed by 40 cycles at 55°C for 30 sec and 70°C for 30 sec. The reaction was run in the 7,900 Sequence Detection System 2.3 (Applied Biosystem). The Livak method was preferred to analyze the change in fold expression of miR-21 via calculating 2-ΔΔCt. The Ct mean cycle threshold is predicted as a total number of cycles to generate a fluorescent signal to cross threshold value in real time quantitative PCR (Applied Biosystem).
Statistical Analysis
All statistical analysis was done using Excel and SPSS Softwares. Expression level findings of serum miR-21 was subjected to statistical t-test and ANOVA taking p-value <0.05 as statistically significant.
Results
miR-21 expression level in OSMF and OSCC patients
The fold change for OSMF and OSCC patients was calculated by comparing with healthy controls by means of Livak method. Additionally, when relative mean fold increase in OSMF serum were compared with OSCC serum sample by statistical t-test. It was observed that mean fold increase in OSCC cases was comparatively more significant than OSMF (p=0.001). Therefore, a significant association of miR-21 expression was found between OSMF and OSCC patients (Figure: 1, 2; Table 2).
Figure 1.
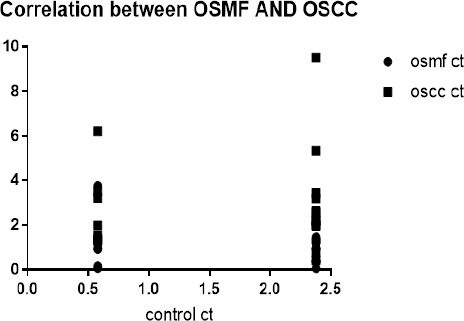
Correlation between Ct Value of Oral Sub-Mucous Fibrosis and Oral Squamous Cell Carcinoma
Figure 2.
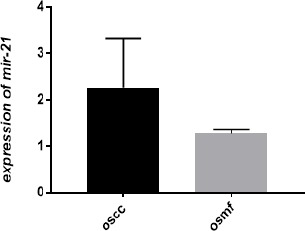
Correlation between Fold Change in Oral Sub-Mucous Fibrosis and Oral Squamous Cell Carcinoma Cases Showing Higher Fold Change Increase in OSCC Cases in Comparison to OSMF Cases
Table 2.
miRNA-21 Expression in Clinical Parameters of OSMF and OSCC Cases
| Mean | Standard deviation | p-value | |
|---|---|---|---|
| Sex | |||
| Male | 1.9803 | 1.8721 | 0.738 |
| Female | 1.6033 | 1.7027 | |
| Clinical stage | Mean | Standard deviation | |
| I | 0.6409 | 0.5658 | |
| II | 1.3043 | 0.1303 | 0 |
| III | 2.5116 | 1.5221 | |
| IV | 4.9047 | 4.0335 | |
| CASE | |||
| OSCC | 2.8281 | 2.1187 | 0.001 |
| OSMF | 1.0284 | 0.8173 |
Correlation of miR-21 with demographical parameters such as sex, tobacco, smoking, pan-masala, alcohol in OSMF/OSCC patient
The miR-21 expression profiling of 20 blood serum samples of OSCC, 20 of OSMF and 40 of healthy control subjects were analyzed using qRT-PCR. Oral mucosal lesions such as inflammation, hyperplasia and dysplasia was not observed in the healthy controls. The fold change of miR-21 in OSMF and OSCC group were calculated by comparison with healthy subjects by using Livak method. The demographical and histopathological features of participants are mentioned in Table 1. OSMF patients group were taken for validation of test to assess the significant expression of miR-21 in OSMF and OSCC subjects using t-test. Demographical parameters such as sex have shown slightly higher mean fold increase in males as compared to females, but the difference was not significant (p=0.738), whereas correlation of miR-21 expression comparison with other factors like tobacco, smoking and alcohol was not significant in OSCC in comparison OSMF. The non significant ratio for tobacco (p=0.503), smoking (p=0.166), alcohol (p=0.201). Therefore the data represented that there were no significant differences in expression of miR-21 and many demographical parameters like age, sex, tobacco, alcohol and smoking.
On the other hand one demographic parameter pan masala has shown significant relationship with expression of miR-21 in the OSCC as compared with OSMF participants of the study. The standard deviation was higher (2.310) in non chewers in comparison to chewers (0.9015) in OSCC and OSMF cases. The mean fold increase in the pan-masala chewers was significantly higher (.000) in comparison to pan-masala non chewers (p =0.005) in OSCC cases in comparison to pre-cancerous stages (Table 1).
MiR-21 expression and clinical parameters of OSCC
The purpose of our study was to investigate the correlation between clinical stages of OSCC and expression of miR-21 and the result were subjected to statistical test- One way ANOVA. The comparison has shown a significant value (p=0.000; Table 2) in OSCC clinical stages and miR-21 expression. Thus indicating a significant relationship between expression of miR-21 and clinical stages of OSCC patients.
Discussion
Various methods for the detection of cancer such as cytological analysis of sputum, endoscopic ultrasound-guided fine needle aspiration and computed tomography have demonstrated many type of limitation along with low accuracy of diagnosis, diagnosis at later stage and invasion in the human body. Hence, an urgent need is felt to recognize proper and noninvasive tumor biomarkers for early stage diagnosis. The CEA and CA19-9 are non invasive biomarkers for detection of cancer and are extensively used in clinics, but they possess low sensitivity and specificity for screening of cancer procedures (Locker et al., 2006). Many studies have been reported that cancer is not only affected by intrinsic but also by exogenous genetic alterations of cells and miRs act as a regulator of gene expression has been seen to be involved in tumor progression (Hutvagner et al., 2002).
Molecular studies, mainly expression profiles of genes are usually used to classify OSCC and to improve the diagnosis, therapeutic and prognosis management of patients (Kroh et al., 2010; Jacobsen et al., 2011). Detection of tumor at their early stage improves the overall survival rate of OSCC patients, which emerges an urgent requirement to discover more specific and sensitive molecular biomarkers for diagnosis of oral cancer at their early stage (Huang et al., 2010; Toiyama et al., 2013). Additionally, recent studies have demonstrated that ruptured cells release miRs in circulatory system including body fluids such as blood and urine etc. The discovery of the circulating miRs in blood serum has emerged as an early biomarker in cancer (Mitchel et al., 2008).
The main feature of tumor biology is an abnormal cell proliferation and miRs are known to regulate cellular processes such as proliferation, differentiation and apoptosis (Bartel et al., 2004). Many studies have reported that miRs may act as oncogenes or tumor suppressor genes (Hammod et al., 2006). Tumor tissues show significantly different expression pattern of miR as compared to normal tissues (Bishop et al., 2010; Corsini et al., 2012). Hence abnormal expression pattern of miRs are more likely to yield valuable information as important biomarkers for the diagnosis, treatment and prognosis. Michael etal., (2003), have reported 28 different miRs in normal mucosa and colonic adenocarcinoma and also noted down-regulation of two miR, miR-143 and miR-145 in adenomatous and cancer stages of colorectal neoplasia. Xie et al., (2010); and Iorio et al., (2005) reported miR-21 and miR-155 as diagnostic biomarker for non small cell lung cancer. Other studies also uggested about the downregulation of miR-125b miR-145, miR-155 in neoplastic breast cancer tissues in comparison to normal tissue.
Expression of miR-21 is mainly associated with development of tumor (Huang et al., 2009; Huang et al., 2013). Up-regulation of miR-21 has been seen in lung cancer, gastric cancer, pancreatic cancer, breast cancer, glioblastoma, neuroendocrine tumor, colon cancer, bile duct cancer and prostate cancer (Chan et al., 2008). A single gene, present on fragile site FRA17B on 17p23.2 that is overlapped with the gene encoding trans-membrane protein 49 (TMEM49) also known as VMP1, encodes miR-21 (Krichevsky et al., 2009). It contains 22 nucleotides that are processed from a 3,400 nucleotide pre-transcript (Cai et al., 2004). The miR-21 regulates several cellular processes such as cell proliferation, differentiation and apoptosis. It is also involved in tumor cell invasion, vascular infiltration and metastasis via targeting several genes such as tropomyosin 1, methyladenosine and programmed cell death gene4 in p53 mediated pathway and transforming growth factor beta pathway (Slaby et al., 2007). Many studies have revealed that miR-21 plays an important role in diagnosis of tumor and their evaluation. Schetter et al., (2008) in his study observed that over-expression of miR-21 in colon cancer is related to poor accuracy of prognosis and treatment. Yan et al., (2008) noted high expression of miR-21 in breast cancer along with lymph node metastasis and poor prognosis. Gao et al., (2011) found that up-regulation of miR-21 in lung squamous cell carcinoma is related to poor prognosis. Additionally, recent studies have shown that the broken cells release miRs that enter into circulatory system including blood and other fluids of the body (Mitchell et al., 2008).
Currently, Circulating miRs have become a potential molecular biomarker for early diagnosis of cancer due to their abundance in tissues and body fluids (Imperial et al., 2004; Wu et al., 2014; Schee et al., 2012). In present study, qRT- PCR was used to detect the expression level of serum miR-21 in OSCC as compared to OSMF patients and to determine its feasibility as potential biomarker of OSCC and OSMF. The present study revealed that expression of miR-21 is up-regulated in serum of OSCC as compared to pre-cancer stage (OSMF). Hence, the miR-21 expression was higher in OSCC patients as compared to OSMF patients. Additionally, in the present study no significant relationship was observed between expression of miR-21 and demographical parameters age, gender, smoking, alcohol, tobacco but a significant relationship was observed between pan–masala chewers in OSCC/OSMF patients. A study on colorectal cancer has shown up-regulation of miR-21 in different stages of cancer from early to later stage (Toiyama et al., 2013; Liu et al., 2013). In the present study, a significant high expression of miR-21 was observed from stages I-IV, they were correlated with different stages from early to later stage. The results of the present study indicate that expression of miR-21 in serum could be employed as novel non invasive biomarker for diagnosis of cancer at their early stage. However, more studies are needed on larger sample size to validate the findings. The qRT-PCR requires an appropriate internal control for serum for validation of results of the miR. The miR-16, C. elegans-39 and RNU6B has been used as important internal controls in earlier studies and in the present study, C. elegans-39 was used as reference control due to their greater stability in serum.
In conclusion, the findings of this study clearly demonstrate that serum miR-21 expression is higher in OSCC patients than OSMF patients and the clinicopathological correlation of clinical stages of OSCC patients have shown a significant increase in expression of miR-21 from stage I-IV. The results of the present study does not reveal any positive correlation between demographical parameters, age, sex, alcohol, smoking, tobacco except pan-masala and miR-21 expression in oral cancer. However, there is an urgent need of well designed studies with larger sample size to ascertain the diagnostic role of miR-21 in OSCC and their OSMF. Many studies on circulating miR-21 application in clinical practice are still needed to validate the miR-21 as reliable diagnostic and prognostic biomarker in future research.
Conflict of interest statement
There is no conflict of interest.
Acknowledgements
Authors acknowledge and thanks to Era’s Lucknow Medical College and Hospital for providing facilities for experimental work and Integral University, Lucknow for providing Ph.D. registration (Manuscript number- IU/R and D/2017-MCN000212) to Ms. Pooja Singh.
References
- 1.Aziz SR. Coming to America:betel nut and oral submucous fibrosis. JADA. 2010;141:423–8. doi: 10.14219/jada.archive.2010.0194. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs:genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Beckwith-Hall BM, Thompson NA, Nicholson JK, et al. A metabonomic investigation of hepatotoxicity using diffusion-edited H-1 NMR spectroscopy of blood serum. Analyst. 2003;128:814–18. doi: 10.1039/b302360p. [DOI] [PubMed] [Google Scholar]
- 4.Bishop JA, Benjamin H, Cholakh H, et al. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16:610–19. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 5.Brindle JT, Nicholson JK, Schofield PM, et al. Application of chemometrics to H-1 NMR spectroscopic data to investigate a relationship between human serum metabolic profiles and hypertension. Analyst. 2003;128:32–6. doi: 10.1039/b209155k. [DOI] [PubMed] [Google Scholar]
- 6.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped polyadenylated transcripts that Call also function as mRNAs. RNA. 2004;10:1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carraro S, Rezzi S, Reniero F, et al. Metabolomics applied to exhaled breath condensate in childhood asthma. Am J Respir Crit Care Med. 2007;175:986–90. doi: 10.1164/rccm.200606-769OC. [DOI] [PubMed] [Google Scholar]
- 8.Chan SH, Wu CW, Li AF, et al. miR21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res. 2008;28:907–11. [PubMed] [Google Scholar]
- 9.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31:4550–9. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng CJ, Slack FJ. The duality of oncomiR addiction in the maintenance and treatment of cancer. Cancer J. 2012;18:232. doi: 10.1097/PPO.0b013e318258b75b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corsini LR, Bronte G, Terrasi M, et al. The role of microRNAs in cancer:diagnostic and prognostic biomarkers and targets of therapies. Expert Opin Ther Targets. 2012;2:103–9. doi: 10.1517/14728222.2011.650632. [DOI] [PubMed] [Google Scholar]
- 12.Duarte IF, Goodfellow BJ, Barros A, et al. Metabolic characterisation of plasma in juveniles with glycogen storage disease type 1a (GSD1a) by high-resolution H-1 NMR spectroscopy. NMR Biomed. 2007;20:401–12. doi: 10.1002/nbm.1073. [DOI] [PubMed] [Google Scholar]
- 13.Elango JK, Gangadharan P, Sumithra S, et al. Trends of head and neck cancers in urban and rural India. Asian Pac J Cancer Prev. 2006;7:108–12. [PubMed] [Google Scholar]
- 14.Gao W, Shen H, Liu LX, et al. MiR21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. Cancer Res Clin Oncol. 2011;137:557–66. doi: 10.1007/s00432-010-0918-4. [DOI] [PubMed] [Google Scholar]
- 15.Gupta S, Singh R, Gupta OP, et al. Prevalence of oral cancer and pre-cancerous lesions and the association with numerous risk factors in North India:A hospital based study. Natl J Maxillofac Surg. 2014;5:142–8. doi: 10.4103/0975-5950.154816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haider SM, Merchant AT, Fikree FF, et al. Clinical and func¬tional staging of oral submucous fibrosis. Br J Oral Maxillofac Surg. 2000;38:12–5. doi: 10.1054/bjom.1999.0062. [DOI] [PubMed] [Google Scholar]
- 17.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 18.harma M, Madan M, Manjari M, et al. Prevalence of head and neck squamous cell carcinoma (HNSCC) in our population:The clinicpathological and morphological description of 198 cases. Int J Adv Res. 2015;3:827–33. [Google Scholar]
- 19.Hazarey VK, Erlewad DM, Mundhe KA, et al. Oral submucous fibrosis:a study of 1000 cases from central India. J Oral Pathol Med. 2007;36:12–7. doi: 10.1111/j.1600-0714.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- 20.Huang GL, Zhang XH, Guo GL. Clinical significance of miR-21 expression in breast cancer:SYBRGreen I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep. 2009;21:673–9. [PubMed] [Google Scholar]
- 21.Huang Y, Yang YB, Zhang XH, et al. MicroRNA-21 gene and cancer. Med Oncol. 2013;30:376. doi: 10.1007/s12032-012-0376-8. [DOI] [PubMed] [Google Scholar]
- 22.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–26. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 23.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 24.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Colorectal cancer study group. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–14. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 25.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 26.Jacobsen N, Andreasen D, Mouritzen P. Profiling microRNAs by real-time PCR. Methods in molecular biology. 2011;732:39–54. doi: 10.1007/978-1-61779-083-6_4. [DOI] [PubMed] [Google Scholar]
- 27.Jornet PL, Garcia FJ, Berdugo ML, et al. Mouth self-examination in a population at risk of oral cancer. Aust Dent J. 2015;60:59–64. doi: 10.1111/adj.12274. [DOI] [PubMed] [Google Scholar]
- 28.Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21:a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544–51. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 29.Krichevsky AM, Gabriely G. miR2l:a small multifaceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroh EM, Parkin RK, Mitchell PS, et al. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin WJ, Jiang RS, Wu SH, et al. Smoking, alcohol and betel quidand oral cancer:A prospective cohort study. J Oncol. 2011;2011:525976. doi: 10.1155/2011/525976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu GH, Zhou ZG, Chen R, et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumor Biol. 2013;34:2175–81. doi: 10.1007/s13277-013-0753-8. [DOI] [PubMed] [Google Scholar]
- 33.Liu MF, Jiang S, Lu Z, et al. Physiological and pathological functions of mammalian microRNAs, miRNA biology. Elsevier Ltd, TXCC; 2010. p. 223. [Google Scholar]
- 34.Locker GY, Hamilton S, Harris J, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–27. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 35.Makinen VP, Soininen P, Forsblom C, et al. Diagnosing diabetic nephropathy by H-1 NMR metabonomics of serum. Magn Reson Mat Phys Biol Med. 2006;19:281–96. doi: 10.1007/s10334-006-0054-y. [DOI] [PubMed] [Google Scholar]
- 36.Michael MZ, O'Connor SM, Pellekaan NG, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 37.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Nat Acad Sci. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mudduluru G, George-William J, Muppala S, et al. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Biosci Rep. 2011;31:185–97. doi: 10.1042/BSR20100065. [DOI] [PubMed] [Google Scholar]
- 40.Odunsi K, Wollman RM, Ambrosone CB, et al. Detection of epithelial ovarian cancer using H-1-NMR–based metabonomics. Int J Cancer. 2005;113:782–8. doi: 10.1002/ijc.20651. [DOI] [PubMed] [Google Scholar]
- 41.Pindborg JJ, Mehta FS, Gupta PC, et al. Prevalence of oral submucous fibrosis among 50,915 Indian villagers. Br J Cancer. 1968;22:646–54. doi: 10.1038/bjc.1968.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ranganathan K, Devi MU, Joshua E, et al. Oral submucous fibrosis:a case control study in Chennai, South India. J Oral Pathol Med. 2004;33:274–7. doi: 10.1111/j.0904-2512.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 43.Schee K, Boye K, Abrahamsen TW, et al. Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer. 2012;12:505. doi: 10.1186/1471-2407-12-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schetter AJ, Leung SY, Sohn JJ, et al. MiR expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 46.Siegel RL, Sahar L, Robbins A, et al. Where can colorectal cancer screening interventions have the most impact? Cancer Epidemiol Biomarkers Prev. 2015;15:0082. doi: 10.1158/1055-9965.EPI-15-0082. [DOI] [PubMed] [Google Scholar]
- 47.Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR21, miR31 , miR14 3 and miR145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 48.Stubbs M, Robinson SP, Hui C, et al. The importance of tumor metabolism in cancer prognosis and therapy;pre-clinical studies on rodent tumors with agents that improve tumor oxygenation. Adv Enzyme Regul. 2002;42:131–41. doi: 10.1016/s0065-2571(01)00027-9. [DOI] [PubMed] [Google Scholar]
- 49.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Nat Cancer Inst. 2013;105:849–59. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsang TM, Huang JTJ, Holmes E, et al. Metabolic profiling of plasma from discordant schizophrenia twins:correlation between lipid signals and global functioning in female schizophrenia patients. J Proteome Res. 2006;5:756–60. doi: 10.1021/pr0503782. [DOI] [PubMed] [Google Scholar]
- 51.Whitehead TL, Kieber-Emmons T. Applying in vitro NMR spectroscopy and H-1 NMR metabonomics to breast cancer characterization and detection. Prog Nucl Magn Reson Spectrosc. 2005;47:165–74. [Google Scholar]
- 52.Wu CW, Ng SC, Dong Y, et al. Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res. 2014;20:2994–02. doi: 10.1158/1078-0432.CCR-13-1750. [DOI] [PubMed] [Google Scholar]
- 53.Xie Y, Todd NW, Liu Z, et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170–6. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan LX, Huang XF, Shao Q, et al. MicroRNA miR21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]


