Abstract
Aim:
VEGF gene polymorphisms can induce either increase or inhibition of VEGF secretion, with altered promoter activity. The VEGF rs699947 SNP is located in the promoter region and is associated with susceptibility to breast carcinoma development. Here, we investigated the association of the -2578C>A polymorphism in the VEGF gene with breast cancer risk in Saudi women.
Methodology:
Genotyping of the VEGF-gene variation (-2578A>C) was performed using the amplification refractory mutation system PCR. We investigated the association of VEGF gene variants with different clinicopathological features of breast cancer patients.
Results:
A significant difference was observed in genotype distribution among the breast cancer cases and sex matched healthy controls (p=0.03). The frequencies of the three genotypes CC, CA, AA found in the patient samples were 37%, 45% and 18% and in the healthy controls were 54%,37%, and 09% respectively. An increased risk of developing breast cancer in Saudi women was associated with the VEGF −2578 AA genotype (OR = 2.91, 95 % CI, 1.18-7.20; p = 0.01; RR 1.78 (1.01-3.11 p=0.01), the VEGF −2578 A allele (OR = 1.79, 95 % CI, 1.17-2.73; p = 0.004: RR 1.35 1.07-1.71) and the VEGFR-(CA+ AA) (OR 1.99 1.13-3.51; RR 1.401.0-1.85). Thus the A allele increased the risk of BC when compared with C allele. When we stratified groups of patients according to the status of tumor markers, stage, age and metastasis, statistically significant associations with −2578 C/A SNP were revealed.
Conclusion:
Our data showed a significant association of the VEGF -2578C>A polymorphism with BC susceptibility in Saudi women. The VEGF -2578AA homozygote significantly increases the risk and can be useful as a predisposing genetic marker. Further studies with larger sample sizes are necessary to confirm our findings.
Keywords: VEGF-Vascular endothelial growth factor, SNP, Single-nucleotide polymorphism, UTR-Untranslated region
Introduction
Breast cancer is the most frequently diagnosed noncutaneous malignancy in women worldwide, which affects more than 1 million women annually (Bray, 2013; Babu, 2013). Saudi Arabia has the lowest rate of breast cancer incidence in the Arab world. The nationwide average of incidence in the Kingdom is 22 patients for every 100,000 women. In the UAE, 23 patients, Kuwait 46 patients, Jordan 49 patients, Qatar 48 patients and Bahrain 53 patients, for every 100,000 women Chouchane et al., 2014). There is a substantial rise in the incidence of breast cancer in Saudi Arabia in recent years, particularly among younger females compared to affected females’ in western countries (AlJohani et al., 2016). Although the etiology of Breast cancer is entirely unknown, there is abundant evidence that genetic factors play key roles in the pathogenesis and progression of Breast cancer (Hashemi et al., 2013). The human VEGF gene consists of eight exons separated by seven introns that exhibit alternative splicing to form a family of proteins (Vincenti et al., 1996) and plays a key role in a number of pathological processes including angiogenesis, tumor growth, and metastasis. Angiogenesis is a vital step in the development of cancer and is necessary for primary tumor growth, invasiveness, and metastases. Overexpression of VEGF was found in several tumor tissues (Nakamura et al., 2002). Breast cancer is involving lymph angiogenesis, which is the recruitment of blood and lymphatic vessels, to a growing tumor (Schoppmann et al., 2002). Large number of evidences from in vitro and in vivo experiments has shown that increased VEGF expression is associated with tumor growth and metastasis (Ferrara et al., 2002). Furthermore, the inhibition of VEGF signaling results in suppression of both tumor-induced angiogenesis and tumor growth (Ferrara et al., 2003). Pharmacogenomics studies have reported associations between single nucleotide polymorphisms (SNPs) in VEGF and VEGFR-2 and bevacizumab response in metastatic breast cancer Thus, genetic variability in VEGF and VEGFR-2 represents a logical candidate to study as a potential biomarker for bevacizumab (Schneider et al., 2012). Previous data have suggested that single nucleotide polymorphisms within VEGF gene have biologic importance in predicting risk and prognosis of cancer including breast cancer. It has been indicated that gene polymorphism in the promoter reagion, intron, exon, or untranslated regions (3’- and 5’-UTR) may affect the production or function of the corresponding protein (Ruggiero et al., 2011).
Till date some VEGF gene polymorphisms have been reported some of them are depicted in Figure 1. The -2578C/A (rs699947) SNP in its promoter region and the +405G/C (rs2010963) SNP in the 5’-untranslated region of the VEGF gene are associated with altered VEGF secretion (Almawi et el., 2013). Accordingly, these polymorphisms have been suspected to correlate with the progression and prognosis of cancer. Angiogenesis and inflammation are implicated in breast cancer prognosis; however, the role of individual germline variation in related genes is unknown. Studies assessing the effect of antiangiogenesis drug bevacizumab (BEV) on breast cancer (BC) outcome have shown different effects on progression-free and overall survival, suggesting that a subgroup of patients may benefit from this treatment (Hein et al., 2015).
Figure 1.

Nomenclature of VEGFA SNPs
The high levels of circulating VEGF have been observed in individuals with various malignancies including breast (Heer et el., 2001), uterine (Moon et al., 2000), gastrointestinal (Karayiannakis et al., 2002), lung (Kishiro et el., 2002) and prostate (Li et al., 2005). Several studies investigated the VEGF genetic polymorphisms in Breast cancer in different ethnic groups and led to different conclusions (Rahoui et al; 2014; James et al 2014; Chen et al., 2014; Luo et al., 2013; Rodrigue et al., 2012; Wang et al., 2011; Kapahi et al., 2015) however, the results have been inconsistent, suggesting that the association between the VEGF -2578C/A polymorphism and cancer requires further investigation.
To the best of our knowledge, there are no reports concerning the impact of VEGF -2578C>A gene polymorphism on Breast Cancer risk in Saudi Arabian women. Hence, this study aimed to evaluate the possible association between VEGF -2578C>A polymorphism with risk or protection of Breast Cancer women of Saudi Arabian.
Materials and Methods
The study included clinically, histologically, pathologically and radiologically confirmed cases of Breast cancer. This population-based case–control study was done on 100 cases and 100 gender matched healthy women with no history of any types of cancer and not related to the patients.
The exclusion criteria include Patients unwilling or unable to comply with the protocol. Patients with a history of previous cancer or metastasized cancer from other organs except Breast were excluded. After assessing the clinicopathological findings, a 4ml sample of peripheral blood was collected by venipuncture in EDTA tubes from each patient and healthy control.
DNA extraction
The DNA was extracted by using DNeasy Blood Kit (cat 69506) from Qiagen (Germany) as per the manufactures instructions. The extracted DNA was dissolved in nuclease-free water and stored at 4°C until use. Quality and integrity of DNA were checked by NanoDrop™ (Thermo Scientific, USA).
VEGF -2578C/A (rs699947) genotyping
VEGF -2578C>A genotyping was detected by using amplification-refractory mutation system –PCR (ARMS-PCR). ARM Systems are based on the use of sequence-specific PCR primers that allow amplification of test DNA only when the target allele is contained within the sample. Following an ARMS reaction the presence or absence of a PCR product is diagnostic for the presence or absence of the target allele. The VEGF -2578C>A genotyping primers were designed by using primer3 software as depicted inTable 1.
Table 1.
Amplification- Refractory Mutation System –PCR Primers for VEGF -2578C>A Gene Polymorphism
| Direction | Primer Sequence | AT | Product size | |
|---|---|---|---|---|
| FO- VEGF | 5-CCTTTTCCTCATAAGGGCCTTAG-3 | 58oC | 353bp | |
| RO- VEGF | 5-AGGAAGCAGCTTGGAAAAATTC-3 | |||
| FI –VEGF (Fwt) | A allele | 5-TAGGCCAGACCCTGGCAA-3 | 149bp | |
| RI- VEGF (Rmt) | C allele | 5-GTCTGATTATCCACCCAGATCG-3 | 243bp |
Fo-outer forward primer, Ro-Reverse outer primer; AT-annealing temperature; FI-Inner forward primer, RI-Inner Reverse primer
Table 2.
Preparation of PCR Cocktail for VEGF -2578C>A Polymorphism
| 1x | |
|---|---|
| PCR master mix | 10ul |
| Forward primer FO | 0.30 ul |
| Reverse primer RO | 0.30 ul |
| Forward primer FI A | 0.20 ul |
| Reverse primer RI C | 0.20 ul |
| Nuclease free water | 12ul |
| Total volume | 23ul |
| DNA (50ng) | 2ul |
The ARMS-PCR was performed in a reaction volume of 25uL containing template DNA (50ng), FO -0.30uL, RO -0.30uL, RI -0.20uL, RI -0.20uL of 25pmol of each primers and 10uL from GoTaq® Green Master Mix (cat no M7122) (Promega, USA). The final volume of 25uL was adjusted by adding nuclease free ddH2O. Finally, the 2ul of DNA was added from each patient.
Thermocycling conditions
The amplification conditions used were at 95 °C for 10 minutes followed by 40 cycles of 94oC for 35sec, 58 °C for 40 sec, 72 °C for 45 sec followed by the final extension at 72 °C for 10 minutes.
The amplification products were separated by electrophoresis through 2% agarose gel stained with 0.5μg/mL ethidium bromide and visualized on a UV transilluminator. Primers FO and RO flank the exon of the VEGF -2578C>A gene, resulting a band of 353bp to act as a control for DNA quality and quantity. Primers Fwt and RO amplify a wild-type allele (C allele), generating a band of 229 bp, and primers FO and Rmt generate a band of 149bp from the mutant allele (A allele) as depicted in Figure 2.
Figure 2.
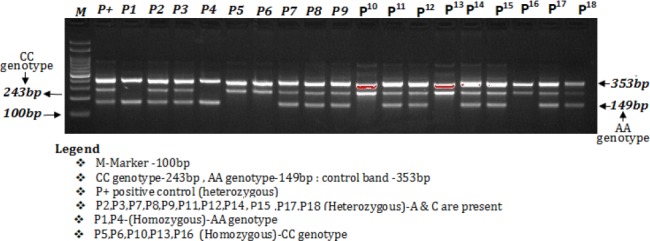
Detection of VEGF-2578C/A (rs699947) Genotyping by ARMS-PCR in Breast Cancer Patients and Healthy Controls
The best temperature was determined to be 58°C in the temperature range of 55°C to 63°C tested with a gradient PCR thermocycler. The annealing temperature was lowered from 60 to 58°C to favor the binding of both forward wild and reverse mutant primers that contain mismatches to the templates. The number of cycles was increased from 30 to 40 cycles, significantly enhancing the yields of all three PCR products. Together, these changes resulted in a more robust amplification of the mutant allele and a less competing reaction from the control, as shown by the relative intensities of the corresponding bands on agarose gel electrophoresis.
Statistical analysis
Deviations from Hardy-Weinberg disequilibrium (HWD) was calculated by Chi-square (χ2) goodness-of-fit test. Group differences were compared using Student’s two-sample t-test or one-way analysis of variance (ANOVA) for continuous variables and Chi-squared for categorical variables. Differences in the VEGF gene allele and genotype frequencies between groups were evaluated using Chi-square test. The associations between VEGF -2578C>A genotypes and risk of breast cancer were estimated by computing the odds ratios (ORs), risk ratios (RRs) and risk differences (RDs) with 95 % confidence intervals (CIs). Allele frequencies among cases as well as controls were evaluated by using the Chi–square Hardy-Weinberg equilibrium test. A p-value < 0.05 was considered significant. All statistical analyses were performed using Graph Pad Prism 6.0 and SPSS 16.0.
Results
The Hardy-Weinberg Equilibrium Analysis: The genotype distributions and allele frequencies of the SNPs rs699947 located in the VEGFR gene showed no deviation from HWE (χ2 = 0.44 P=0.612) in the patient group similarly the genotype distributions and allele frequencies of the SNPs rs699947 showed no deviation from HWE (χ2 = 0.52 p=0.712) in the control group. Thus, we chose 10% samples from normal control group randomly to review genotyping results, showing that the accuracy rate was more than 99%.
Study population
All demographic features of the subjects are depicted inTable 3. In brief, a total of 100 Breast cancer patients and the same number of gender matched healthy control were analyzed. This research was approved by the Research ethics committee, University of Tabuk and written informed consent was obtained from all the subjects before enrollment. Blood (4ml) samples were collected from participants in EDTA tubes.
Table 3.
Clinicopathological Characteristics of Breast Cancer Patients
| Parameters | No | % | Healthy Controls % |
|---|---|---|---|
| Patients | 100 | 100% | 100 (100%) |
| Age Group | |||
| Age >40 | 68 | 68% | |
| Age<40 | 32 | 32% | |
| Stage | |||
| Early (I & II) | 37 | 37% | |
| Advanced (III & IV) | 63 | 63% | |
| Grading | |||
| Grade I | 14 | 14% | |
| Grade II | 33 | 33% | |
| Grade III | 53 | 53% | |
| Estrogen receptor | |||
| Positive | 67 | 67% | |
| Negative | 33 | 33% | |
| Progesterone Receptor | |||
| Positive | 64 | 64% | |
| Negative | 36 | 36% | |
| Her2/neu | |||
| Positive | 48 | 48% | |
| Negative | 52 | 52% | |
| Distant Metastasis | |||
| Positive | 65 | 65% | |
| Negative | 35 | 35% |
Of 100 consecutive breast cancer patients, 32 (32%) patients were below or equal to 40 years age and 68 (68%) were above 40 years of age. Of breast cancer cases 37 (37%) were in early (I and II) stage and 63 (63%) cases were in advanced stages (III and IV). Histological grading of the patients tumor showed that 14 (14%), 33 (33%) and 53 (53%) were in grade I, II and III respectively. Out of 100 cases, how distant metastasis. Metastasis status of patients showed that 65 (65%) patients had distant metastasis and 35 (35%) do not show distant metastasis. Based on the receptor status, out of 100 Breast cancer cases 48 (48%) were positive for Her2/neu, 67 (67%) were carrying estrogen receptor and 64(64%) were +ve for progesterone receptor.
Case-control genotype distribution
This study observed that high percentage of CA (45%) and AA (18%) genotype was found in patients compared to controls CA (37%) and TT (9%) genotype while lower CC (37%) genotype in patients compared to control CC (54%) genotype as depicted in Table 4. We observed a statistically significant difference in the frequencies of VEGF rs699947 genotypes among patients and gender matched healthy controls (P=0.030). The frequency of A allele (fA) was found to be higher among breast cancer patients (0.41) whereas, the lower frequency of A allele (fA) was observed among healthy controls (0.28). However the frequency of C allele (fC) was found to be lower among breast cancer patients (0.59) than the healthy controls (0.72) as depicted inTable 3.
Table 4.
Allelic Frequencies of VEGF-(-2578 C>A) Polymorphism in Cases and Controls
| Subjects | N= | CC | AA | CA | A allele | C allele | P-Value |
|---|---|---|---|---|---|---|---|
| Cases | 100 | 37 (37%) | 18 (18%) | 45 (45%) | 0.41 | 0.59 | p=0.0303 |
| Controls | 100 | 54 (54%) | 09 (9%) | 37 (37%) | 0.28 | 0.72 | |
| Significance | X2=6.96 , df:2 | ||||||
Correlation between VEGF rs699947C>A gene variation and age
As depicted in Table 4, statistical analysis of the correlation between VEGF rs699947C>A gene variation in breast cancer patients revealed highly significant associations with age status (p=0.042). The distribution of AA genotype increased significantly among younger patients (<40 years of age) (37.5% vs 8%) whereas the frequency of heterozygosity CA increased significantly among older cases (>40 years of age) (48% Vs 3%).
Correlation between VEGF gene variation (rs699947C>A) and stage status
As depicted in Table 4, statistical analysis of the correlation between VEGF rs699947C>A gene variation in breast cancer patients revealed highly significant associations with stage status (p=0.042). The distribution of the CA and AA genotype increased significantly with increasing the breast stage like CA (53% Vs 29.72%) but AA genotype remains almost the same among the different stages (18.91% vs 17.46%).
Correlation between VEGF rs699947C>A gene variation and histological grade status
As depicted in Table 4, statistical analysis of the correlation between VEGF rs699947C>A gene variation in breast cancer patients with histological grade status revealed no significant associations in different grades (p=0.094).
Correlation between VEGF rs699947C>A gene variation and receptor status
As depicted in Table 4, statistical analysis of the correlation between VEGF rs699947C>A gene in breast cancer patients with estrogen receptor status revealed highly significant associations (p=0.02) except for PR and her2/neu status revealed non significant associations. VEGF rs699947 polymorphism was associated with ER expression (p=0.02) as manifested by a higher distribution of (CA) genotypes in ER- positive than in ER- negative patients (56% versus 30%).
Correlation between VEGF rs699947C>A gene variation and metastasis status
As depicted in Table 4, statistical analysis of the correlation between VEGF rs699947C>A gene in breast cancer patients with metastasis status of breast cancer patients revealed a strong significant association (p =0.008). The distribution of heterozygosity (CA) was higher in distant metastatic cases (56.92%).
Risk of Breast Cancer with VEGF rs699947 C>A gene polymorphism in BC patients
A multivariate analysis based on logistic regression like odds ratio, risk ratio and risk difference with 95% confidence intervals were calculated for each group to estimate the association between the VEGF rs699947 variant and risk of Breast cancer in Saudi patients. Odds ratio and risk ratio with 95% confidence intervals were calculated for each group to estimate the degree of association between the VEGF rs699947 variant and risk of Breast cancer risk in Saudi patients as depicted in tab 5.
Table 5.
Correlation between VEGF (-2578C>A) Polymorphisms and Clinicopathological Characteristics of Breast Cancer (BC) Patients.
| Parameters | N= | CC % | CA % | AA % | X2 | |
|---|---|---|---|---|---|---|
| Age Group | ||||||
| Age >40 | 68 (68%) | 29 (42.60) | 33 (48.52) | 6 (8.82) | 12.36 | p=0.002 |
| Age<40 | 32 (32%) | 08 (25) | 12 (3.84) | 12 (37.5) | ||
| Stage status | ||||||
| Early (I & II) | 37 (37%) | 19 (51.35) | 11 (29.72) | 07 (18.91) | 6.34 | p=0.042 |
| Advanced (III & IV) | 63 (63%) | 18 (50) | 34 (53.96) | 11 (17.46) | ||
| Grading status | ||||||
| Grade I | 14 (14%) | 6 (42.85) | 5 (35.71) | 03 (21.42) | 0.66 | p=0.94 |
| Grade II | 33 (33%) | 11 (33.33) | 16 (48.48) | 06 (18.18) | ||
| Grade I | 14 (14%) | 6 (42.85) | 5 (35.71) | 03 (21.42) | 0.74 | p=0.69 |
| Grade III | 53 (53%) | 20 (37.73) | 24 (45.28) | 09 (16.98) | ||
| Estrogen receptor status | ||||||
| Positive | 67 (67%) | 26 (38.80) | 38 (56.70) | 06 (8.95) | 12.45 | p=0.002 |
| Negative | 33 (33%) | 11 (33) | 10 (30.30) | 12 (36.36) | ||
| Progesterone Receptor status | ||||||
| Positive | 64 (64%) | 23 (35.93) | 30 (46.87) | 11 (17.18) | 0.61 | p=0.80 |
| Negative | 36 (36%) | 14 (38.88) | 15 (41.66) | 07 (19.44) | ||
| Her2/neu status | ||||||
| Positive | 48 (50%) | 15 (31.25) | 27 (56.25) | 08 (16.66) | 3.35 | p=0.18 |
| Negative | 52 (52%) | 22 (42.30) | 18 (34.61) | 10 (19.23) | ||
| Distant Metastasis status | ||||||
| Positive | 65 (65%) | 22 (33.84) | 37 (56.92) | 06 (9.23) | 14.3 | p=0.008 |
| Negative | 35 (35%) | 15 (42.85) | 08 (22.85) | 12 (34.28) | ||
The findings indicated that VEGF rs699947 variant increased the risk of Breast cancer in codominant (CA vs CC) OR=2.91, 95% CI = 1.18–7.20, P = 0.021) but non-significant for AA vs CC (OR =1.77, 95% CI = 0.97–3.24, P = 0.06) and dominant (CA+AA vs CC) OR = 1.99, 95% CI = 1.13–3.51, P = 0.016) inheritance models tested. During the allelic comparison, the A allele increased the risk of Breast cancer with odd ratio (OR = 1.66, 95% CI = 1.10–2.51, P = 0.020) and risk ratio RR= 1.31(1.04-1.65) P= 0.020) as depicted in Table 5.
Discussion
The study was conducted in a prospective manner, based on a single-institution cohort of Saudi Arabia (Tabuk) women diagnosed with breast cancer. The VEGF rs699947 C/A gene polymorphism was revealed to be associated with an overall increased risk of Breast cancer in different studies. The prevalence of rs699947C/A gene polymorphism was analyzed in the different healthy populations from different and countries of the world as depicted in Table 6. Similarly The prevalence of rs699947 C/A gene polymorphism was analyzed in the Breast cancer women of different populations of the world as depicted in Table 7. The prevalence of rs699947 CA genotype was 45% and AA genotype 18% in Breast cancer patients were identified as significantly higher than that in the healthy individuals (37% and 09%, respectively) and the A allele of rs699947 was found to be as more frequent in patients with Breast cancer than that of controls (0.41 vs 0.28 respectively). We observed a statistically significant difference in the frequencies of VEGF rs699947 genotypes among patients and gender matched healthy controls (p=0.030).
Table 6.
Association of VEGF rs699947 C>A Gene Variation with Breast Cancer
| Genotypes | Healthy controls | Breast cancer patients | OR (95% CI) | Risk Ratio (RR) | P-Value | ||
|---|---|---|---|---|---|---|---|
| (N=100) | % | (N=100) | % | ||||
| Codominant | |||||||
| VEGF-CC | 54 | 54% | 37 | 37% | 1 (ref.) | 1 (ref.) | |
| VEGF-CA | 37 | 37% | 45 | 45% | 1.77 (0.97-3.24) | 1.31 (0.98-1.76) | 0.06 |
| VEGF-AA | 9 | 9% | 18 | 18% | 2.91 (1.18-7.20) | 1.78 (1.01-3.11) | <0.021 |
| Dominant | |||||||
| VEGF-CC | 54 | 54%) | 37 | 37% | 1 (ref.) | 1 (ref.) | |
| VEGF- (CA+ AA) | 46 | 46% | 63 | 63% | 1.99 (1.13-3.51) | 1.40 (1.0-1.85) | <0.016 |
| Recessive | |||||||
| VEGF- (CC+ CA) | 101 | 91.80% | 100 | 84.74% | 1 (ref.) | 1 (ref.) | |
| VEGF-AA | 9 | 8.20% | 18 | 15.25% | 2.02 (0.86-4.7) | 1.50 (0.86-2.61) | 0.1 |
| Allele | |||||||
| VEGF-C | 155 | 73.80% | 137 | 31% | 1 (ref.) | 1 (ref.) | |
| VEGF-A | 55 | 26.20% | 81 | 69% | 1.66 (1.10-2.51) | 1.31 (1.04-1.65) | <0.020 |
Table 7.
Distribution of VEGFA rs699947 C>A Polymorphism Genotypes in Healthy Controls
| Country | Ethnicity | Controls | CC | AC | AA | Author |
|---|---|---|---|---|---|---|
| UK | European | 66 | 13 | 38 | 15 | Yang, 2003 |
| Japan | East Asian | 203 | 93 | 91 | 19 | Awata, 2005 |
| Poland | European | 91 | 29 | 43 | 19 | Buraczynska,2007 |
| Australia | European | 93 | 26 | 43 | 24 | Abhary, 2009 |
| Australia | European | 181 | 45 | 91 | 45 | Abhary, 2009 |
| Japan | East Asian | 292 | 163 | 107 | 22 | Nakamura, 2009 |
| Korea | East Asian | 134 | 92 | 36 | 6 | Chun, 2010 |
| China | East Asian | 138 | 82 | 51 | 5 | Yang, 2011 |
| Saudi Arabia | Middle east | 100 | 54 | 37 | 9 | Our study |
The prevalence of VEGF rs699947 AA genotype in our study group was smaller than that reported in USA (28%), Polish (28%), Swedish (26%), England (25%), and German (22%) except in Morocco (4%) and Thailand (6%) as depicted in Figure 3, whereas the frequency of VEGF rs699947 CA genotype reported in our study group was 45% that was similar to that reported in USA (49%), Polish (47%), Swedish (47%), England (50%), Thailand and German (49%) as depicted in Figure 3.
Figure 3.
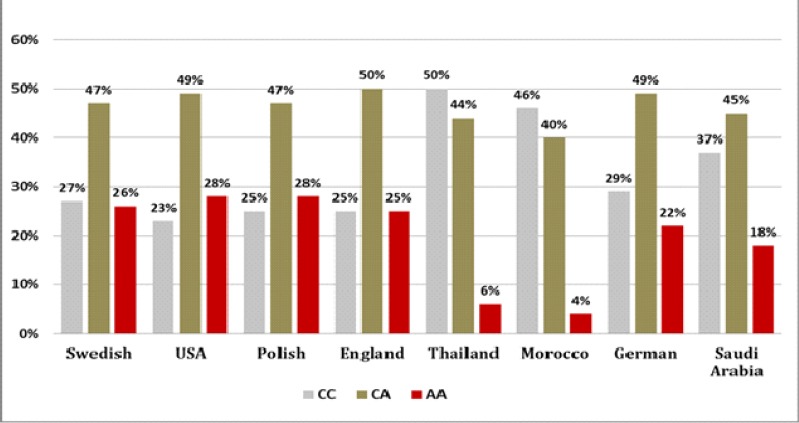
Distribution of VEGFA rs699947C> A Gene Polymorphism in Breast Cancer in the World
VEGF is believed to serve as an important factor for angiogenesis through various mechanisms (Yoshiji et al., 1996). Some of studies identified several functional polymorphisms of the VEGF gene that might affect serum VEGF expression level, including −634G>C, −1154G>A, 936C>T, −1498C>T, −2578C>A, and −460C>T (García-Closas et al., 2007; Koukourakis et al., 2004). Several previous studies reported that functional genetic polymorphisms could alter mRNA or protein expression, thus generating significant influence on disease development of various diseases including cancer (Flego et al., 2013).
Recently, the relationships between VEGF gene variations and the risk of breast cancer have been extensively studied; however, the reported results were inconsistent. The findings of our study proposed that VEGF rs699947 variant (AA) significantly increased the risk of breast cancer. The stratified analysis was performed by clinicopathological characteristics of breast cancer patients, and our findings proposed that cases with rs699947 AA genotype have increased risk of developing breast cancer in young individuals (age <40 years), as well as in patients with advanced stage. Similarly a significant association was observed with distant metastasis cases. However, patients with CA genotype had a lower risk of developing Progesterone receptor (-ve) and her2-neu negative breast cancer. Similar results were reported by Kapahi et al., (2015) who investigated the impact of VEGF −2578C/A, polymorphisms on Breast cancer in North Indian population. Also Zhang et al., (2015) performed an ethnicity-specific subgroup analysis, but did not find any significant associations between the -2578 C/A polymorphisms in the VEGF gene and breast cancer risk in either Caucasians or Asians. Similarly Chen et al., (2014) performed a meta-analysis revealed no significant association between VEGF −2578C/A and the risk of cancer. Subgroup analyses showed that the VEGF −2578C/A polymorphism is not associated with bladder and breast cancers but is associated with colorectal and lung cancers. A Meta-analysis was performed by Wang et al., (2008) between VEGF SNPS like +936C/T, −1154A/G, −2578C/A, −634G/C, and −460T/C and risk of Breast cancer. The overall results of combined analyses revealed that all the five polymorphisms of VEGF were not associated with the risk of Breast cancer. Nasr et al., (2008) has also shown that −2,578 CA variant correlates with disease stages in which angiogenesis plays a critical role. The VEGF −2,578 C/A polymorphism has shown a significant association with BC susceptibility. Similarly our results showed significant associations of VEGF −2,578CA and −2,578AA genotype with stage status (p=0.042). The distribution of CA and homozygous AA genotype increased significantly with increasing the breast stage among the breast cancer cases (53% Vs 29.72%). However, Rahoui et al., (2014) reported that carriers of −2,578 A allele had a reduced risk to develop a Breast cancer therefore their result is contradicted; however, the results obtained by Scheneider et al., (2008) has shown that AA genotype was associated with higher risk of breast cancer in Caucasian and African-American populations. Meanwhile, Jacobs et al., (2006) have reported that the C allele was associated with increased risk of invasive cancer in the American population, but not for in situ or overall breast cancer. Shahbazi et al., (2002) has shown that the −2,578C allele was associated with increased VEGFA production by peripheral blood mononuclear cells (Cuzick et al., 2006). Many well established and widely used genetic variations that predict increased the risk of breast cancer has been reported (Koukourakis et al., 2004). The elucidation of genetic variations has allowed the testing and FDA approval of a drug which actually decreases the future risk of disease in Breast cancer women at high risk. Unfortunately, the reduction in risk to date has been confined to the ER+ subgroup of tumors. In vitro model suggested a haplotypic effect of the polymorphic VEGFA promoter on both basal and stimulated promoter activity however in non-small cell lung cancer; a low VEGFA expression in cancer tissues was significantly associated with the presence of the −2,578 CC, −634 GG and −1,154 AA and GA genotypes (Cui et al., 2013). The nature of breast carcinogenesis pathways is complex; there is no clear reason for the discrepancies in different studies. Ethnic, genetic, and environmental factors may interact in various ways to affect the risk of Breast cancer in different areas. Several studies have been performed to determine the frequency and distribution of VEGF polymorphism −2,578 A/C in different ethnic groups as summarized in Table 7.
This study investigates the association of −2,578 C/A polymorphisms of VEGF-A with Breast cancer susceptibility and aggressiveness among Saudi women. Thus, characterization of VEGF polymorphisms in healthy women gives for the first time the allelic frequencies of VEGF−2578C/A polymorphism which can be used as a reference in others studies. This case–control study demonstrated that VEGF−2,578 C alleles seem to have a protective effect against Breast cancer in Saudi Arabian women. This genetic variant VEGF−2,578 C may act as a low-penetrance Breast cancer risk gene whereas −2,578 A alleles can be used as genetic biomarkers for Breast cancer susceptibility. The overall results of combined analyses revealed that VEGF −2,578 A allele and A carrier genotypes have been shown to be associated with an increased risk of Breast cancer. This study adds to the emerging evidence that VEGF-A polymorphisms play an important role in Breast cancer development. Further large-scale studies in Saudi population are necessary to confirm our results. Moreover, the effect of other functional VEGF polymorphisms on Breast cancer susceptibility and development merits further surveys. The main limitation of the present study is that it is only of moderate size.
In conclusion, our data showed a significant associations of VEGF -2578C>A polymorphism with BC susceptibility in Saudi women. VEGF -2578AA homozygote significantly increases the risk of Breast cancer and can be useful as predisposing genetic marker for BC. Furthers studies with larger sample sizes are necessary to confirm our findings.
Disclosure
Authors have declared that no competing interests exist.
Consent
All authors hereby declare that all experiments have been examined and approved by the Research ethics committee, University of Tabuk, and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki.
Acknowledgements
We acknowledge the support from the Deanship of Scientific Research, University of Tabuk for funding this research (S-1438-0105). We are grateful to the patients with whose cooperation this study was possible.
References
- 1.Abdelilah L, Yassir S, Nadia T, et al. Investigating the association of vascular endothelial growth factor polymorphisms with breast cancer:a Moroccan case–control study. Med Oncol. 2014;31:193. doi: 10.1007/s12032-014-0193-3. [DOI] [PubMed] [Google Scholar]
- 2.Abhary S, Burdon KP, Gupta A, et al. Common sequence variation in the VEGFA gene predicts risk of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2009;50:5552–8. doi: 10.1167/iovs.09-3694. [DOI] [PubMed] [Google Scholar]
- 3.AlJohani B, AlMalik O, Anwar E, et al. Impact of surgery on survival in stage IV breast cancer. Breast J. 2016;22:678–82. doi: 10.1111/tbj.12662. [DOI] [PubMed] [Google Scholar]
- 4.Almawi WY, Saldanha FL, Mahmood NA, et al. Relationship between VEGFA polymorphisms and serum VEGF protein levels and recurrent spontaneous miscarriage. Hum Reprod. 2013;28:2628–35. doi: 10.1093/humrep/det308. [DOI] [PubMed] [Google Scholar]
- 5.Awata T, Kurihara S, Takata N, et al. Functional VEGF C-634G polymorphism is associated with development of diabetic macular edema and correlated with macular retinal thickness in type 2 diabetes. Biochem Biophys Res Commun. 2005;333:679–85. doi: 10.1016/j.bbrc.2005.05.167. [DOI] [PubMed] [Google Scholar]
- 6.Babu GR, Samari G, Cohen SP, et al. Breast cancer screening among females in Iran and recommendations for improved practice:a review. Asian Pac J Cancer Prev. 2011;12:1647–55. [PubMed] [Google Scholar]
- 7.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–45. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 8.Buraczynska M, Ksiazek P, Baranowicz-Gaszczyk I, Jozwiak L. Association of the VEGF gene polymorphism with diabetic retinopathy in type 2 diabetes patients. Nephrol Dial Transplant. 2007;22:827–32. doi: 10.1093/ndt/gfl641. [DOI] [PubMed] [Google Scholar]
- 9.Chen Q, Zhou Z, Shan L, et al. Association of the vascular endothelial growth factor -2578C/A polymorphism with cancer risk:a meta-analysis update. Biomed Rep. 2014;2:823–30. doi: 10.3892/br.2014.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Q, Zhou Z, Shan L, et al. Association of the vascular endothelial growth factor -2578C/A polymorphism with cancer risk:A meta-analysis update. Biomed Rep. 2014;2:823–30. doi: 10.3892/br.2014.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chouchane L, Boussen H, Sastry KSR. Breast cancer in Arab populations:molecular characteristics and disease management implications. Lancet Oncol. 2013;14:417–24. doi: 10.1016/S1470-2045(13)70165-7. [DOI] [PubMed] [Google Scholar]
- 12.Chun MY, Hwang HS, Cho HY, et al. Association of vascular endothelial growth factor polymorphisms with nonproliferative and proliferative diabetic retinopathy. J Clin Endocrinol Metab. 2010;95:3547–51. doi: 10.1210/jc.2009-2719. [DOI] [PubMed] [Google Scholar]
- 13.Cuzick J, Powles T, Veronesi U, et al. Overview of the main outcomes in breast-cancer prevention trials. Lancet. 2003;361:296–300. doi: 10.1016/S0140-6736(03)12342-2. [DOI] [PubMed] [Google Scholar]
- 14.Eskandari-Nasab E, Hashemi M, Rezaei D, et al. Evaluation of UDP-glucuronosyltransferase 2B17 (UGT2B17) and dihydrofolate reductase (DHFR) genes deletion and the expression level of NGX6 mRNA in breast cancer. Mol Biol Rep. 2012;39:10531–9. doi: 10.1007/s11033-012-1938-8. [DOI] [PubMed] [Google Scholar]
- 15.Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer. 2002;2:795–803. doi: 10.1038/nrc909. [DOI] [PubMed] [Google Scholar]
- 16.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–76. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 17.Flego V, Ristić S, DevićPavlić S, et al. Tumor necrosis factor-alpha gene promoter -308 and -238 polymorphisms in patients with lung cancer as a second primary tumor. Med Sci Monit. 2013;11:846–51. doi: 10.12659/MSM.889554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Closas M, Malats N, Real FX, et al. Large-scale evaluation of candidate genes identifies associations between VEGF polymorphisms and bladder cancer risk. PLoS Genet. 2007;23:e29. doi: 10.1371/journal.pgen.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashemi M, Eskandari-Nasab E, Fazaeli A, et al. Association between polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and breast cancer risk in a sample Iranian population. Biomark Med. 2012;6:797–803. doi: 10.2217/bmm.12.61. [DOI] [PubMed] [Google Scholar]
- 20.Hashemi M, Eskandari-Nasab E, Fazaeli A, et al. Bi-directional PCR allele-specific amplification (bi-PASA) for detection of caspase-8 -652 6N ins/del promoter polymorphism (rs3834129) in breast cancer. Gene. 2012;505:176–9. doi: 10.1016/j.gene.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 21.Hashemi M, Fazaeli A, Ghavami S, et al. Functional polymorphisms of FAS and FASL gene and risk of breast cancer-pilot study of 134 cases. PLoS One. 2013;8:e53075. doi: 10.1371/journal.pone.0053075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heer K, Kumar H, Read JR, et al. Serum vascular endothelial growth factor in breast cancer:its relation with cancer type and estrogen receptor status. Clin Cancer Res. 2001;7:3491–4. [PubMed] [Google Scholar]
- 23.Hein A, Lambrechts D, von Minckwitz G, et al. Genetic variants in VEGF pathway genes in neoadjuvant breast cancer patients receiving bevacizumab:Results from the randomized phase III Gepar Quinto study. Int J Cancer. 2015;137:2981–8. doi: 10.1002/ijc.29656. [DOI] [PubMed] [Google Scholar]
- 24.Hyodo I, Doi T, Endo H, et al. Clinical significance of plasma vascular endothelial growth factor in gastrointestinal cancer. Eur J Cancer. 1998;34:2041–5. doi: 10.1016/s0959-8049(98)00282-2. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs EJ, Feigelson HS, Bain EB, et al. Polymorphisms in the vascular endothelial growth factor gene and breast cancer in the cancer prevention study II cohort. Breast Cancer Res. 2006;8:R22. doi: 10.1186/bcr1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James R, Ramesh G, Krishnamoorthy L, et al. Prevalence of +405G>C, -1154G>A vascular endothelial growth factor polymorphism in breast cancer. Indian J Clin Biochem. 2014;29:21–8. doi: 10.1007/s12291-013-0307-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Q, Hemminki K, Enquist K, et al. Vascular endothelial growth factor polymorphisms in relation to breast cancer development and prognosis. Clin Cancer Res. 2005;11:3647–53. doi: 10.1158/1078-0432.CCR-04-1803. [DOI] [PubMed] [Google Scholar]
- 28.Kapahi R, Guleria K, Sambyal V, et al. Association of VEGF and VEGFR1 polymorphisms with breast cancer risk in North Indians. Tumour Biol. 2015;36:4223–34. doi: 10.1007/s13277-015-3059-1. [DOI] [PubMed] [Google Scholar]
- 29.Karayiannakis AJ, Syrigos KN, Polychronidis A, et al. Circulating VEGF levels in the serum of gastric cancer patients:correlation with pathological variables, patient survival, and tumor surgery. Ann Surg. 2002;236:37–42. doi: 10.1097/00000658-200207000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishiro I, Kato S, Fuse D, et al. Clinical significance of vascular endothelial growth factor in patients with primary lung cancer. Respirology. 2002;7:93–8. doi: 10.1046/j.1440-1843.2002.00376.x. [DOI] [PubMed] [Google Scholar]
- 31.Koukourakis MI, Papazoglou D, Giatromanolaki, et al. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer. 2004;46:293–8. doi: 10.1016/j.lungcan.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 32.Koukourakis MI, Papazoglou D, Giatromanolaki R, et al. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer. 2004;46:293–8. doi: 10.1016/j.lungcan.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Kantoff PW, Ma J. Prediagnostic plasma vascular endothelial growth factor levels and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1557–61. doi: 10.1158/1055-9965.EPI-04-0456. [DOI] [PubMed] [Google Scholar]
- 34.Luo T, Chen L, He P, et al. Vascular endothelial growth factor (VEGF) gene polymorphisms and breast cancer risk in a Chinese population. Asian Pac J Cancer Prev. 2013;14:2433–37. doi: 10.7314/apjcp.2013.14.4.2433. [DOI] [PubMed] [Google Scholar]
- 35.Moon HS, Kim SC, Ahn JJ. Concentration of vascular endothelial growth factor (VEGF) and transforming growth factor-beta1 (TGF-beta1) in the serum of patients with cervical cancer:prediction of response. Int J Gynecol Cancer. 2000;10:151–6. doi: 10.1046/j.1525-1438.2000.00013.x. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura M, Abe Y, Tokunaga T. Pathological significance of vascular endothelial growth factor A isoform expression in human cancer. Pathol Int. 2002;52:331–9. doi: 10.1046/j.1440-1827.2002.01367.x. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura S, Iwasaki N, Funatsu H, Kitano S, Iwamoto E. Impact of variants in the VEGF gene on progression of proliferative diabetic retinopathy. Graefes Arch Clin Exp Ophthalmol. 2009;247:21–6. doi: 10.1007/s00417-008-0915-3. [DOI] [PubMed] [Google Scholar]
- 38.Nasr HB, Chahed K, Bouaouina N, Chouchane G. Functional vascular endothelial growth factor -2578 C/A polymorphism in relation to nasopharyngeal carcinoma risk and tumor progression. Clin Chim Acta. 2008;395:124–9. doi: 10.1016/j.cca.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura R, Nagao K, Miyayama H, et al. Higher plasma vascular endothelial growth factor levels correlate with menopause, overexpression of p53, and recurrence of breast cancer. Breast Cancer. 2003;10:120–8. doi: 10.1007/BF02967636. [DOI] [PubMed] [Google Scholar]
- 40.Pharoah PD, Tyrer J, Dunning AM, et al. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet. 2007;3:e42. doi: 10.1371/journal.pgen.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahoui J, Laraqui A, Sbitti Y, et al. Investigating the association of vascular endothelial growth factor polymorphisms with breast cancer:a Moroccan case-control study. Med Oncol. 2014;31:193. doi: 10.1007/s12032-014-0193-3. [DOI] [PubMed] [Google Scholar]
- 42.Rahoui J, Sbitti Y, Touil N, et al. The single nucleotide polymorphism +936 C/T VEGF is associated with human epidermal growth factor receptor 2 expression in Moroccan breast cancer women. Med Oncol. 2014;31:336. doi: 10.1007/s12032-014-0336-6. [DOI] [PubMed] [Google Scholar]
- 43.Rodrigues P, Furriol J, Tormo E. The single-nucleotide polymorphisms +936 C/T VEGF and -710 C/T VEGFR1 are associated with breast cancer protection in a Spanish population. Breast Cancer Res Treat. 2012;133:769–78. doi: 10.1007/s10549-012-1980-1. [DOI] [PubMed] [Google Scholar]
- 44.Ruggiero D, Dalmasso C, Nutile T, et al. Genetics of VEGF serum variation in human isolated populations of cilento:importance of VEGF polymorphisms. PLoS One. 2011;6:e16982. doi: 10.1371/journal.pone.0016982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sa-Nguanraksa D, Chuangsuwanich T, Pongpruttipan T, et al. Vascular endothelial growth factor -634G/C polymorphism is associated with increased breast cancer risk and aggressiveness. Mol Med Rep. 2013;8:1242–50. doi: 10.3892/mmr.2013.1607. [DOI] [PubMed] [Google Scholar]
- 46.Schneider BP, Miller KD. Angiogenesis of breast cancer. J Clin Oncol. 2005;23:1782–90. doi: 10.1200/JCO.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 47.Schneider BP, Radovich M, Sledge GW, et al. Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res Treat. 2008;111:157–63. doi: 10.1007/s10549-007-9755-9. [DOI] [PubMed] [Google Scholar]
- 48.Schneider BP, Shen F, Miller KD. Pharmacogenetic biomarkers for the prediction of response to antiangiogenic treatment. Lancet Oncol. 2012;13:427–36. doi: 10.1016/S1470-2045(12)70275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoppmann SF, Horvat R, Birner G. Lymphatic vessels and lymphangiogenesis in female cancer:mechanisms, clinical impact and possible implications for anti-lymphangiogenic therapies (Review) Oncol Rep. 2002;9:455–60. [PubMed] [Google Scholar]
- 50.Shahbazi M, Fryer AA, Pravica B, et al. Vascular endothelial growth factor gene polymorphisms are associated with acute renal allograft rejection. J Am Soc Nephrol. 2002;13:260–4. doi: 10.1681/ASN.V131260. [DOI] [PubMed] [Google Scholar]
- 51.Vincenti V, Cassano C, Rocchi M, Persico Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation. 1996;93:1493–5. doi: 10.1161/01.cir.93.8.1493. [DOI] [PubMed] [Google Scholar]
- 52.Wang K, Liu L, Zhu ZM. Five polymorphisms of vascular endothelial growth factor (VEGF) and risk of breast cancer:a meta-analysis involving 16,703 individuals. Cytokine. 2011;56:167–73. doi: 10.1016/j.cyto.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 53.Wang K, Liu L, Zhu ZM, et al. Five polymorphisms of vascular endothelial growth factor (VEGF) and risk of breast cancer:a meta-analysis involving 16,703 individuals. Cytokine. 2011;56:167–73. doi: 10.1016/j.cyto.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 54.Yang B, Cross DF, Ollerenshaw N, et al. Polymorphisms of the vascular endothelial growth factor and susceptibility to diabetic microvascular complications in patients with type 1 diabetes mellitus. J Diabetes Complications. 2003;17:1–6. doi: 10.1016/s1056-8727(02)00181-2. [DOI] [PubMed] [Google Scholar]
- 55.Yang X, Deng Y, Gu H, et al. Polymorphisms in the vascular endothelial growth factor gene and the risk of diabetic retinopathy in Chinese patients with type 2 diabetes. Mol Vis. 2011;17:3088–96. [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshiji H, Gomez D, Shibuya M, Thorgeirsson S. Expression of vascular endothelial growth factor, its receptor, and other angiogenic factors in human breast cancer. Cancer Res. 1996;56:2013–16. [PubMed] [Google Scholar]
- 57.Zhang Y, Yu YF, Wang JZ, Jia H. Vascular endothelial growth factor +405G/C and -2578C/A polymorphisms and breast cancer risk:a meta-analysis. Genet Mol Res. 2015;14:8909–18. doi: 10.4238/2015.August.3.14. [DOI] [PubMed] [Google Scholar]


