Abstract
Salmonella typhi and Helicobacter infections have been shown to increase risk of gallbladder cancer (GBC), but findings have been inconsistent. Other bacterial infections may also be associated with GBC. However, information on microbial pathogens in gallbladder bile of GBC patients is scarce. We aimed to investigate the microbial communities in gallbladder bile of patients with GBC and cholelithiasis (CL). Seven GBC patients and 30 CL patients were enrolled in this study. Genomic DNA was extracted from bile and the V3-V4 region of 16S rRNA was amplified. The sequencing results were compared with the 16S database, and the bacteria were identified by homology searches and phylogenetic analysis. DNA was detected in the bile of three GBC (42.9%; Bolivia, 1; Chile, 2) and four CL patients (13.3%; Bolivia, 1; Chile, 3). Of the 37 patients, 30 (81.1%) were negative and unable to analyze. Salmonella typhi and Helicobacter sp. were not detected in bile from any GBC patients. As the predominant species, Fusobacterium nucleatum, Escherichia coli, and Enetrobacter sp. were detected in bile from GBC patients. Those in bile from CL patients were Escherichia coli, Salmonella sp., and Enerococcus gallinarum. Escherichia coli was detected in bile samples from both GBC and CL patients. Whether the bacteria detected in bile from GBC patients would associated with the development of GBC warrant further investigation.
Keywords: Microbial pathogens, next generation sequencing, 16S rRNA, V3-V4 region, gallbladder cancer
Introduction
The important role of cholelithiasis (CL) and the presence of gallstones in the development of gallbladder cancer (GBC) has been established in several previous studies (Randi et al., 2006; Roa et al., 2006; Hsing et al., 2007; Shrikhande et al., 2010; Dutta et al., 2013). Kanthan et al., (2015) divided the risk factors for GBC into four broad categories: patient demographics, gallbladder abnormality, patient exposure, and infections. They concluded that not only gallstones, but also Salmonella and Helicobacter infections, were important risk factors for GBC. Several studies (Kumar et al., 2006; Walawalkar et al., 2013) have demonstrated an association between bacterial infections and increased risk of GBC. However, although associations between Salmonella typhi (Andia et al., 2008; Nagaraja and Eslick, 2014) and Helicobacter (Martel et al., 2009; Pandey et al., 2010; Yakoob et al., 2011) infections and GBC risk have been investigated by many researchers, the findings remain inconsistent. Apparent discrepancies may be explained by differences in the samples used (gallbladder bile, gallbladder tissue, and serum) and/or in the diagnostic procedures used (culture method, serological examination, and polymerase chain reaction [PCR] analysis). More detailed studies are therefore needed to ascertain if these bacterial infections are indeed related to GBC risk.
A previous study demonstrated that PCR analysis was the most sensitive of the above methods for detecting Salmonella typhi in bile samples (Zhou and Pollard, 2010). Next generation sequencing (NGS) technology can be used to detect and identify microbial organisms directly in the natural environment or in human biological samples, without the need for isolation and confirmation by culture (Salipante et al., 2013). This technology also enables the detection and identification of unculturable microorganisms in biological samples.
We hypothesized that tumours in organs containing body fluid may be caused by microbes in the fluid; more specifically GBC may be caused by Salmonella typhi, Helicobacter sp., or other unknown bacteria in gallbladder bile. Previous studies have investigated the bacterial communities in bile collected from CL patients or healthy individuals usin 16S rRNA gene sequencing (Wu et al., 2013; Shen et al., 2015; Ye et al., 2016). However, to the best of our knowledge, no previous study has examined the bacteria in gallbladder bile collected from patients with both GBC and CL using NGS technology.
We therefore conducted a preliminary case-control study to reveal the association between bacterial bile infection and GBC risk, and compare gallbladder bile bacteria between patients with GBC and those with CL.
Materials and Methods
Subjects
A total of seven GBC patients (Bolivia, 2; Chile, 5), and 30 CL patients (Bolivia, 22; Chile, 8) were enrolled in this study. Each patient had a diagnosis of GBC or CL at the Instituto de Gastroenterologia Boliviano-Japones in La Paz, Bolivia, from August 2014 to August 2015, or at the Sotero del Rio Hospital in Santiago, Chile, from December 2014 to May 2016.
Bile sample collection
Gallbladder bile samples were collected from all patients at the hospital, after they were diagnosed with GBC or CL. At least 1 mL of bile was collected from each patient by aspiration from the gallbladder after laparoscopic cholecystectomy, injected into a sterile 1.5 mL Eppendorf safe-lock tubes, and covered with a lid. All samples were stored at −80 °C and transported from Bolivia or Chile to Japan under frozen conditions. The samples remained stored at −80 °C until DNA extraction.
DNA extraction from bile samples
Genomic DNA was extracted from the bile samples using NucleoSpin Soil (Macherey-Nagel GmbH and Co. KG, Duren, Germany). The extracted DNA was purified using an AMPure XP kit (Beckman Coulter, Inc., CA, USA), and the concentrations were quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., MA, USA). Double-stranded DNA concentrations were measured using a Quant-iT dsDNA BR Assay Kit (Thermo Fisher Scientific Inc.).
Library preparation
Library preparation was performed according to the instructions (Illumina, 2015) provided by Illumina, Inc. (San Diego, CA, USA). The devices, reagents, and primers used are noted below.
Amplicon PCR
A 460-bp region of DNA was amplified, including the 16S V3-V4 region, using NGS with Tks Gflex DNA Polymerase (Takara Bio Inc., Tokyo, Japan) and a Nextera XT Index Kit (Illumina, Inc.). The 16S amplicon PCR forward and reverse primers were as follows:
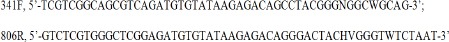
The PCR amplification conditions were as follows: 94 °C for 60 s for 1 cycle; 98 °C for 10 s, 50 °C for 15 s, and 68 °C for 15 s for 28 cycles; 4 °C hold.
PCR clean-up
The 16S V3 and V4 amplicon was purified from free primers and primer dimer species using AMpure XP beads (Beckman Coulter, Inc.).
Index PCR
This step was performed using a Nextera XT Index Kit (Illumina, Inc.). The amplification conditions were as follows: 94 °C for 60 s for 1 cycle; 98 °C for 10 s, 60 °C for 15 s, and 68 °C for 15 s for 8 cycles; 4 °C hold. The PCR products were purified using AMpure XP beads (Beckman Coulter, Inc.).
Library quantification, normalization, and pooling
The libraries were evaluated using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA), quantified using a fluorometric method, and diluted with 10 mM Tris pH 8.5 buffer solution to 4 nM. Each 5 μL sample was then pooled for one MiSeq run.
Library denaturing and MiSeq sample loading
Library denaturing was performed using a MiSeq Sequencing System (Illumina, Inc.), MiSeq Reagent Kit v3 (Illumina, Inc.), and Phix Control Kit v3 (Illumina, Inc.). Prior to cluster generation and sequencing, pooled libraries were denatured with NaOH, diluted with hybridization buffer solution, and heat denatured. A minimum of 5% Phix was included in each run.
Metagenomics workflow
The metagenomics was conducted using the following programs, database, and methods.
Clustering: CD-HIT-OUT (ver. 0.0.1). Homology: BLAST (Ver. 2.2.20), DDBJ 16S rRNA database (Ver. 2016_08_17). Systematic taxonomy: RDP classifier (Ver. 2.2), GreemGenes (the latest version in Aug of 2016). Phylogenetic trees data preparation: PyNest (Ver. 1.2), FastTree (Ver. 2.1.3), GreenGenes (the latest version). Comparative metagenomics: principal coordinate analysis based on unifrac distance matrix, cluster samples analysis by unweigted pair group method with arithmetic mean method.
Ethics statement
The study procedure was approved by the Ethics Committee of Niigata University of Health and Welfare (No.17540-141030), and was conducted according to the principles expressed in the 1964 Helsinki Declaration. Prior to the start of the study, Bolivian and Chilean researchers informed the subjects of the purpose, procedure, and anticipated results of the study, as well as the risks and benefits. All participants provided informed consent.
Statistical analysis
All statistical analyses were performed using Stata Data Analysis and Statistical Software (STATA 14; StataCorp, College Station, TX, USA). Differences between two groups (GBC and CL, Bolivia and Chile) were analysed by χ2 or Fisher’s exact tests. A p value < 0.05 (two-tailed) was considered to indicate statistical significance.
Results
The characteristics of seven patients with GBC from Bolivia and Chile are shown in Table 1. Two Bolivian patients (1 woman, 1 man) with GBC were enrolled (mean age 48.0 years), both of whom had gallstones and chronic cholecystitis according to histopathological diagnosis. Only the male patient had bacteria in the bile, giving a detection rate of bile bacteria of 50% (1/2). Five Chilean patients (3 women, 2 men) were enrolled in this study (mean age 62.4 years), four of whom had gallstones. Four patients had chronic cholecystitis, and bile bacteria were detected in two female patients, giving a detection rate of 40% (2/5).
Table 1.
Characteristics of Bolivian and Chilean Patients with Gallbladder Cancer
| Country Gender | Age, y | Histological diagnosis | Presence of gallstones | Type of cholecystitis | Bacteria in the bile |
|---|---|---|---|---|---|
| Bolivia | |||||
| Woman | 51 | Poorly differentiated invasive adenocarcinoma | Positive | Chronic | Negative |
| Man | 45 | Bile duct invasive adenocarcinoma | Positive | Chronic | Positive |
| Chile | |||||
| Woman | 53 | Adenosquamous carcinoma | Positive | Negative | Negative |
| Woman | 68 | Poorly differentiated tubular adenocarcinoma | Positive | Acute on chronic | Positive |
| Woman | 71 | Poorly differentiated adenocarcinoma | Positive | Chronic | Positive |
| Man | 52 | Poorly differentiated carcinoma with signet cells component | Positive | Chronic | Negative |
| Man | 68 | Moderatlely differentiated tubular adenocarcinoma | Negative | Chronic | Negative |
The characteristics of the Bolivian and Chilean patients with CL are shown in Table 2. There were no significant differences in gender, mean age, presence of cholecystitis, rate of cholesterol gallstones, or bile-bacteria-detection rate between Bolivia and Chile.
Table 2.
Characteristics of Bolivian and Chilean Patients with Cholelithiasis
| Bolivia (n = 22) | Chile (n = 8) | P value | |
|---|---|---|---|
| Gender | |||
| Women (%) | 12 (55) | 7 (88) | 0.2 |
| Men (%) | 10 (45) | 1 (12) | |
| Mean Age | |||
| Women (SD) | 49.4 (6.1) | 55.4 (14.2) | 0.32 |
| Men (SD) | 44.9 (5.8) | 45 | |
| Total (SD) | 47.1 (6.2) | 54.3 (13.5) | 0.054 |
| Cholecystitis (%) | 22 (100) | 8 (100) | 1 |
| Cholesterol gallstone (%) | 12 (55) | 3 (38) | 0.45 |
| Bacteria detected bile (%) | 3 (14) | 1 (13) | 1 |
The detection rates of bile bacteria in Bolivian and Chilean patients with GBC and CL are shown in Table 3. The bile-bacteria-detection rates among Bolivian patients were 50% (1/2) for GBC patients and 14% (3/22) for CL patients, with no significant difference in bile-bacteria-detection rates between the GBC and CL patients (p = 0.31). The detection rates in the Chilean patients were 40% (2/5) for GBC patients and 13% (1/8) for CL patients, with no significant difference in bile-bacteria-detection rates between the GBC and CL patients (p = 0.51). Bolivian and Chilean patients with GBC and CL thus had similar detection rates of bile bacteria.
Table 3.
Detection Rates of Bile Bacteria in Bolivian and Chilean Patients with Gallbladder Cancer and Cholelithiasis
| Positive (%) | Negative (%) | OR | 95% CI | P value | |
|---|---|---|---|---|---|
| Bolivia | |||||
| CL | 3 (14) | 19 (86) | 1 | ||
| GBC | 1 (50) | 1 (50) | 6.3 | 0.5-80.3 | 0.31 |
| Chile | |||||
| CL | 1 (13) | 7 (87) | 1 | ||
| GBC | 2 (40) | 3 (60) | 4.7 | 0.4-50.1 | 0.51 |
| Total | |||||
| CL | 4 (13) | 26 (87) | 1 | ||
| GBC | 3 (43) | 4 (57) | 4.9 | 0.9-28.2 | 0.11 |
CL, cholelithiasis; GBC, gallbladder cancer; OR, odds ratio; CI, confidence interval; Infection rates are shown in parentheses
The bacteria detected in bile samples of Bolivian patients with GBC and CL are shown in Table 4. A total of 24 and 12 types of bacteria were found in bile from GBC and CL patients, respectively. The predominant species were Fusobacterium nucleatum subsp. animals ATCC 51,191 (Fusobacterium nucleatum) in GBC patients and Escherichia coli (E. coli), Enterococcus gallinarum, and Salmonella sp. in CL patients. E. coli, bacterium 28W412, and Pseudomonas pseudoalcaligenes were found in both GBC and CL patients.
Table 4.
Bacteria Detected in Bile of Bolivian Patients with Gallbladder Cancer or Cholelithiasis
| Consensus Lineage | GBC | CL-1 | CL-2 | CL-3 | #OTU ID |
|---|---|---|---|---|---|
| Escherichia coli | + | ++ | + | Cluster 0 | |
| Enterococcus gallinarum | ++ | Cluster 1 | |||
| Salmonella sp. ATK1 | + | ++ | Cluster 2 | ||
| Bacteroides fragilis | ++ | Cluster 3 | |||
| Fusobacterium mucleatum subsp.* | + | Cluster 4 | |||
| Pyramidobacter piscolens | + | Cluster 5 | |||
| bacterium NLAE-zl-H528 | + | Cluster 6 | |||
| Odoribcter splanchnicus DSM 20712 | + | Cluster 7 | |||
| Anaeroglobus sp. S4-A15 | + | Cluster 8 | |||
| bacterium IARFR1475 | + | Cluster 9 | |||
| Streptococcus sp. ChDC B623 | + | Cluster 10 | |||
| Veillonellaceae bacterium oral taxon 150 | + | Cluster 11 | |||
| Mythylobacterium zatmanii | + | + | Cluster 12 | ||
| Clostridium sp. | + | Cluster 13 | |||
| Selenomonas sputigena | + | Cluster 14 | |||
| Enterococcus faecalis | + | Cluster 15 | |||
| Bacteroides uniformis | + | Cluster 16 | |||
| bacterium NLAE-zl-H504 | + | Cluster 17 | |||
| bacterium 28W412 | + | + | + | Cluster 18 | |
| Clostridium perfringens | + | Cluster 19 | |||
| *** no hit *** | + | + | + | Cluster 20 | |
| Dialister pneumosintes | + | Cluster 21 | |||
| Selenomonas artemidis | + | Cluster 22 | |||
| Klebsiella sp. | + | + | Cluster 23 | ||
| Agrobacterium larrymoorei | + | Cluster 24 | |||
| Prevotella denticola | + | Cluster 25 | |||
| bacterium P1C8 | + | Cluster 26 | |||
| Pseudomonas pseudoalcaligenes | + | + | + | Cluster 27 | |
| Pseudomonas koreensis | + | + | Cluster 28 | ||
| Slackia sp. CM382 | + | Cluster 29 | |||
| Bacteroides salyersiae | + | Cluster 30 |
Bacteria detected in bile samples from Chilean patients with GBC and CL are shown in Table 5. A total of eight and two types of bacteria were found in the bile from GBC and CL patients, respectively. The predominant species were E. coli and Enterobacter sp. B10 (2014) in GBC patients, and E. coli in the CL patient. E. coli was detected in bile samples from both GBC and CL patients. Enterobacter sp. B10 (2014) and Klebsiella oxytoca were detected in bile samples from two GBC patients.
Table 5.
Bacteria Detected in Bile of Chilean Patients with Gallbladder Cancer or Cholelithiasis
| Consensus Lineage | GBC-1 | GBC-2 | CL | #OTU ID |
|---|---|---|---|---|
| Escherichia coli | ++ | ++ | Cluster 0 | |
| Enterobacter sp. B10 (2014) | ++ | + | Cluster 1 | |
| Klebsiella oxytoca | + | + | Cluster 2 | |
| bacterium NLAE-zl-P344 | + | Cluster 3 | ||
| Streptococcus sanguinis | + | Cluster 4 | ||
| Enterococcus durans | + | Cluster 5 | ||
| Propionibacterium acidifaciens | + | Cluster 6 | ||
| Klebsiella pneumoniae | + | Cluster 7 | ||
| Citrobacter sp. AL7 | + | Cluster 8 |
OTU, an operational taxonomic unit; ++, predominant species; +, detected species.
Discussion
The results of this study demonstrated bacterial infection rates in bile of 42.9% in combined Bolivian and Chilean patients with GBC, and 13.3% in patients with CL. Salmonella sp. and Helicobacter sp., previously suggested to be associated with the development of GBC, were not detected in the bile of GBC patients, while Fusobacterium nucleatum, E. coli, and Enetrobacter sp. were the predominant species in Bolivian and Chilean GBC patients.
Gallstones with attendant chronic cholecystitis has been reported to be an important risk factor for GBC (Hundal and Shaffer, 2014). However, although several studies have examined the role of bacterial infection in GBC risk, inconsistent findings have been reported. There are several possible explanations for the apparent discrepancy in results regarding the relationship between bacterial infection and GBC risk. The use of different samples and methods may have led to differences among results. There are various ways of checking for bacterial infections, e.g., culture methods, serological examination, and PCR, of which PCR has been reported to be the most sensitive for detecting bacteria in bile samples (Zhou and Pollard, 2010). Culture methods are commonly used to detect bacterial infection, but specific media are needed to allow the development of anaerobic or acid-fast bacteria, making it difficult to culture and identify multiple types of bacteria simultaneously. NGS-based 16S rRNA sequencing has recently been developed and used for microbiological research. This technology provides a culture-free method that permits the analysis of the whole microbial community in a sample (Salipane et al., 2013). Previous studies have mainly examined the associations between Salmonella typhi or Salmonella paratyphi (Andia et al., 2008; Nagaraja and Eslick, 2014), or Helicobacter sp. (Martel et al., 2009; Pandey et al., 2010; Yakoob et al., 2011) infections and GBC risk, but other unknown bacteria may also be related to GBC risk. We therefore aimed to clarify the characteristics of the entire microbial communities in gallbladder bile from GBC and CL patients using NGS-based 16S rRNA sequencing.
Previous studies found detection rates of 44.8% (Roa et al., 1999) and 81.0% (Csendes et al., 1994) for bile bacteria in Chilean patients with GBC, compared with a detection rate of 40.0% in the current study, using NGS technology. The reasons for the lower detection rate were unclear, but it is possible that the chance of bacterial infection may have decreased in recent years as a result of antibiotic administration. The predominant species of bile bacteria in Bolivian GBC patients was Fusobacterium nucleatum, which is known to be isolated from the oral cavity (Gharbia et al., 1990). This bacterium is known to be an opportunistic pathogen, but since 2011, many researchers has reported to be associated with colorectal cancer risk (Ray, 2011; Castellarin et al., 2012; Keku and McCoy, 2013; Miwa et al., 2015; Nosho et al., 2016; Repass et al., 2016). Primary sclerosing cholangitis and ulcerative colitis (UC) patients are recognized as a high-risk group for cholangiocarcinoma and GBC (Herzog and Goldblum, 1996; Yamamoto et al., 2003; Lewis et al., 2007; Noda et al., 2009; Pastogi et al., 2012). Fusobacterium and other bacteria were detected in the UC patients (Andoh et al., 2007). Our finding that Fusobacterium nucleatum was detected in the bile from the GBC patients might be explained within the association among GBC risk, UC, and Fusobacterium infection. In contrast, the predominant bacteria of Chilean GBC patients were E. coli and Enterobacter sp. B10 (2014). Given that these bacteria were also found in previous studies in Chile (Csendes et al., 1994; Roa et al., 1999), the bacteria may live in the bile of some GBC patients. A previous study has demonstrated that Fusobacterium nucteatum, E. coli, and Enterobacter sp. infections contribute to promoting colon cancer (Gagnaire et al., 2017). The presence of these bacteria in the bile might also increase the risk of GBC by the same mechanism as colon cancer. However, further studies are needed to confirm the relationship between these bacrterial infections and the development of GBC.
Contrary to our expectations, Salmonella sp. was not detected in bile from the patients with GBC in the present study, though this may have been because of the small number of Bolivian GBC patients. On the other hand, the failure to detect Salmonella sp. in the bile of Chilean patients with GBC may be related to a decrease in infection rate of Salmonella typhi. Typhoid fever is known to be caused by Salmonella typhi, and was a common infection in Chile with an incidence of 100–121 per 100,000 population from 1976–1985. The incidence rate subsequently decreased sharply to the current status of 3.3 per 100,000 population years (Andia et al., 2008). The lack of Salmonella sp. detection in the Chilean GBC patients may thus reflect the decreased infection rate of Salmonella typhi. Furthermore, most Chilean patients with GBC were diagnosed at an advanced stage, and bile collection from the gallbladder was thus difficult and the collected volume was small. Salmonella typhi infection prior to GBC diagnosis was treated with antibiotics, which may thus have affected the development of the bacteria. A report from India, which has a higher incidence of GBC in the north compared with the south, found higher Salmonella typhi infection rates in the north (Banerjee et al., 2014). The detection rate of Salmonella sp. from the bile might thus parallel the Salmonella typhi infection rate among people living in high-GBC-incidence areas.
Nine studies have reported on the associations between Helicobacter infection and GBC risk since 2000 (Martel et al., 2009; Pandey et al., 2010; Yakoob et al., 2011), of which eight detected Helicobacter sp. in bile or gallbladder tissue from GBC patients. These nine studies used various different methods (PCR, serological examination, and culture) to detect Helicobacter sp., including PCR in three, PCR or serological examination in four, serological examination in one, and all three methods in one. A study in Germany using all three methods failed to detect Helicobacter sp. (Bohr et al., 2007), and the authors suggested that these negative results may reflect the low incidence of GBC in Germany. Overall, these studies suggest that Helicobacter infection may be related to the development of GBC, particularly in areas of high GBC incidence.
To the best of our knowledge, no previous study has examined the association between Helicobacter infection and GBC risk in Chileans. We found no Helicobacter in any bile samples from Bolivian or Chilean patients with GBC or CL. However, Fox et al., (1998) detected Helicobacter sp. in bile and gallbladder tissue from Chilean patients with cholecystitis using PCR. The detected bacteria were Helicobacter bilis, Flexispira rappini (ATCC 49317), and Helicobacter pullorum based on the sequencing data. They suggested an association between bile-resistant Helicobacter infections and gallbladder disease, and pointed out the need to confirm the roles of these bacteria in the development of GBC. Chile has a high incidence of GBC, and further studies are therefore needed to clarify the association between Helicobacter infection and GBC. Wang et al., (2015) reported that Helicobacter pylori was rapidly induced into Helicobacter pylori L-form in human bile, which is difficult to isolate using routine bacteriological methods and difficult to identify even by PCR. It may therefore be necessary to pay specific attention to the L-form, as well as wild-type Helicobacter sp. in order to increase its detection in bile.
This study had some limitations. The sample sizes, especially in Bolivian GBC patients and Chilean patients with GBC and CL, were small, and the low bacterial detection rates in the bile may not reflect the accurate detection rates. Furthermore, the absence of a significant difference in bacterial infection rates between GBC patients and CL patients may also have been caused by a lack of statistical power associated with the small number of patients analysed. This is reflected by the wide 95% CI. More bacterial strains were detected in bile from Bolivian compared with Chilean patients. The reasons for this difference are unclear, but may be related to differences in ethnicity or daily life environments between the populations. Despite these limitations, this study provides the first evidence regarding the characteristics of the microbial communities in bile from Bolivian and Chilean patients with GBC or CL using NGS technology.
In summary, we clarified the nature of the microbial communities present in bile in Bolivian and Chilean patients with GBC and CL using 16S rRNA metagenomics. The predominant species in Bolivian GBC patients was Fusobacterium sp., but those in Chilean GBC patients were E. coli and Enterobacter sp. B10 (2014), while Salmonella sp. and Helicobacter sp. were not detected in bile from GBC patients. These findings provide fundamental data regarding the pathogenesis of GBC in Bolivia and Chile, but further studies are needed to ascertain the roles of the bacteria detected in the present study in the development of GBC.
Acknowledgments
This work was supported by JSPS KAKENHI, grant number JP26460812. We are indebted to Ms. Kayo Okuda of Takara Bio Inc. for providing expert technical skills for metagenomics analysis, and to Ms. Ryoko Nozaki for technical assistance.
References
- 1.Andoh A, Sakata S, Koizumi Y, et al. Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis. 2007;13:955–62. doi: 10.1002/ibd.20151. [DOI] [PubMed] [Google Scholar]
- 2.Andia ME, Hsing AW, Andreotti G, Ferreccio C. Geographic variation of gallbladder cancer mortality and risk factors in Chile:a population-based ecologic study. Int J Cancer. 2008;123:1411–6. doi: 10.1002/ijc.23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee T, Shukla BN, Filgona J, et al. Trends of typhoid fever seropositivity over ten years in north India. Indian J Med Res. 2014;140:310–3. [PMC free article] [PubMed] [Google Scholar]
- 4.Bohr UR, Kuester D, Meyer F, et al. Low prevalence of Helicobacteraceae in gall-stone disease and gall-bladder carcinoma in the German population. Clin Microbiol Infect. 2007;13:525–31. doi: 10.1111/j.1469-0691.2007.01690.x. [DOI] [PubMed] [Google Scholar]
- 5.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Csendes A, Becerra M, Burdiles P, et al. Bacteriological studies of bile from the gallbladder in patients with carcinoma of the gallbladder, cholelithiasis, common bile duct stones and no gallstones disease. Eur J Surg. 1994;160:363–7. [PubMed] [Google Scholar]
- 7.Dutta U, Nagi B, Garg PK, et al. Patients with gallstones develop gallbladder cancer at an earlier age. Eur J Cancer Prev. 2013;14:381–5. doi: 10.1097/00008469-200508000-00011. [DOI] [PubMed] [Google Scholar]
- 8.Fox JG, Dewhirst FE, Shen Z, et al. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology. 1998;114:755–63. doi: 10.1016/s0016-5085(98)70589-x. [DOI] [PubMed] [Google Scholar]
- 9.Gagnaire A, Nadel B, Raoult D, et al. Collateral damage:insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 2017;15:109–28. doi: 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- 10.Gharbia SE, Shah HN, Lawson PA, Haapasalo M. Distribution and frequency of Fusobacterium nucleatum subspecies in the human oral cavity. Oral Microbiol Immunol. 1990;5:324–7. doi: 10.1111/j.1399-302x.1990.tb00434.x. [DOI] [PubMed] [Google Scholar]
- 11.Herzog K, Goldblum JR. Gallbladder adenocarcinoma and acalculous chronic lymphoplasmacytic cholecystitis associated with ulcerative colitis. Mod Pathol. 1996;9:194–8. [PubMed] [Google Scholar]
- 12.Hsing AW, Gao YT, Han TQ, et al. Gallstones and the risk of biliary tract cancer:a population-based study in China. Br J Cancer. 2007;97:1577–82. doi: 10.1038/sj.bjc.6604047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hundal R, Shaffer EA. Gallbladder cancer:epidemiology and outcome. Clin Epidemiol. 2014;6:99–109. doi: 10.2147/CLEP.S37357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Illumina Inc. 16S metagenomic sequencing library preparation. Part#15044223 Rev. B. 2015 [Google Scholar]
- 15.Kanthan R, Senger JL, Ahmed S, Kanthan SC. Gallbladder Cancer in the 21st Century. J Oncol. 2015;2015:967472. doi: 10.1155/2015/967472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keku TO, McCoy AN, Azcarate-Peril AM. Fusobacterium spp and colorectal cancer:cause or consequence? Trends Microbiol. 2013;21:506–8. doi: 10.1016/j.tim.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S, Kumar S, Kumar S. Infection as a risk factor for gallbladder cancer. J Surg Oncol. 2006;93:633–9. doi: 10.1002/jso.20530. [DOI] [PubMed] [Google Scholar]
- 18.Lewis JT, Talwalkar JA, Rosen CB, et al. Prevalence and risk factors for gallbladder neoplasia in patients with primary sclerosing cholangitis:evidence for a metaplasia-dysplasia-carcinoma sequence. Am J Surg Pathol. 2007;31:907–13. doi: 10.1097/01.pas.0000213435.99492.8a. [DOI] [PubMed] [Google Scholar]
- 19.de Martel C, Plummer M, Parsonnet J, et al. Helicobacter species in cancers of the gallbladder and extrahepatic biliary tract. Br J Cancer. 2009;100:194–9. doi: 10.1038/sj.bjc.6604780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653–61. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagaraja V, Eslick GD. Systematic review with meta-analysis:the relationship between chronic Salmonella typhi carrier status and gall-bladder cancer. Aliment Pharmacol Ther. 2014;39:745–50. doi: 10.1111/apt.12655. [DOI] [PubMed] [Google Scholar]
- 22.Noda H, Chiba F, Toyama N, Konishi F. Mucin-producing carcinoma of the gallbladder associated with primary sclerosing cholangitis and ulcerative colitis. J Hepatobiliary Pancreat Surg. 2009;16:83–5. doi: 10.1007/s00534-008-0001-6. [DOI] [PubMed] [Google Scholar]
- 23.Nosho K, Sukawa Y, Adachi Y, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22:557–66. doi: 10.3748/wjg.v22.i2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey M, Mishra RR, Dixit R, et al. Helicobacter bilis in human gallbladder cancer:results of a case-control study and a meta-analysis. Asian Pac J Cancer Prev. 2010;11:343–7. [PubMed] [Google Scholar]
- 25.Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide:geographical distribution and risk factors. Int J Cancer. 2006;118:1591–602. doi: 10.1002/ijc.21683. [DOI] [PubMed] [Google Scholar]
- 26.Ray K. Colorectal cancer:Fusobacterium nucleatum found in colon cancer tissue--could an infection cause colorectal cancer? Nat Rev Gastroenterol Hepatol. 2011;8:662. doi: 10.1038/nrgastro.2011.208. [DOI] [PubMed] [Google Scholar]
- 27.Repass J, Maherali N, Owen K. Registered report:Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Elife. 2016;5:e10012. doi: 10.7554/eLife.10012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roa I, Ibacache G, Carvallo J, et al. Microbiological study of gallbladder bile in a high risk zone for gallbladder cancer. Rev Med Chil. 1999;127:1049–55. (In Spanish) [PubMed] [Google Scholar]
- 29.Roa I, Ibacache G, Roa J, et al. Gallstones and gallbladder cancer-volume and weight of gallstones are associated with gallbladder cancer:a case-control study. J Surg Oncol. 2006;93:624–8. doi: 10.1002/jso.20528. [DOI] [PubMed] [Google Scholar]
- 30.Rastogi A, Bihari C, Singh S, et al. Adenoendocrinecarcinoma of gallbladder in a patient with primary sclerosing cholangitis and ulcerative colitis. Trop Gastroenterol. 2012;33:158–60. doi: 10.7869/tg.2012.40. [DOI] [PubMed] [Google Scholar]
- 31.Salipante SJ, Sengupta DJ, Rosenthal C, et al. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS One. 2013;8:e65226. doi: 10.1371/journal.pone.0065226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen H, Ye F, Xie L, et al. Metagenomic sequencing of bile from gallstone patients to identify different microbial community patterns and novel biliary bacteria. Sci Rep. 2015;5:17450. doi: 10.1038/srep17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shrikhande SV, Barreto SG, Singh S, et al. Cholelithiasis in gallbladder cancer:coincidence, cofactor, or cause! Eur J Surg Oncol. 2010;36:514–9. doi: 10.1016/j.ejso.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Walawalkar YD, Gaind R, Nayak V. Study on Salmonella Typhi occurrence in gallbladder of patients suffering from chronic cholelithiasis-a predisposing factor for carcinoma of gallbladder. Diagn Microbiol Infect Dis. 2013;77:69–73. doi: 10.1016/j.diagmicrobio.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Wang DN, Ding WJ, Pan YZ, et al. The Helicobacter pylori L-form:formation and isolation in the human bile cultures in vitro and in the gallbladders of patients with biliary diseases. Helicobacter. 2015;20:98–105. doi: 10.1111/hel.12181. [DOI] [PubMed] [Google Scholar]
- 36.Wu T, Zhang Z, Liu B, et al. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics. 2013;14:669. doi: 10.1186/1471-2164-14-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yakoob J, Khan MR, Abbas Z, et al. Helicobacter pylori:association with gall bladder disorders in Pakistan. Br J Biomed Sci. 2011;68:59–64. doi: 10.1080/09674845.2011.11730324. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto T, Uki K, Takeuchi K, et al. Early gallbladder carcinoma associated with primary sclerosing cholangitis and ulcerative colitis. J Gastroenterol. 2003;38:704–6. doi: 10.1007/s00535-002-1126-z. [DOI] [PubMed] [Google Scholar]
- 39.Ye F, Shen H, Li Z, et al. Influence of the biliary system on biliary bacteria revealed by bacterial communities of the human biliary and upper digestive tracts. PLoS One. 2016;11:e0150519. doi: 10.1371/journal.pone.0150519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou L, Pollard AJ. A fast and highly sensitive blood culture PCR method for clinical detection of Salmonella enterica serovar Typhi. Ann Clin Microbiol Antimicrob. 2010;9:14. doi: 10.1186/1476-0711-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


