Abstract
Background:
Alteration in the biotransformation of exogenous compounds can result in production of reactive oxygen species (ROS), which can predispose cells to malignant transformation in the head and neck. This study aimed to evaluate the expression of genes involved in antioxidant metabolism in the oral squamous cell carcinoma (OSCC).
Methods:
The expression of eighty-four genes was evaluated in OSCC and non-tumor tissues by quantitative real-time polymerase chain reaction using the TaqMan Gene Expression Array. The biological mechanisms related to the differentially expressed genes were investigated using Gene – NCBI, KEGG, UNIPROT and REACTOME databases.
Results:
Twenty-one genes encoding enzymes involved in antioxidant metabolism were differentially expressed in the OSCC case. Four genes (ATOX1, PRDX4, PRNP, and SOD2) were up-regulated, and seventeen (ALOX12, CAT, CSDE1, DHCR24, DUOX1, DUOX2, EPHX2, GLRX2, GPX3, GSR, GSTZ1, MGST3, PRDX1, OXR1, OXSR1, SOD1, and SOD3) were down-regulated. We identified 14 possible novel biomarkers for OSCC. The differentially expressed genes appeared related to important biological processes involved in carcinogenesis, such as inflammation, angiogenesis, apoptosis, genomic instability, invasion, survival, and cell proliferation.
Conclusions:
Our study identified novel biomarkers which might warrant further investigation regarding OSCC pathogenesis since the altered expression in the genes can modulate biological processes related to oxidative stress and predispose cells to malignant transformation in the oral cavity.
Keywords: Oxidative stress, biomarker, carcinoma squamous cell, oral cavity, carcinogens
Introduction
Oxidative stress is associated with the imbalance of intracellular reactive oxygen species (ROS) / antioxidants, and ROS that are not efficiently neutralized or removed (Bentz, 2007). This imbalance leads to the increase of ROS, formation of DNA adducts, activation of proto-oncogenes, and inhibition of tumor suppressor genes (Matsui et al., 2000). ROS are produced as a result of the consumption of nicotinamide adenine dinucleotide phosphate (NADPH) by microsomal cytochrome P450 (CYP450) (Zangar et al., 2004). CYP450 is the main enzyme family which plays a role in the oxidative biotransformation of drugs and other lipophilic exogenous chemical compounds (Guengerich, 2008). Redox cycling catalyzed by CYP enzymes results in the release of superoxide radicals, an important ROS. Intra- and extracellular levels of superoxide radicals are controlled by antioxidant enzymes, such as the superoxide dismutase (SOD) family. SOD reduces superoxide to hydrogen peroxide and molecular oxygen (Young and Woodside, 2001; Buettner et al., 2011). Despite the presence of SOD, other enzymes and agents compose an antioxidant system able to eliminate the reactive species formed in the redox reactions. Catalase (CAT), peroxiredoxin (PRDX), glutathione peroxidase (GPx), and glutathione reductase (GR) are the main enzymatic antioxidants directly involved in the neutralization of hydrogen peroxide (Halliwell, 2007).
The lack of antioxidants and the increase of lipid peroxidation and nitric oxide have been observed in cases of oral cancer (Rasheed et al., 2007). Oxidative stress-related enzymes can participate in the carcinogenesis and play an essential role in individuals’ susceptibility to this disease (Masood et al., 2011). Therefore, alteration in the expression of these enzymes can indicate possible mechanisms involved in head and neck cancer. This study aimed to investigate the contribution of ROS metabolism in the development of oral squamous cell carcinoma (OSCC). We evaluated the expression profile of 84 genes involved in the antioxidant metabolism in OSCC and identified the biological mechanisms related to the differentially expressed genes.
Materials and Methods
Tissue samples
This study was approved by the Research Ethics Committee of Sao Jose do Rio Preto Medical School – FAMERP (no. 752.570). Sixteen tissue samples (eight OSCC and eight adjacent non-tumor tissues) were collected from 2013 to 2015 during surgical resection procedure at the Otolaryngology and Head and Neck Surgery Service, Hospital de Base, Sao Jose do Rio Preto, Brazil. Fresh tissues were immediately frozen in liquid nitrogen and stored at -80ºC until processing. All cases were histologically confirmed by a pathologist.
Table 1 shows the clinicopathological characteristics of the OSCC patients. The tumor staging was performed according to the parameters of the Union International Control Cancer and American Joint Committee for Cancer (Sobin and Wittekind, 2010). T1N0 tumors were classified as stage I; T1N1 and T2N0-1 tumors were classified as stage II; T3N0-1 and T1-3N2 tumors were classified as stage III; and T4N0-3, T1-3N3 and T1-4N0-3M1 tumors were classified as stage IV. For statistical analysis, the tumors were grouped into non-advanced (stage I and II) and advanced (stage III and IV) categories.
Table 1.
Clinicopathological Characteristics of the OSCC Patients
| Patients (n=08) | |
|---|---|
| Characteristics | n (%) |
| Age (Mean ± SD) | 66.38 (±12.56) |
| Gender (Male / Female) | 5 (62.5 %) / 3 (37.5 %) |
| Tobacco consumption | 7 (87.5 %) |
| Alcohol consumption | 6 (75 %) |
| T classification | |
| T1 – T2 | 4 (50 %) |
| T3 – T4 | 4 (50 %) |
| N classification | |
| N0 | 7 (87.5 %) |
| N1, N2, N3 | 1 (12.5 %) |
| Tumor Stage | |
| I e II | 4 (50 %) |
| III e IV | 4 (50 %) |
SD, standard deviation
Quantitative Polymerase Chain Reaction (qPCR)
RNA samples were extracted from tissues using TRIzol Reagent (Ambion, Austin, TX). Synthesis of complementary DNA (cDNA) was performed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Expression of genes involved in the antioxidant metabolism was quantified using the TaqMan Array Human Antioxidant Mechanisms (Applied Biosystems, Foster City, CA). The array contains probes for quantification of the expression of 84 genes encoding antioxidant proteins. The relative expression (fold change) was calculated using the ΔΔCt method (Livak and Schmittgen, 2001), using non-tumor samples as calibrators. Amplification reaction was performed according to manufacturer’s instructions on the StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA).
Identification of biological mechanisms related to the differentially expressed genes
The investigation of the biological processes related to the differentially expressed genes was performed using Gene – NCBI, KEGG, UNIPROT and REACTOME databases.
Statistical analyses
Boxplots were performed to check the normally distributed data for outliers. D’Agostino & Pearson’s omnibus normality test was used to test for normal distribution. Gene expression analyses were performed using the One Sample T test or Wilcoxon Signed Rank test. Also, the Mann-Whitney test was used to evaluate the gene expression in relation to the tumor progression. Statistical analyses were performed using GraphPad Prism version 5.1. P-value < 0.05 was considered significant.
Results
Twenty-one genes were differentially expressed in OSCC (P < 0.05) compared to non-tumor tissues (Table 2). Four genes were up-regulated (fold change > 1), and 17 were down-regulated (fold change < 1) in OSCC (Figure. 1). Fourteen genes (ALOX12, CSDE1, DHCR24, DUOX1, DUOX2, EPHX2, GRLX2, GPX3, GSR, GSTZ1, MGTS3, OXR1, OXSR1, and SOD1) differentially expressed in OSCC were investigated for the first time by this study. After bioinformatics analysis, the oxidative stress-related genes were associated with fifteen important biological processes for carcinogenesis, including inflammation, angiogenesis, apoptosis, genomic instability, invasion, survival, and cell proliferation (Table 3). The differential expression of these genes first investigated by our study suggests an important participation of Glutathione metabolism in oral cancer since four of these genes participate in this metabolic pathway (GSR, MGST3, GPX3, and GSTZ1).
Table 2.
Differentially Expressed Oxidative Stress-related Genes in Oral Squamous Cell Carcinoma
| Gene Symbol | Gene Name | Chromosome | Gene ID | Fold change (median) | Minimum | Maximum | P-value |
|---|---|---|---|---|---|---|---|
| ALOX12* | Arachidonate 12-lipoxygenase,12S type | 17p13.1 | 239 | 0.029 | 0.0007 | 0.223 | 0.015 |
| ATOX1 | Antioxidant 1copper chaperone | 5q32 | 475 | 1.555 | 0.904 | 5.086 | 0.031 |
| CAT | Catalase | 11p13 | 847 | 0.639 | 0.184 | 1.111 | 0.009 |
| CSDE1* | Cold shock domain containing E1 | 1p22 | 7812 | 0.769 | 0.336 | 1.252 | 0.043 |
| DHCR24* | 24-dehydrocholesterol reductase | 1p32.3 | 1718 | 0.393 | 0.140 | 1.097 | 0.031 |
| DUOX1* | Dual oxidase 1 | 15q15.3 | 53905 | 0.160 | 0.065 | 0.645 | 0.031 |
| DUOX2* | Dual oxidase 2 | 15q15.3 | 50506 | 0.254 | 0.086 | 0.907 | 0.031 |
| EPHX2* | Epoxidehidrolase 2 | 8p21 | 2053 | 0.167 | 0.043 | 0.417 | 0.031 |
| GLRX2* | Glutaredoxin 2 | 1q31.2 | 51022 | 0.663 | 0.348 | 0.790 | 0.031 |
| GPX3* | Glutathione peroxidase 3 (plasma) | 5q33.1 | 2878 | 0.318 | 0.021 | 0.913 | 0.015 |
| GSR* | Glutathione-disulfide reductase | 8p21.1 | 2936 | 0.599 | 0.125 | 0.937 | 0.006 |
| GSTZ1* | Glutathiona S-transferase zeta 1 | 14q24.3 | 2954 | 0.693 | 0.500 | 0.921 | 0.031 |
| MGST3* | Microsomal glutathione S-transferase 3 | 1q23 | 4259 | 0.533 | 0.364 | 1.234 | 0.031 |
| OXR1* | Oxidation Resistance 1 | 8q23 | 55074 | 0.411 | 0.151 | 0.982 | 0.031 |
| OXSR1* | Oxidative-stress responsive 1 | 3p22.2 | 9943 | 0.643 | 0.206 | 0.942 | 0.015 |
| PRDX1 | Peroxiredoxin 1 | 1p34.1 | 5052 | 0.511 | 0.134 | 1.064 | 0.001 |
| PRDX4 | Peroxiredoxin 4 | Xp22.11 | 10549 | 1.610 | 1.169 | 3.385 | 0.031 |
| PRNP | Prion protein | 20p13 | 102460857 | 2.74 | 0.961 | 4.823 | 0.031 |
| SOD1* | Superoxide dismutase 1, soluble | 21q22.11 | 6647 | 0.528 | 0.226 | 0.977 | 0.008 |
| SOD2 | Superoxide dismutase 2, mitochondrial | 6q25.3 | 6648 | 1.79 | 0.571 | 3.151 | 0.022 |
| SOD3 | Superoxidedismutase 3, extracellular | 4p15.2 | 6649 | 0.178 | 0.055 | 0.948 | 0.031 |
Biomarkers not yet evaluated in oral squamous cell carcinoma. Fold change > 1 – overexpressed genes. Fold change < 1 – down-expressed genes.
Figure 1.
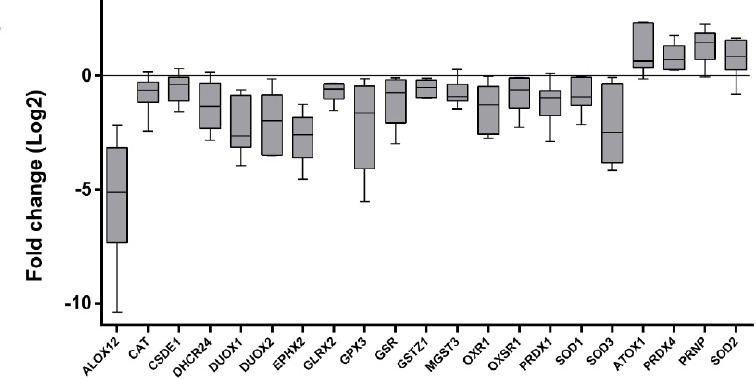
Relative Expression of Oxidative Stress-related Genes in OSSC. Fold Change were Log2 transformed (y-axis). Whiskers plot (min. to max.). Calibrator (non-tumor tissues) log fold change = 0. Overexpressed genes: ATOX1, PRDX4, PRPN, and SOD2 (fold change > 0). Down-expressed genes: ALOX12, CAT, CSDE1, DHCR24, DUOX1, DUOX2, EPHX2, GLRX2, GPX3, GSR, GSTZ1, MGST3, OXR1, OXSR1, PRDX1, SOD1, and SOD3 (fold change < 0).
Table 3.
Biological Mechanisms Related to the Differentially Expressed Genes
| Biological process | Differentially expressed genes |
|---|---|
| Chemical carcinogenesis | MGTS3 |
| Drug metabolism - Cytochrome P450 | MGTS3; GSTZ1 |
| Glutathione metabolism | GSR, MGST3, PRDX4, GPX3, GSTZ1 |
| Xenobiotic metabolism - Cytochrome P450 | MGST3; GSTZ1 |
| Arachidonic acid metabolism | EPHX2, GPX3, ALOX12 |
| Tyrosine metabolism | GSTZ1 |
| Thyroid hormone synthesis | GSR; DUOX1, DUOX2, GPX3 |
| FoxO signaling pathway | SOD2, CAT |
| Tryptophan metabolism | CAT, DHCR24 |
| Steroid biosynthesis | DHCR24 |
| Detoxification of reactive oxygen species | SOD3, SOD2, SOD1, ATOX1, PRDX1, CAT, OXR1 |
| Prion diseases | PRPN |
| Nucleotide metabolism | GLRX2 |
| Regulation of gene transcription | CSDE1 |
| Phosphorylation protein | OXSR1 |
The analysis of gene expression according to the tumors’ clinical parameters (tumor extension or tumor progression) is presented in the Tables 4 and 5. Only one patient presented nodal metastasis. Gene expression was not associated with tumor extent (P > 0.05). Regarding the tumor progression (TNM stage), only the PRDX1 gene was significantly down-expressed in advanced tumors (P = 0.036).
Table 4.
Down-Regulated Genes According to the Clinical Parameters of Oral Tumors
| Genes | Expression pattern in OSCCa | Tumor extent | P-value | TNM stage | P-value | ||
|---|---|---|---|---|---|---|---|
| T1-T2 | T3-T4 | I-II | III-IV | ||||
| Fold change (median)a | Fold change (median) | Fold change (median) | Fold change (median) | ||||
| ALOX12 | Down-regulated | 0.052 | 0.019 | 0.857 | 0.075 | 0.012 | 0.228 |
| CAT | Down-regulated | 0.568 | 0.733 | 0.486 | 0.651 | 0.629 | 1.0 |
| CSDE1 | Down-regulated | 0.793 | 0.646 | 0.486 | 0.798 | 0.542 | 0.143 |
| DHCR24 | Down-regulated | 0.688 | 0.269 | 0.229 | 0.796 | 0.234 | 0.057 |
| DUOX1 | Down-regulated | 0.158 | 0.389 | 0.533 | 0.186 | 0.135 | 1.0 |
| DUOX2 | Down-regulated | 0.164 | 0.358 | 0.533 | 0.241 | 0.269 | 1.0 |
| EPHX2 | Down-regulated | 0.113 | 0.327 | 0.133 | 0.131 | 0.236 | 0.700 |
| GLRX2 | Down-regulated | 0.599 | 0.723 | 0.800 | 0.665 | 0.662 | 1.0 |
| GPX3 | Down-regulated | 0.390 | 0.318 | 0.857 | 0.723 | 0.194 | 0.4 |
| GSR | Down-regulated | 0.599 | 0.626 | 0.800 | 0.691 | 0.453 | 0.393 |
| GSTZ1 | Down-regulated | 0.590 | 0.823 | 0.267 | 0.661 | 0.725 | 1.0 |
| MGST3 | Down-regulated | 0.635 | 0.534 | 0.629 | 0.773 | 0.498 | 0.229 |
| OXR1 | Down-regulated | 0.496 | 0.316 | 0.800 | 0.627 | 0.175 | 0.200 |
| OXSR1 | Down-regulated | 0.519 | 0.928 | 0.400 | 0.643 | 0.651 | 0.857 |
| PRDX1 | Down-regulated | 0.615 | 0.419 | 0.200 | 0.646 | 0.391 | 0.036* |
| SOD1 | Down-regulated | 0.529 | 0.706 | 0.886 | 0.556 | 0.466 | 0.571 |
| SOD3 | Down-regulated | 0.168 | 0.522 | 0.533 | 0.259 | 0.096 | 1.0 |
OSCC, oral squamous cell carcinoma;
, Compared to non-tumor tissue;
, Statistically significant
Table 5.
Up-Regulated Genes According to the Clinical Parameters of Oral Tumors
| Genes | Expression pattern in OSCCa | Tumor extent | P-value | TNM stage | P-value | ||
|---|---|---|---|---|---|---|---|
| T1-T2 | T3-T4 | I-II | III-IV | ||||
| Fold change (median)a | Fold change (median) | Fold change (median) | Fold change (median) | ||||
| ATOX1 | Up-regulated | 1.425 | 5.010 | 0.229 | 1.294 | 3.989 | 0.114 |
| PRDX4 | Up-regulated | 1.376 | 2.548 | 0.237 | 1.243 | 2.166 | 0.100 |
| PRNP | Up-regulated | 2.961 | 2.244 | 0.629 | 2.744 | 2.711 | 0.857 |
| SOD2 | Up-regulated | 1.663 | 2.344 | 0.886 | 1.806 | 1.790 | 1.0 |
OSCC, oral squamous cell carcinoma;
, Compared to non-tumor tissue.
Discussion
Our results showed twenty-one differentially expressed oxidative stress-related genes in OSCC. Fourteen of these genes were analyzed for the first time in OSCC, highlighting possible novel biomarkers for OSCC development. These genes play a role in the control of oxidative stress, mediating the neutralization of the deleterious effects of oxygen byproducts by removing free radical intermediates. Among the differentially expressed genes first investigated in our samples, GSR (Glutathione reductase), MGST3 (Microsomal glutathione S-transferase 3), GPX3 (Glutathione peroxidase 3), and GSTZ1 (Glutathiona S-transferase zeta 1) participate in Glutathione metabolism. The deregulation of these four genes involved in the same pathway could suggest an important role for this metabolism in oral carcinogenesis. GSTZ1 and MGST3 genes encode GST enzymes and were negatively regulated in this study. MGST3 genes encode a membrane-associated protein, which reduces the GSH and serves antioxidant functions (Yates et al., 2006). Reduced expression of MGST3, as observed in our study, can contribute to the inefficient detoxification of carcinogens and malignant cell transformation (Strid et al., 2008).
In our study, Superoxide dismutase 1 (SOD1) gene expression was reduced in oral tumors. Superoxide is an important ROS and plays a central role in oxidative stress. The SOD enzyme converts superoxides to hydrogen peroxide and oxygen, and it prevents the accumulation of ROS and altered proteins in the membranes of mitochondria, avoiding oxidative damage and cancer, so reduced amounts of this enzyme appear to be crucial in oral carcinogenesis. Low expression of SOD1 was also observed in gastric (Monari et al., 2006) and endometrial adenocarcinoma (Pejić et al., 2008). In epidermoid carcinoma cells, the inhibition of SOD1 resulted in both pro-oxidant and antioxidant effects caused by the increase of superoxide levels and the decrease of hydrogen peroxide levels (Juarez et al., 2008). The reduction of hydrogen peroxide levels resulted in the protection of protein tyrosine phosphatases from the effects of oxidation, leading to the inhibition of growth factor-mediated phosphorylation of ERK 1 and 2 (extracellular signal-regulated kinase 1 and 2). The ERK signaling pathway is important for the survival and proliferation of tumor cells (Mehdizadeh et al., 2016). However, the excess of superoxides is toxic and could contribute to tumor development. On the other hand, increased levels of SOD1 were observed in lung (Lee et al., 2010) and breast (Papa et al., 2014) adenocarcinoma cell lines, suggesting a differential activity of SOD1 in different tumor types.
The OXR1 (Oxidation Resistance 1) gene also plays an important role in protection against oxidative stress. In OXR1-deficient HeLa cells, the oxidative stress increases in response to hydrogen peroxide (Yang et al., 2014), contributing to cell damage and malignant transformation. As in our results, OXR1 gene expression was negatively regulated in breast tumor cells (Sirchia and Luparello, 2007).
The reduced expression of DUOX1 and DUOX2 (Dual oxidase 1 and 2) genes observed in our study suggests deficiency of the antioxidant system and the increase of superoxide levels, which consequently increased oxidative stress in the tumor microenvironment. DUOX1 and DUOX2 genes participate in thyroid hormone synthesis through the release of hydrogen peroxide (De Deken et al., 2000). Among the NADPH oxidases, only DUOX1 and DUOX2 catalyze the production of hydrogen peroxide instead of extracellular superoxide.
Corroborating our results, evidence indicates that DUOX1 genes can act as tumor suppressors in the initiation and progression of cancer (Ling et al., 2014). DUOX1 is frequently silenced by promoter hyper-methylation in lung cancer (Luxen et al., 2008). DUOX1 expression was also reduced in liver cancer and hepatocellular carcinoma (Ling et al., 2014). Furthermore, the high expression of DUOX1 was associated with a reduced risk of death in thyroid carcinoma (Pulcrano et al., 2007). In this context, the reduced expression of DUOX1 genes in OSCC corroborates its role in suppressing the increase of ROS and tumor growth related to the arrest of the G2/M cell cycle (Ling et al., 2014). Regarding the DUOX2 gene, although its elevated expression has been associated with stomach and colon tumors (Qi et al., 2016), its reduced expression in oral carcinoma can be related to the increase of ROS production and promotion of tumor growth.
GPX3, ALOX12 (Arachidonate 12-lipoxygenase, 12S type), and EPHX2 (Epoxide Hidrolase 2) genes participate in arachidonic acid metabolism and were down-regulated in our study. This metabolism is responsible for the formation of lipid mediators termed eicosanoids, which are associated with inflammation related to the carcinogenesis (Chen et al., 2004). ALOX12 enzymes metabolize arachidonic acid into 12(S)-HETE ((5Z,8Z,10E,14Z)-(12S)-12-Hydroxyicosa-5,8,10,14-tetraenoic acid), which is associated with tumor progression and metastasis. 12(S)-HETE acts in the modulation of integrin, regulates the secretion of proteinases, increases the motility and invasion of tumor cells, and modulates angiogenesis (Dilly et al., 2013). High expression of ALOX12 genes was detected in several tumor types, such as breast (Natarajan and Nadler, 1998), kidney (Yoshimura et al., 2004), and prostate cancer (Gao et al., 1995). Interestingly, ALOX12 expression was reduced in oral tumors in our study. This finding suggests that the function of ALOX12 may be non-essential for the oral tumorigenesis. Reduced expression of GPX3 suggests an increase of inflammatory response in oral tumors. GPX3 enzymes can prevent the activation of lipoxygenases like ALOX12, thereby avoiding excessive tissue inflammation (Coussens and Werb, 2002). Although oral tumors showed reduced expression of ALOX12 genes in our study, other enzymes regulated by GPX3 could act on oral cancer. Also down-regulated in oral tumor samples, EPHX2 genes encoded soluble epoxide hydrolase (sEH) and also presented reduced expression in liver (Zhang et al., 2010) and kidney (Grigo et al., 2008) tumors. However, the increase in EPHX2 expression was observed in other tumor types, such as ovarian (Enayetallah et al., 2006). In head and neck cells, EPHX2 was overexpressed in response to the administration of N’-nitrosonornicotine, a subproduct of tobacco metabolism (Khammanivong et al., 2016). However, there are insufficient data in the literature to clarify the role of EPHX2 in head and neck tumorigenesis.
The metabolism of amino acids plays an important role in carcinogenesis because they are precursors for protein synthesis. CAT and GSTZ1 enzymes participate in amino acid metabolism, and reduced gene expression can be associated with the alteration of cell-signaling pathways related to the cancer. Amino acids are necessary for mTOR (mechanistic target of rapamycin) signaling, which is functionally activated in cancer and responsible for the regulation of growth, proliferation, motility, survival, autophagy, and transcription (Hay and Sonenberg, 2004).
The DHCR24 (24-dehydrocholesterol reductase) gene, which was down-regulated in this study, participates in steroid biosynthesis (Mitsche et al., 2015). In contrast to the findings of this study, increased expression of the DHCR24 gene was observed in breast (Nagai et al., 2003) and prostate tumors (Nelson, 2002). Furthermore, high levels of DHCR24 were associated with tumor progression in melanomas and prostate and breast cancer, possibly due to the increase of cholesterol synthesis. Thus, the DHCR24 gene seemingly has no influence on oral carcinogenesis by cholesterol metabolism, but it does do by other mechanisms, such as oxidative stress control, cell apoptosis, or inflammation (Greeve et al., 2000; 64. Wu et al., 2013).
The GRLX2 gene is also involved in apoptosis and showed reduced expression in oral tumors in this study. The protein Grx2 is responsible for cellular homeostasis and deglutathionylation of protein-mixed disulfides with GSH (Allen and Mieyal, 2012). In cellular processes, Grx2 serves important processes, such as sulfur assimilation, DNA biosynthesis, apoptosis, redox regulation, intermediation of the activity of transcriptional ligation factors, cell differentiation, and defense against oxidative stress (Hashemy et al., 2007). Members of the Glutaredoxin family regulate through deglutathionylation the activity of several enzymes, such as Ras (Rat Sarcoma), Fas (TNF Receptor 6), ASK1 (Apoptosis Signal-Regulating Kinase 1), NF-κB, and Pro-Caspase-3, which play a role in the control of cell apoptosis (Allen and Mieyal, 2012). Deglutathionylation of Ras can result in activation of the MEKK (Mitogen-activated protein kinase/Extracellular signal regulated kinase kinase kinase) pathway, inhibition of Akt, and activation of apoptotic effector proteins (Allen and Mieyal, 2012). In this context, the reduction of GRLX2 expression could avoid oral tumor cell death, contributing to tumor development.
In this study, the CSDE1 gene, which encodes the protein Unr (upstream of N-ras), presented reduced expression in oral tumors. CSD (cold shock domain) are nucleic-acid-binding proteins that act in the regulation of gene transcription, protein translation, and cell proliferation. The expression of these proteins is induced by cellular stress. CSD proteins are constantly secreted and bind to specific receptors on a cell’s surface, influencing cell proliferation and migration (Lindquist et al., 2014). Unr protein expression can be regulated by auto regulation and inhibited by such genes as c-myc (Anderson and Catnaigh, 2015), an oncogene related to tumor cell proliferation. Although c-myc is up-regulated in OSCC, according to the literature (Pai et al., 2009), there are no consistent data on the role of Unr in the mechanism involved in head and neck cancer development.
The OXSR1 gene, meanwhile, was down-regulated in this study. OSR1 protein is responsible for the regulation of ion transportation and downstream signaling in response to environmental stress. OSR1 is activated in response to oxidative stress (Kozlov et al., 2016), and it is involved in angiogenesis by activation of WNK 1 (With no lysine (K)) protein (Dbouk et al., 2014). According to our knowledge, there are no data in the literature on gene and protein expression of OSR1 in tumor cells. However, the OXSR1 gene has presented hyper-methylation of the promoter region in lung squamous cell carcinoma (Rauch et al., 2012), leading to the inhibition of gene transcription. This finding could indicate inhibition of this gene by mechanisms of gene expression regulation that could influence tumor development. Furthermore, it was observed that OSR1 participates in molecular pathways associated with protection mechanisms. A study in human embryonic kidney cells showed that OSR1 is responsible for the phosphorylation of the tumor necrosis factor receptor RELT (Receptor expressed in lymphoid tissues) (Cusick et al., 2006). Tumor necrosis factors are involved in tumor cell death, proliferation, apoptosis, inflammation, and stress response (Locksley et al., 2001). Therefore, RELT activation mediated by OSR1 is an important mechanism for the apoptosis induction and inhibition of tumor development. Reduced expression of OXSR1 in OSCC corroborates its role in cellular protection and inhibition of the malignant transformation.
According to our knowledge, there are no data in the literature on SOD1, OXR1, MGTS3, GSTZ1, GSR, GPX3, DUOX1, DUOX2, ALOX12, EPHX2, DHCR24, GRLX2, CSDE1, and OXSR1 expression in OSCC. In this study, we performed a screening test to evaluate the expression profile of oxidative stress-related genes in OSCC. The identification of novel biomarkers for OSCC contributes to the thorough investigation of the role of these genes in a larger sample and in vitro analysis to elucidate the biological processes involved in oral carcinogenesis. Studies on the actuation of these genes in the biological processes involved in tumor development and progression are promising for drug and therapy discoveries for oral cancer treatment.
Acknowledgments
The authors are thankful for the grant #2013/049236, São Paulo Research Foundation (FAPESP), the Coordination for the Improvement of Higher Education Personnel (CAPES) and the collaboration of Hospital de Base / Sao Jose do Rio Preto Medical School Foundation (FUNFARME).
References
- 1.Allen EM, Mieyal JJ. Protein-thiol oxidation and cell death:regulatory role of glutaredoxins. Antioxid Redox Signal. 2012;17:1748–63. doi: 10.1089/ars.2012.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson EC, Catnaigh PÓ. Regulation of the expression and activity of Unr in mammalian cells. Biochem Soc Trans. 2015;43:1241–6. doi: 10.1042/BST20150165. [DOI] [PubMed] [Google Scholar]
- 3.Bentz B. Head and neck squamous cell carcinoma as a model of oxidative-stress and cancer. J Surg Oncol. 2007;96:190–1. doi: 10.1002/jso.20817. [DOI] [PubMed] [Google Scholar]
- 4.Buettner GR. Superoxide dismutase in redox biology:the roles of superoxide and hydrogen peroxide. Anticancer Agents Med Chem. 2011;11:341–6. doi: 10.2174/187152011795677544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Wang S, Wu N, Yang CS. Leukotriene A4 hydrolase as a target for cancer prevention and therapy. Curr Cancer Drug Targets. 2004;4:267–83. doi: 10.2174/1568009043333041. [DOI] [PubMed] [Google Scholar]
- 6.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cusick JK, Xu LG, Bin LH, Han KJ, Shu HB. Identification of RELT homologues that associate with RELT and are phosphorylated by OSR1. Biochem Biophys Res Commun. 2006;340:535–43. doi: 10.1016/j.bbrc.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 8.Dbouk HA, Weil LM, Perera GK, et al. Actions of the protein kinase WNK1 on endothelial cells are differentially mediated by its substrate kinases OSR1 and SPAK. Proc Natl Acad Sci U S A. 2014;111:15999–16004. doi: 10.1073/pnas.1419057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Deken X, Wang D, Many MC, et al. Cloning of two human thyroid cDNAs encoding new members of the NADPH oxidase family. J Biol Chem. 2000;275:23227–33. doi: 10.1074/jbc.M000916200. [DOI] [PubMed] [Google Scholar]
- 10.Dilly AK, Ekambaram P, Guo Y, et al. Platelet-type 12-lipoxygenase induces MMP9 expression and cellular invasion via activation of PI3K/Akt/NF-kappaB. Int J Cancer. 2013;133:1784–91. doi: 10.1002/ijc.28165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enayetallah AE, French RA, Grant DF. Distribution of soluble epoxide hydrolase, cytochrome P450 2C8, 2C9 and 2J2 in human malignant neoplasms. J Mol Histol. 2006;37:133–41. doi: 10.1007/s10735-006-9050-9. [DOI] [PubMed] [Google Scholar]
- 12.Gao X, Grignon DJ, Chbihi T, et al. Elevated 12-lipoxygenase mRNA expression correlates with advanced stage and poor differentiation of human prostate cancer. Urology. 1995;46:227–37. doi: 10.1016/s0090-4295(99)80198-8. [DOI] [PubMed] [Google Scholar]
- 13.Greeve I, Hermans-Borgmeyer I, Brellinger C, et al. The human DIMINUTO/DWARF1 homolog seladin-1 confers resistance to Alzheimer's disease-associated neurodegeneration and oxidative stress. J Neurosci. 2000;20:7345–52. doi: 10.1523/JNEUROSCI.20-19-07345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grigo K, Wirsing A, Lucas B, Klein-Hitpass L, Ryffel GU. HNF4 alpha orchestrates a set of 14 genes to down-regulate cell proliferation in kidney cells. Biol Chem. 2008;389:179–87. doi: 10.1515/BC.2008.011. [DOI] [PubMed] [Google Scholar]
- 15.Guengerich FP. Cytochrome P450 and chemical toxicology. Chem Res Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 16.Halliwell B. Biochemistry of oxidative stress. Biochem Soc Trans. 2007;35:1147–50. doi: 10.1042/BST0351147. [DOI] [PubMed] [Google Scholar]
- 17.Hashemy SI, Johansson C, Berndt C, Lillig CH, Holmgren A. Oxidation and S-nitrosylation of cysteines in human cytosolic and mitochondrial glutaredoxins:effects on structure and activity. J Biol Chem. 2007;282:14428–36. doi: 10.1074/jbc.M700927200. [DOI] [PubMed] [Google Scholar]
- 18.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–45. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 19.Juarez JC, Manuia M, Burnett ME, et al. Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc Natl Acad Sci U S A. 2008;105:7147–52. doi: 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khammanivong A, Anandharaj A, Qian X, et al. Transcriptome profiling in oral cavity and esophagus tissues from (S)-N'-nitrosonornicotine-treated rats reveals candidate genes involved in human oral cavity and esophageal carcinogenesis. Mol Carcinog. 2016;55:2168–82. doi: 10.1002/mc.22459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozlov SV, Waardenberg AJ, Engholm-Keller K, et al. Reactive oxygen species (ROS)-activated ATM-dependent phosphorylation of cytoplasmic substrates identified by large-scale phosphoproteomics screen. Mol Cell Proteomics. 2016;15:1032–47. doi: 10.1074/mcp.M115.055723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SY, Jeon HM, Kim CH, et al. CuZnSOD and MnSOD inhibit metabolic stress-induced necrosis and multicellular tumour spheroid growth. Int J Oncol. 2010;37:195–202. doi: 10.3892/ijo_00000667. [DOI] [PubMed] [Google Scholar]
- 23.Lindquist JA, Brandt S, Bernhardt A, Zhu C, Mertens PR. The role of cold shock domain proteins in inflammatory diseases. Mol Med (Berl) 2014;92:207–16. doi: 10.1007/s00109-014-1136-3. [DOI] [PubMed] [Google Scholar]
- 24.Ling Q, Shi W, Huang C, et al. Epigenetic silencing of dual oxidase 1 by promoter hypermethylation in human hepatocellular carcinoma. Am J Cancer Res. 2014;4:508–17. [PMC free article] [PubMed] [Google Scholar]
- 25.Livak K, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-??Ct method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies:integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 27.Luxen S, Belinsky SA, Knaus UG. Silencing of DUOX NADPH oxidases by promoter hypermethylation in lung cancer. Cancer Res. 2008;68:1037–45. doi: 10.1158/0008-5472.CAN-07-5782. [DOI] [PubMed] [Google Scholar]
- 28.Masood N, Malik FA, Kayani MA. Expression of xenobiotic metabolizing genes in head and neck cancer tissues. Asian Pac J Cancer Prev. 2011;12:377–82. [PubMed] [Google Scholar]
- 29.Matsui A, Ikeda T, Enomoto K, et al. Increased formation of oxidative DNA damage, 8-hydroxy-2'-deoxyguanosine, in human breast cancer tissue and its relationship to GSTP1 and COMT genotypes. Cancer Lett. 2000;151:87–95. doi: 10.1016/s0304-3835(99)00424-3. [DOI] [PubMed] [Google Scholar]
- 30.Mehdizadeh A, Somi MH, Darabi M, Jabbarpour-Bonyadi M. Extracellular signal-regulated kinase 1 and 2 in cancer therapy:a focus on hepatocellular carcinoma. Mol Biol Rep. 2016;43:107–16. doi: 10.1007/s11033-016-3943-9. [DOI] [PubMed] [Google Scholar]
- 31.Mitsche MA, McDonald JG, Hobbs HH, Cohen JC. Flux analysis of cholesterol biosynthesis in vivo reveals multiple tissue and cell-type specific pathways. Elife. 2015;4:e07999. doi: 10.7554/eLife.07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monari M, Trinchero A, Calabrese C, et al. Superoxide dismutase in gastric adenocarcinoma:is it a clinical biomarker in the development of cancer? Biomarkers. 2006;11:574–84. doi: 10.1080/13547500600899134. [DOI] [PubMed] [Google Scholar]
- 33.Nagai MA, Ros N, Bessa SA, et al. Differentially expressed genes and estrogen receptor status in breast cancer. Int J Oncol. 2003;23:1425–30. [PubMed] [Google Scholar]
- 34.Natarajan R1, Nadler J. Role of lipoxygenases in breast cancer. Front Biosci. 1998;3:81–8. doi: 10.2741/a369. [DOI] [PubMed] [Google Scholar]
- 35.Nelson PS. Identifying immunotherapeutic targets for prostate carcinoma through the analysis of gene expression profiles. Ann N Y Acad Sci. 2002;975:232–46. doi: 10.1111/j.1749-6632.2002.tb05955.x. [DOI] [PubMed] [Google Scholar]
- 36.Pai R, Pai S, Lalitha R, et al. Over-expression of c-Myc oncoprotein in oral squamous cell carcinoma in the South Indian population. Ecancermedicalscience. 2009;3:128. doi: 10.3332/ecancer.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papa L, Manfredi G, Germain D. SOD1, an unexpected novel target for cancer therapy. Genes Cancer. 2014;5:15–21. doi: 10.18632/genesandcancer.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pejić S, Todorović A, Stojiljković V, et al. Superoxide dismutase and lipid hydroperoxides in blood and endometrial tissue of patients with benign, hyperplastic and malignant endometrium. An Acad Bras Cienc. 2008;80:515–22. doi: 10.1590/s0001-37652008000300011. [DOI] [PubMed] [Google Scholar]
- 39.Pulcrano M, Boukheris H, Talbot M, et al. Poorly differentiated follicular thyroid carcinoma:Prognostic factors and relevance of histological classification. Thyroid. 2007;17:639–46. doi: 10.1089/thy.2007.0029. [DOI] [PubMed] [Google Scholar]
- 40.Qi R, Zhou Y, Li X, et al. DUOX2 Expression Is Increased in Barrett Esophagus and Cancerous Tissues of Stomach and Colon. Gastroenterol Res Pract. 2016;2016:1835684. doi: 10.1155/2016/1835684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasheed MH, Beevi SS, Rajaraman R, Bose SJ. Alleviation of oxidative and nitrosative stress following curative resection in patient with oral cavity cancer. J Surg Oncol. 2007;96:194–9. doi: 10.1002/jso.20818. [DOI] [PubMed] [Google Scholar]
- 42.Rauch TA, Wang Z, Wu X, et al. DNA methylation biomarkers for lung cancer. Tumour Biol. 2012;33:287–96. doi: 10.1007/s13277-011-0282-2. [DOI] [PubMed] [Google Scholar]
- 43.Sirchia R, Luparello C. Mid-region parathyroid hormone-related protein (PTHrP) and gene expression of MDA-MB231 breast cancer cells. Biol Chem. 2007;388:457–65. doi: 10.1515/BC.2007.059. [DOI] [PubMed] [Google Scholar]
- 44.Sobin LH, Wittekind CH. Head and neck tumors. In: Sobin LH, Gospodarowicz MK, Wittekind Ch, editors. “TNM classification of malignant tumors”. Berlin: Springer Verlag; 2010. pp. 22–45. [Google Scholar]
- 45.Strid T, Söderström M, Hammarström S. Leukotriene C4 synthase promoter driven expression of GFP reveals cell specificity. Biochem Biophys Res Commun. 2008;366:80–5. doi: 10.1016/j.bbrc.2007.11.097. [DOI] [PubMed] [Google Scholar]
- 46.Wu BJ, Chen K, Shrestha S, et al. High-density lipoproteins inhibit vascular endothelial inflammation by increasing 3β-hydroxysteroid-Δ24 reductase expression and inducing heme oxygenase-1. Circ Res. 2013;112:278–88. doi: 10.1161/CIRCRESAHA.111.300104. [DOI] [PubMed] [Google Scholar]
- 47.Yang M, Luna L, Sørbø JG, et al. Human OXR1 maintains mitochondrial DNA integrity and counteracts hydrogen peroxide -induced oxidative stress by regulating antioxidant pathways involving p21. Free Radic Biol Med. 2014;77:41–8. doi: 10.1016/j.freeradbiomed.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Yates MS, Kwak MK, Egner PA, et al. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 2006;66:2488–94. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 49.Yoshimura R, Inoue K, Kawahito Y, et al. Expression of 12-lipoxygenase in human renal cell carcinoma and growth prevention by its inhibitor. Int J Mol Med. 2004;13:41–6. [PubMed] [Google Scholar]
- 50.Young IS, Woodside JV. Antioxidants in health and disease. J Clin Pathol. 2001;54:176–86. doi: 10.1136/jcp.54.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zangar RC, Davydov DR, Verma S. Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicol Appl Pharmacol. 2004;199:316–31. doi: 10.1016/j.taap.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 52.Zhang D, Ai D, Tanaka H, Hammock BD, Zhu Y. DNA methylation of the promoter of soluble epoxide hydrolase silences its expression by an SP-1-dependent mechanism. Biochim Biophys Acta. 2010;1799:659–67. doi: 10.1016/j.bbagrm.2010.09.006. [DOI] [PubMed] [Google Scholar]


