Abstract
The human cytoplasmic protein tyrosine phosphatase (HCPTP) has been identified as a potential target for inhibition in order to downregulate metastatic transformation in several human epithelial cancers such as breast, prostate and colon cancer. Docking with two scoring functions on both isoforms of HCPTP was employed as an initial virtual screen to identify potential inhibitors. Compounds identified as potential inhibitors via this in silico screen were subjected to kinetic analysis in order to validate their selection as improved inhibitors. Eleven compounds with IC50’s of less than 100 μM were identified in a single concentration screen. Five of these compounds were determined to have IC50 of less than 10 μM; however, all but one of these compounds inhibited via non-specific aggregation. The validated effective inhibitor, which is based on a naphthyl sulfonic acid, strongly resembles a previously synthesized rationally designed azaindole phosphonic acid. This similarity suggests subsequent inhibitor optimization based on this scaffold may generate effective inhibitors of HCPTP. The structural elements of the computationally identified inhibitors are discussed to analyze the combined use of rationally design and virtual screening to reduce false negatives in the identification of multiple strong inhibitors of HCPTP.
Keywords: HCPTP, Human Cytoplasmic Protein Tyrosine Phosphatase, computational screening, rational design
1) INTRODUCTION
In many cell signaling systems, increased phosphorylation is correlated with an increased transformation phenotype in cells. It follows that kinases have been regarded as tumor activators while phosphatases function as tumor suppressors, and most cancer therapeutic discovery has therefore focused on kinase inhibitors. The current cancer drugs Dasatinib and Iressa target Src/Abl kinase and the epidermal growth factor receptor, respectively. 1–2 However, decreased phosphorylation is also capable of acting as a signal leading to increased invasiveness in cell cultures. The receptor tyrosine kinase EphA2 has been identified as a cancer therapeutic target and specifically has been shown to be hypophosphorylated in several different types of breast cancer. 3–5 Under-phosphorylation of EphA2 has been correlated with the degree of invasivity, the distribution of EphA2, and EphA2 turnover. 5 The increased invasivity is thought to occur by disruption of the cell-cell interactions in the hypophosphorylated state. Another phosphatase, PTP1B, has been shown to be involved in breast cancer progression pathways through a similar tyrosine kinase receptor. 6 The human cytoplasmic protein tyrosine phosphatase (HCPTP) is a known interaction partner for EphA2 and is found overexpressed in cells that have hypophosphorylated EphA2. 7 Application of the non-specific phosphatase inhibitor H2O2 is known to return test cells to a normal phenotype, and it is expected that a more targeted inhibitor of HCPTP will counteract the metastatic phenotype. 7–8 This suggests that the inhibition of the HCPTP may also be a viable method for future cancer therapeutics that could control cancer cell invasivity and thus metastasis. Analysis of this system has prompted several recent research studies whose goal is to develop inhibitors of HCPTP.9–11
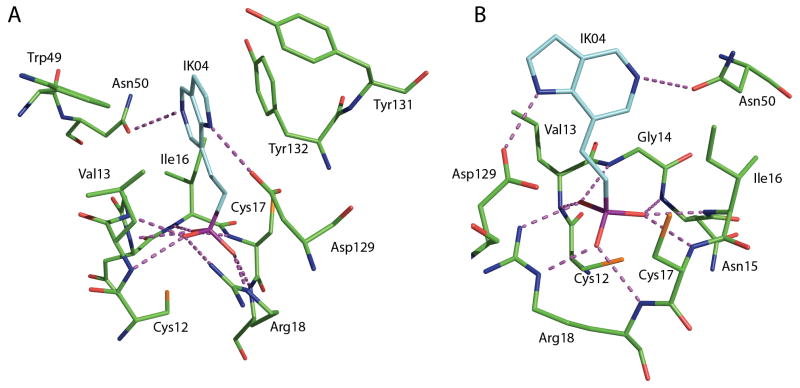
The development of inhibitors can proceed from a structural perspective with the availability of x-ray crystallographic structures of both human isoforms, HCPTP-A and HCPTP-B. 12–13 The structures of the two HCPTP isoforms, which are formed by differential RNA splicing resulting in changes between residues 40 and 73, have revealed a highly conserved overall fold with an average Cα RMSD of 0.79 Å. These structures contain elements unique to this family of phosphatases that may enable the generation of inhibitors that are specific for HCPTP or even its individual isoforms. 13 Previous studies on lead compound identification for inhibitors of HCPTP were based on the observed arrangement of inorganic phosphate, water and adenine present in the active site of the yeast low molecular phosphatase, which is highly homologous to HCPTP-A. 14–16 A rationally designed 5-azaindole IK04 (Figure 1) made specific use of three interactions believed to be critical in binding for both isoforms of HCPTP. First, an interaction between the charged phosphonate and the P-loop provides the opportunity for six hydrogen bonds. Next, the nitrogen in the five-membered ring of the azaindole is predicted to interact with the catalytically important aspartic acid 129, which provides both an additional opportunity for hydrogen bonding as well as a conserved orientation in the active site. Finally, the active site pocket is filled by the azaindole ring system, which takes advantage of interactions with both the hydrophobic residues in the P-loop as well as the aromatic residues that surround the opening to the active site. After experimental kinetic analysis, this synthetic azaindole was found to be a better inhibitor than any of the previously identified compounds.
Figure 1.
A) Rationally designed azaindole inhibitor (IK04) shown in its predicted binding orientation to HCPTP-B. Panels A and B illustrate the three motifs common among inhibitors, which were inherent in the design of the rationally designed azaindole: hydrogen binding to the phosphate binding loop, hydrogen bonding with Asp 129, and hydrophobic interaction with the cavity of the active site and the aromatic residues that line the entrance to the active site. The hydrogen bonding opportunities (<3.5 Å) for compounds to bind in the active site - amine backbone of the phosphate binding loop (residues 13–18), the side chain of Arg 18, the side chain of Asp 129 and the side chain of Asn 50 - are shown as purple dashed lines. B) A 90° rotation of the same predicted binding orientation of IK04 in HCPTP-B.
One method for the identification of inhibitors of phosphatases is to employ a fragment based screening methodology. 17 However, this approach requires significant synthetic organic chemistry support that may not be available to all projects of interest. An alternative and very popular approach to identify novel inhibitors is the use of in silico docking to evaluate the relative binding strength of a set of compounds. 18 Computational docking is frequently extended to rank entire libraries of small molecules to identify several potential compounds. Since significant optimization of the rationally designed compound is still required to obtain a potent, specific inhibitor for the HCPTP’s, we decided to use in silico screening independently and then compare the resulting structures and activities to the rational design concepts.
In order to rapidly and inexpensively explore alternative lead compound motifs, the in silico screening of publically available small molecule libraries has been performed as the next step of HCPTP inhibitor development. For the purposes of this investigation the Diversity Set, a part of the Developmental Therapeutics Program of the National Cancer Institute, was selected for study because of its chemical diversity and accessible compounds. Two docking programs, AutoDock and Glide, were utilized to determine their relative effectiveness for use with HCPTP. Compounds identified in the in silico screening were obtained from NCI and subjected to in vitro analysis. Predicted binding orientations of effective inhibitors were examined in detail to establish pharmacophore contacts that maximize the chemical diversity of functional groups that the active site of HCPTP can accommodate and to compare the structural features to the ones obtained from the rational design. 11, 16 The combined information can then be incorporated into future iterations of scaffold design and inhibitor optimization.
2) RESULTS
This investigation utilized an initial in silico screen of the National Cancer Institute’s Diversity Set to increase the speed of lead compound identification through the generation of an enriched subset of inhibitors that could then be analyzed in vitro. The 52 compounds that were selected for detailed in vitro analysis were identified in the in silico screening by selecting 27 compounds with the lowest predicted binding energy from each of the AutoDock and Glide studies, which is approximately 1.5% of the library. These two programs yielded nearly unique 27-member compound lists; only 2 compounds were indicated by both programs. Thirteen of the 52 compounds were insoluble, thus 39 compounds were analyzed by an in vitro phosphatase assay. All of the 39 compounds that were analyzed are shown in the Supplementary Information along with their predicted binding energies. Interestingly, most of the selected compounds contained many of the previously discussed structural elements from the rational inhibitor design.16
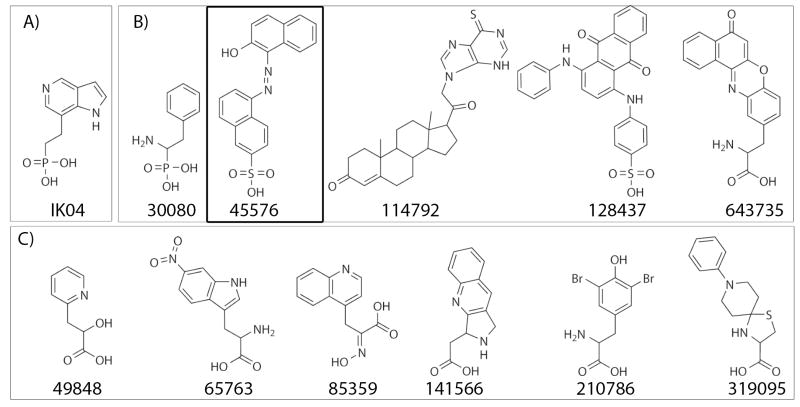
Each of the selected compounds was analyzed as an inhibitor in vitro at a single concentration of 100 μM in order to identify compounds that have IC50 values of at least 100 μM. Assays were conducted at pH 5 for both HCPTP-A and HCPTP-B as well as at pH 7 for HCPTP-A. The percent changes in activity when compared to assays containing no inhibitor are shown in Figure 2 for each enzyme. There were 11 compounds identified as inhibiting both of the HCPTP isoforms by at least 10% at pH 5: 27895, 29870, 30080, 45576, 85359, 114792, 128437, 133811, 141566, 320308, and 643735. Furthermore, 4 compounds elevated the activity of HCPTP by at least 10% at pH 5: 65763, 109816, 143099, and 210786 (for structures, see Supporting Information). Taking into account both the activators and inhibitors of HCPTP, more than 38% of the computationally identified compounds had at least a 10% effect on the activity of HCPTP at pH 5. Five compounds had IC50 values less than or equal to 100 μM, which is approximately 12% of the enzymatically screened compounds. The structures of these strong inhibitors (30080, 45576, 114792, 128437, and 643735) are shown in Figure 3. These compounds were further analyzed at several concentrations in order to determine an accurate IC50 for each compound against each of the enzymes. The determined IC50 are summarized in Table I.
Figure 2.
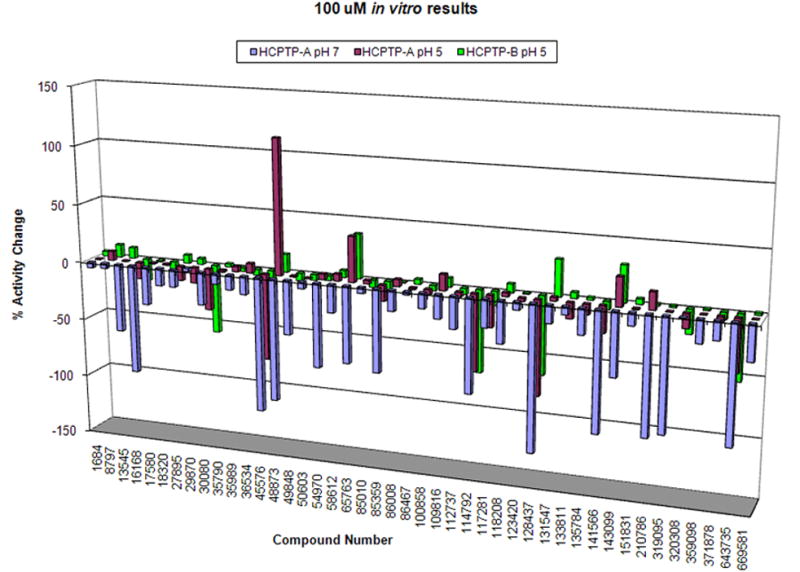
Percent changes of activity of HCPTP-A at pH 7 (blue) and HCPTP-A at pH 5 (magenta) and HCPTP-B at pH 5 (green) in the presence of 100 μM of each of the potential inhibitors. At pH 5 compounds 27895, 29870, 30080, 45576, 85359, 114792, 128437, 133811, 141566, 320308, and 643735 inhibit one isoform by at least 10%. At pH 7, 15 compounds reduced the activity of HCPTP-A by at least 50%: 13545, 16168, 45576, 48873, 49848, 54970, 65763, 85359, 114792, 128437, 141566, 143099, 210786, 319095, and 643735.
Figure 3.
A) Structure of the rationally designed azaindole (IK04). B) The 5 inhibitors of both HCPTP isoforms identified as having IC50 values less than 100 μM at both pH 5 and 7: 30080, 45576, 114792, 128437, and 643735. C) The structures of the additional 6 active inhibitors of HCPTP-A at pH 7: 49848, 65763, 85359, 141566, 210786, and 319095. Compound 45576, which has been determined not to cause aggregation of HCPTP, is boxed for additional emphasis.
Table I.
Experimentally determined IC50 values of compounds for HCPTP-A and HCPTP-B
| Compound | IC50 HCPTP-A (μM) | IC50 HCPTP-B (μM) |
|---|---|---|
| 128437 | 5.5 | 4.5 |
| 45576 | 5.5 | 3.9 |
| 643735 | 108 | 31 |
| 114792 | 93 | 135 |
| 30080 | 223 | 139 |
After each of the compounds was evaluated experimentally at pH 5, it was found that AutoDock predicted 4 compounds that reduced the activity of at least one isoform of HCPTP by 10% and 3 compounds that reduced the activity of at least one isoform of HCPTP by 50%, whereas Glide predicted 9 and 3 compounds which reduced the activity of at least one isoform of HCPTP by 10% and 50%, respectively. This is a 15% inhibitor identification success rate for AutoDock and 33% inhibitor identification success rate for Glide at the 10% inhibition level. The disparity between the two programs is most likely due to the relative insolubility of many of the compounds identified by AutoDock. If only soluble compounds are considered for the AutoDock evaluation, 29% (4 of 14) of the compounds showed 10% inhibition and three of these inhibitors showed 50% inhibition at 100 μM. Each program had a success rate of approximately 10%, by identification of 3 inhibitors which reduced activity by 50%.
The kinetic assays at the physiologically relevant pH 7 showed substantial increases in the number of effective inhibitors for HCPTP-A. As shown in Figure 2, a total of 29 compounds were capable of inhibiting the activity of HCPTP-A at pH 7 by at least 10%: 17580, 18320, 27895, 30080, 35989, 36534, 45576, 49848, 58612, 65763, 85359, 86008, 100858, 109816, 112737, 114792, 117281, 128437, 131547, 135784, 141566, 143099, 151831, 210786, 319095, 359098, 371878, 643735, and 669581. Figure 3 also shows the 6 additional compounds that were not strong inhibitors at pH 5, but that reduced the activity of HCPTP-A by at least 50% at pH 7. This represents a success rate of 74% for identification of inhibitors at the 10% level and a 28% success rate for the identification of inhibitors at the 50% level. AutoDock identified 11 compounds while Glide identified 20 compounds capable of inhibiting HCPTP-A by 10% at 100 μM. Glide was also about twice as effective at identifying strong inhibitors of HCPTP-A by identifying 7 compounds whereas AutoDock only identified 4. The calculated IC50 for all of the identified inhibitors of HCPTP-A are listed in Table II. The structures for all the computationally identified compounds are given in the supplemental information.
Table II.
Experimentally determined IC50 values of compounds for HCPTP-A at pH 5 and 7
| Compound | IC50 HCPTP-A pH 7 (μM) | IC50 HCPTP-A pH 5 (μM) |
|---|---|---|
| 128437 | 3.6 | 5.5 |
| 319095 | 4.0 | - |
| 210786 | 4.6 | - |
| 141566 | 4.8 | - |
| 45576 | 8.1 | 5.5 |
| 643735 | 13 | 108 |
| 114792 | 35 | 93 |
| 65763 | 51 | - |
| 85359 | 72 | - |
| 49848 | 88 | - |
| 30080 | - | 223 |
A dash denotes a compound that did not have an IC50 less than 100 μM at the indicated pH and thus did not meet the screening criteria.
Previous investigations have reported that high throughput screening for phosphatase inhibitors frequently is hindered by the identification of compounds that function as non-specific aggregators. 19 To further validate the candidacy of identified small molecule inhibitors as potential models for future inhibitor development efforts, an additional evaluation step was performed on the 11 inhibitors. These inhibitors were reevaluated in the single concentration IC50 screen in the presence of 0.01% Triton X-100, which is a detergent that is added to disrupt the formation of aggregates. 20 Only compounds 45576, 128437, and 319095 maintained an approximately constant level of inhibition in the presence of detergent and reducing agents. These three compounds were further analyzed via dynamic light scattering (DLS) experiments to investigate whether the compound addition resulted in aggregated protein even in the presence of detergent. Only compound 45576 did not lead to the formation of large aggregates, and so remains a candidate for a novel scaffold for HCPTP inhibitors. DLS data for HCPTP-A in the presence of each of these compounds are available in the supplemental information.
3) DISCUSSION
After computational docking of the 1990 compound NCI Diversity Set by AutoDock and Glide with both HCPTP-A and HCPTP-B isoforms, 39 compounds with high binding scores and good poses were subjected to enzymatic screening. Both activators and inhibitors were identified in the 100 μM screen as seen in Figure 2. The identification of activators of HCPTP may seem like an unexpected result. However, it must be kept in mind that the docking programs only predict binding energies, but not the effect that binding might have on the activity. The strength of binding of a molecule in the active site is expected to correlate with its activity as a competitive inhibitor for HCPTP, but this is not necessarily the case. One explanation for the activators may be that the compound could increase the rate of hydrolysis of the phosphoenzyme intermediate, the rate limiting step of the dephosphorylation process. This is a known mechanism for activation of low molecular weight phosphatases by adenine.14 An alternative explanation is that while a compound may be capable of binding to the active site, there may be a preferential binding site at some secondary location. The effect of binding at a secondary site could increase the affinity of HCPTP for the substrate by an allosteric effect. Although it is clear that experimental evaluations are necessary to characterize the practical inhibitors, the computational docking and screening studies on HCPTP have independently identified a common set of pharmacophores for this particular phosphatase. Three motifs are commonly observed: one or more cyclic systems, an anionic charged group, and an amino group. Each of these structural elements was expected as they resemble the native phosphorylated tyrosine ligand and were utilized in the rational design process, as depicted in Figure 1.
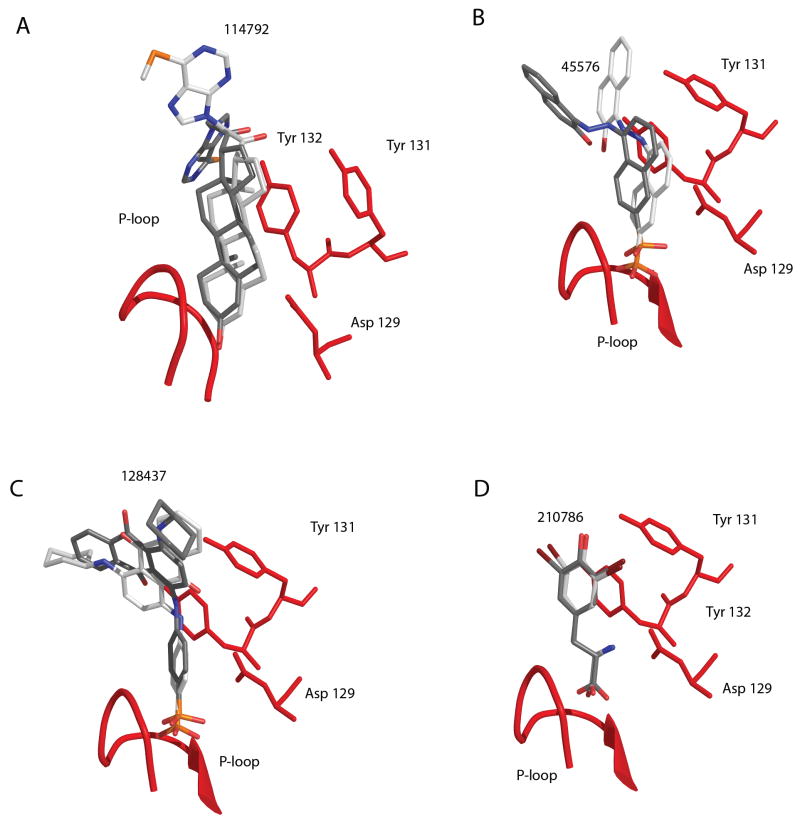
Nearly all of the compounds (38 of 39 compounds) are predicted to place a ring or ring system in the active site. Several ring moieties are present in the inhibitors: isolated rings, two fused rings and larger rings systems. There are 16 inhibitors that insert single rings into the active site, most of these being simple phenyl rings. Eight compounds contain fused ring systems, which are either naphthyl or purine-like rings, also inserted in the active site. The presence of the larger rings and specifically the purine-like rings predicted to bind in the active site support the rationale behind the design of the previously reported azaindole IK04. 16 Surprisingly, there were six steroid-like ring systems, including 114792 (experimental IC50’s of approximately 100 μM for both isoforms at pH 5) which also were predicted to bind in the active site (Figure 4A). This result both illustrates the strong preference for extensive hydrophobic interactions between potential inhibitors and HCPTP as well as suggesting that significantly larger ring systems may be useful in further rounds of inhibitor design.
Figure 4.
Predicted poses of compounds bound to HCPTP. The P-loop of HCPTP is shown as a red ribbon. Additional residues directly implicated in binding are explicitly shown: Asp 129, Tyr 131, and Tyr 132. A) Docked poses of compound 114792 from AutoDock with HCPTP-A in grey and HCPTP-B in white. B) Docked poses of compound 45576 from AutoDock with the prediction for HCPTP-A shown in white and HCPTP-B shown in grey. C) Docked poses of compound 128437 for HCPTP-B with the prediction from AutoDock shown in white and Glide shown in grey. D) Docked poses of compound 210786 from Glide with the prediction for HCPTP-A shown in grey and HCPTP-B shown in white.
Nearly three quarters of the compounds identified in the virtual screening (29 of 39 compounds) were predicted to interact with the P-loop via a charged group in order to form multiple hydrogen bonds between oxygen atoms on the inhibitor and the backbone amines and/or the nitrogens of Arg 18. Seventeen compounds are predicted to utilize carboxylic acid groups to interact with the P-loop. These compounds are unable to make all of the hydrogen bonds available to phosphate or sulfate groups bound to the P-loop as is observed in several crystal structures of low molecular weight phosphatase complexes. 12–15, 21 The carboxylic acid group inhibitors preserved two of the positions of oxygen atoms present in the buffer molecule, phosphate, or pNPP observed in the crystal structures, however two hydrogen bonding opportunities are lost. Phosphate/phosphonate and sulfonate containing compounds, which could be expected to utilize all of the hydrogen bonding capacity of the P-loop, are present in 7 and 5 of the predicted inhibitors, respectively.
The final commonly observed motif is the presence of an amine situated between the charged group and the ring system, positioned to hydrogen bond with Asp 129. Approximately 30% of the compounds identified as potential inhibitors of either isoform of HCPTP have this amine group and half of these were amine substituted carboxylic acids. A likely explanation for this observation is that the additional binding energy supplied by the hydrogen bond of the amine helps to offset the lost hydrogen bonding opportunities available for the carboxylic acid derivatives.
The hydrophobic ring systems, charged P-loop interactions, and hydrogen bonding opportunities for Asp 129 comprise a set of pharmacophores which were also identified as original rational design elements. 11, 16 Thus, the in silico screening confirms that the hypotheses on which the initial structure-based inhibitor design efforts were centered were correct. However, the number of possible structural implementations of the three binding motifs and the even larger number of permutations for interactions that are not directly in the active side are too large for rational design followed by directed synthesis of a small number of compounds. Instead, the many structural elements that best represent the pharmacophore model that are the basis for the rational design are best explored through virtual screening of lead compounds that is followed by experimental determination of the IC50’s. It is very encouraging to note that both methods, rational design and virtual screening, have led to an identical set of structural elements necessary for an optimized small molecule scaffold. The potential utility of this virtual screening approach is demonstrated for the identification a HCPTP inhibitor from a deliberately small virtual library, which covered a large chemical space. The identification of a novel low micromolar inhibitor for HCPTP that is a naphthyl sulfonic acid strongly validates the rational design approach, which utilized a azaindole phosphonic acid, and underline the importance of these structural elements for the development of inhibitors of HCPTP.
The chemical structures for the rationally designed azaindole IK04 and the inhibitors experimentally determined to have IC50 values less than 100 μM are shown in Figure 3. These compounds were initially selected as the among the best known inhibitors reported to date due to their ability to inhibit both isoforms at each pH tested. The predicted binding conformations for several of the strongest inhibitory molecules are illustrated in Figure 4. Each compound places hydrophobic rings in the active site: compound 128437 accomplishes this with a single phenyl ring, compound 45576 utilizes a naphthyl ring, compound 114792 places either a steroid-like or purine-like ring in the active site, and compound 210786 is unique among identified inhibitors with a 2,6-dibromophenol group in the active site. Two compounds employ sulfonate or phosphonate groups to hydrogen bond with the P-loop: 45576 and 128437. Compound 210786 hydrogen bonds with the P-loop via a carboxylic acid moiety. Interestingly, compound 114792 is predicted to utilize hydrophobic interactions to contact the P-loop residues rather than the commonly observed hydrogen bonding pattern with the P-loop backbone. This computational study has identified common binding elements that characterize the interactions with HCPTP’s active site, which are exemplified by the validated inhibition by compound 45576.
Each of these strong inhibitors occupies a highly conserved planar orientation in the active site, which is dominated by the interaction between the rings of the compound and the hydrophobic residues that line the sides of the active site (Val 13 and Ile16) as well as the aromatic residues at the mouth of the active site (Tyr/Trp 49, Tyr131, and Tyr 132). The differences in the predicted binding orientations of compounds between HCPTP-A and HCPTP-B may be best explained by the different amino acids at residues 49 and 50, Tyr 49 and Glu 50 versus Trp 49 and Asn 50 in HCPTP-A and HCPTP-B, respectively. These alternate conformations predicted for compounds usually involve a rotation of the distal portion of the inhibitor, although rotations of the entire inhibitor by 180 degrees have also been observed. This rotation preserves the interactions of the central rings with the active site while positioning the terminal ring(s) in opposite orientations for exiting the active site. This alternative positioning is seen in AutoDock’s predictions for compounds 114792 and 45576, shown in Figures 4A and B respectively. Predictions from the two different docking programs at times also show this difference in orientation in a single isoform, as seen for compound 128437 by both AutoDock and Glide in Figure 4C. Figure 4 illustrates that the bases of the inhibitors that interact with the P-loop are nearly superimposable in the predictions made by both programs on both isoforms.
Deviations of oxygen atoms away from the optimal P-loop interaction pattern are found in compounds without a group capable of interacting with the catalytic Asp 129. Compounds 45576 and 128437 (Figure 4B and C), which do not have a group readily available to interact with Asp 129, position a sulfonate oxygen close enough to hydrogen bond with Asp129 rather than interacting with the residues of the P-loop. Compound 210786 shown in Figure 4D, which possesses an amine to interact with Asp 129, does not show this alternative positioning of oxygen atoms. In fact, all of the compounds with an available amine group positioned to contact Asp 129 show this highly conserved hydrogen bond, as demonstrated by compound 210786 for both HCPTP-A and HCPTP-B.
The compounds that only inhibit at pH 7 also possess these design elements, but have a greatly increased chemical diversity. These compounds are obviously enriched in carboxylic acid derivatives rather than sulfonates or phosphonates. Unfortunately, upon subsequent analysis these compounds were shown to be oxidizing agents and/or aggregators of HCPTP. Thus, the roles played by these substituents must be explored further in additional studies.
One puzzling result from the initial screening is the presence of only a single highly effective phosphonate-containing inhibitor (30080) for HCPTP, which was the weakest of the group of strong inhibitors. There is a relatively small percentage of phosphate or phosphonate compounds in the Diversity Set, of which seven were selected as potential inhibitors. The other two phosphate-containing compounds failed as effective competitive inhibitors, probably because they are substrates. The other phosphonate compounds may not be fully deprotonated under our assay conditions, particularly at low pH, and therefore lose their effective binding to the P-loop. Compound 45576, which contains a sulfonate, was shown to be an effective inhibitor through all of the screening evaluations. The elimination of so many false leads further illustrates the need for multiple rounds of experimental screening. Nonetheless, the use of multiple computational screening studies as an initial screen for small molecule inhibitors of low molecular weight phosphatases has produced a novel naphthyl derivative for future inhibitor optimization.
4) CONCLUSIONS
In silico screening of the NCI Diversity Set was conducted by the docking programs AutoDock and Glide for both HCPTP-A and HCPTP-B. Compounds that were identified as potential inhibitors of at least one isoform of HCPTP were analyzed kinetically. After this in vitro kinetic analysis, five compounds were identified to have IC50 on the order of or less than 100 μM in a single concentration screen at pH 5 with an additional six compounds identified as inhibitors of HCPTP-A at pH 7. These compounds were further analyzed in order to determine IC50 values. Both AutoDock and Glide individually identified several sub-100 μM inhibitors of HCPTP. These newly identified compounds appeared to be the most potent inhibitors reported to date, with IC50 lower than the general phosphatase inhibitor orthovanadate (11 μM and 61 μM for HCPTP-A and HCPTP-B, respectively). Enzymatic analysis at both acidic and neutral pH has provided additional information about the most potent inhibitors of HCPTP. In vitro screening with detergent and dynamic light scattering measurements allowed the elimination of compounds that inhibited by aggregation. The computationally and enzymatically identified inhibitor 45576 contains the same characteristic pharmacophores as previous rationally designed compounds and demonstrates that virtual screening of the NCI Diversity Set by multiple computational programs can contribute to effective inhibitor design.
The docking predictions for the NCI Diversity Set converged to a common set of motifs for both docking programs and isoforms used. The most common differences observed between HCPTP-A and HCPTP-B usually resulted from differential positioning of the most distal portions of inhibitors from the P-loop. The overall conformational space occupied by these molecules is highly conserved. This conservation of docking positions further reinforces the original design elements for rational inhibitor design in HCPTP. The presence of a charged tail is preferred for interacting with the P-loop residues 12 through 18. An amino group has been shown to effectively interact with the catalytically important Asp 129. Multiple rings are used to fill the remainder of the active site. Future designed compounds will continue to incorporate these features, as well as exploring the use of expanded ring systems to interact with the aromatic residues at the mouth of the active site and novel contacts outside of the active site.
For the purposes of in silico screening of compounds for HCPTP, neither AutoDock nor Glide has proven to be more efficient than the other as a predictor of experimental inhibition. Instead, each program has provided unique solutions in the identification of inhibitory compounds. Some of the differences in identification of different compounds could be attributed to the use of different mechanisms for ligand preparation. Namely, the compounds docked via AutoDock did not comprise a complete set of protonation states and tautomeric states. Interestingly, the static crystallographic structure for HCPTP-B has functioned as a better predictor for inhibitors than HCPTP-A when using Glide as the screening program. The reason for this remains unclear, but may be because of small differences in the conformations of the enzymes in these crystal structures. This observation underlines the need for further detailed structural information about the binding of small molecules to both isoforms of HCPTP.
5) EXPERIMENTAL
AutoDock Screening
The crystal structures of HCPTP-A13 and HCPTP-B12 were obtained from the Protein Databank22 as 5PNT and 1XWW, respectively. Non-protein atoms were removed, polar hydrogens added, and histidine residues were made neutral before assigning Kollman charges to each enzyme. Residues which were not given a unit charge were manually adjusted by modification of the charge on the backbone nitrogen. Charges of the catalytically important amino acids were also adjusted to generate the proposed protonation state of the enzyme. Specifically, Asp 129 was protonated while the partial charges of Cys 12 were adjusted to give a total charge of −1. 23 Using the program AutoGrid323, the set of affinity grids in a space of 90 × 104 × 104 grid points and a resolution of 0.2 Å were calculated for aromatic and aliphatic carbon, nitrogen, oxygen, polar hydrogen, sulfur, phosphorus, fluorine, chlorine, bromine and iodine. This grid covers the active site and the proposed specificity cleft. 13 The NCI Diversity Set library of 1990 compounds has been converted to a format usable by AutoDock and is available from the AutoDock website at http://autodock.scripps.edu/resources/databases. This library was chosen based on its comprehensive coverage of the chemical space in a relatively modest sized library as well as the availability of compounds for testing from NCI.
Docking simulations with AutoDock 3.0.524 used the Lamarckian genetic algorithm protocol according to default parameters. 25 A total of 20 independent simulations with a population size of 50 members were run for each ligand using AutoDock. During the local search portion of each energy evaluation, 300 minimization iterations were applied. All other parameters were maintained as previously defined. The final population was clustered with a tolerance of 1.0 Å and ranked according to the docked energy between protein and ligand, a summation of internal ligand energy, and intermolecular energy terms. All compounds that exhibited a predicted inhibition constant, Ki, stronger than 10−5 M to both isoenzyme forms under this protocol were selected for kinetic analysis.
Glide Screening
The original MDL SD files for the Diversity Set were reprocessed by the LigPrep module of the Schrodinger suite to allow an unbiased ligand set. 26–27 LigPrep was run according to default parameters which generated all protonation states between pH 5 and 9 as well as all isomers and tautomers of each of the compounds within the Diversity Set. The enzyme structures were prepared for docking through the program Protein Prep and grids were generated to cover a cube of (20 Å)3 centered on the active site. The prepared ligand files were then subjected to a three-tiered docking strategy in which all of the compounds were docked by Glide according to Virtual High-Throughput Screen (VHTS) conditions in which the ligands are not allowed to change conformation. The 10% of the compounds which showed the lowest docked energies were then re-docked under Standard Precision (SP) conditions in which ligand flexibility is allowed. The 10% of the compounds which showed the lowest docked energies were then re-docked under Extra Precision (XP) conditions in which more strict energetic calculations are conducted. After visual inspection of docked structures from the XP docked data, orders were placed with the National Cancer Institute for the compounds (for structures compare Supporting Information) within 6 kcal/mol of the lowest docked energy so that their inhibition characteristics could be confirmed by experimentation. The program Pymol was used for visualization of docked poses of effective inhibitors with figures generated through the use of PovRay. 28–29
Sample Preparation
Each of the isoforms of HCPTP was generated and purified as has previously been described. 30 Prior to enzymatic analysis, any remaining phosphate salts were removed from the protein samples by dialysis in a buffer containing only 100 mM sodium acetate pH 5, which had been adjusted to an ionic strength of 150 mM with NaCl. Most of the compounds were received from the National Cancer Institute in powder form. An appropriate volume of DMSO was added to generate a 10 mM sample. Compounds which did not dissolve in pure DMSO were diluted with water to bring the concentration of the new solution to 5 mM. Small aliquots of concentrated HCl were added to the remaining solutions which still had particulate matter present. Any solid that had not gone into solution when a total volume of HCl equal to 10% of the total volume of the solution was added was considered to be insoluble and was not studied further. Thirteen compounds could not be resolubilized, therefore only 39 of the 52 compounds ordered were analyzed in the enzyme assay.
Enzyme Inhibition Assays
The enzymatic analysis was conducted as previously reported.31 Enzymatic activity was monitored via a single point quenched product reaction with the substrate analog p-nitrophenyl phosphate (pNPP). Enzyme assays were performed at 25 °C in either 100–250 mM sodium acetate pH 5.0 or 100 mM Tris pH 7, which had been adjusted to an ionic strength of 150 mM with NaCl. The concentration of pNPP was adjusted to 5.0 mM for the assays at pH 5. The final concentration for pNPP used at pH 7 initially was 100 mM, in accordance with the previously published Km for HCPTP, reported as 4.4 mM.28 However, it was determined that this Km is only appropriate for HCPTP-A. HCPTP-B loses considerably more activity at pH 7, showing a Km of 122 mM. In order to saturate the enzyme with a substrate concentration of 5 times higher than the Km, a concentration of 600 mM pNPP would need to be added to the reactions for HCPTP-B. As a result of this requirement in the assays, inhibition of HCPTP-B at pH 7 was undetectable and thus does not appear in this analysis. The enzyme concentrations were 11 nM HCPTP-A and 22 nM HCPTP-B. The assays were initiated by the addition of pNPP, after incubation with the inhibitor for 10 min, and were quenched after 10 min by the addition of 1N NaOH. The p-nitrophenol product was measured at 405 nm with an extinction coefficient of 18000 M−1cm−1. Inhibitors were initially evaluated at a single concentration of 100 μM. Compounds showing at least 50% inhibition at 100 μM were subjected to further assays. A minimum of 5 concentrations were chosen within ±10*IC50. The reactions were carried out in at least triplicate for each isoenzyme and the results were found to be reproducible within ±10%. The IC50 for the inhibitors were then calculated using the DNRPEASY’s Simple Enzyme Inhibition model using the average of the collected inhibition data.32
Dynamic Light Scattering Experiments
The dynamic light scattering (DLS) experiments were performed with the same concentrations of protein and inhibitor as used in the inhibition assays. For these experiments, the enzyme concentration was 11 nM HCPTP-A and 100 μM inhibitor concentrations were used in a total cuvette volume of 1mL in the pH 7 kinetics buffer. Data was collected on a Malvern Zetasizer Nano S. Particle size distributions were calculated for HCPTP-A in the presence and absence of compounds. The apparent hydrodynamic size of HCPTP-A in solution with detergent is approximately 10 nm. Particles greater in size than 50nm were considered to be aggregated inhibitor and/or protein.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the support of the National Cancer Institute (PO3CA23568 to CVS), thte Walther Cancer Institute (AZ postdoctoral fellowship), and the National Cancer Institute for the Biophysics Training Grant for training grant support (T32 GM08296 to KTH). We additionally thank the Markey Center for Structural Biology, the Purdue University Center for Cancer Research, and the Walther Cancer Institute at the University of Notre Dame for providing facilities, equipment, and staff support for this project.
Footnotes
SUPPORTING INFORMATION AVAILABLE: Two-dimensional structures of compounds, Autodock and Glide XP scores, the experimentally observed decreases in activity for the 39 analyzed inhibitors in HCPTP-A and HCPTP-B isoforms as well as the results from the DLS experiments are presented in the supplemental information. (6 pages)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ciardiello F, Caputo R, Bianco R, Damiano V, Fontanini G, Cuccato S, De Placido S, Bianco AR, Tortora G. Clin Cancer Res. 2001;7:1459. [PubMed] [Google Scholar]
- 2.Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM, Fairchild C, Hunt JT, Inigo I, Johnston K, Kamath A, Kan D, Klei H, Marathe P, Pang S, Peterson R, Pitt S, Schieven GL, Schmidt RJ, Tokarski J, Wen ML, Wityak J, Borzilleri RM. J Med Chem. 2004;47:6658. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 3.Landen CN, Kinch MS, Sood AK. Expert Opin Ther Targets. 2005;9:1179. doi: 10.1517/14728222.9.6.1179. [DOI] [PubMed] [Google Scholar]
- 4.Zantek ND, Azimi M, Fedor-Chaiken M, Wang B, Brackenbury R, Kinch MS. Cell Growth Differ. 1999;10:629. [PubMed] [Google Scholar]
- 5.Zelinski DP, Zantek ND, Stewart JC, Irizarry AR, Kinch MS. Cancer Res. 2001;61:2301. [PubMed] [Google Scholar]
- 6.Cortesio CL, Chan KT, Perrin BJ, Burton NO, Zhang S, Zhang ZY, Huttenlocher A. J Cell Biol. 2008;180:957. doi: 10.1083/jcb.200708048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikawa KD, Vidale DR, Van Etten RL, Kinch MS. J Biol Chem. 2002;277:39274. doi: 10.1074/jbc.M207127200. [DOI] [PubMed] [Google Scholar]
- 8.Chiarugi P, Taddei ML, Schiavone N, Papucci L, Giannoni E, Fiaschi T, Capaccioli S, Raugei G, Ramponi G. Oncogene. 2004;23:3905. doi: 10.1038/sj.onc.1207508. [DOI] [PubMed] [Google Scholar]
- 9.Forghieri M, Laggner C, Paoli P, Langer T, Manao G, Camici G, Bondioli L, Prati F, Costantino L. Bioorg Med Chem. 2009;17:2658. doi: 10.1016/j.bmc.2009.02.060. [DOI] [PubMed] [Google Scholar]
- 10.Maccari R, Ottana R, Ciurleo R, Paoli P, Manao G, Camici G, Laggner C, Langer T. ChemMedChem. 2009;4:957. doi: 10.1002/cmdc.200800427. [DOI] [PubMed] [Google Scholar]
- 11.Zabell AP, Corden S, Helquist P, Stauffacher CV, Wiest O. Bioorg Med Chem. 2004;12:1867. doi: 10.1016/j.bmc.2004.01.042. [DOI] [PubMed] [Google Scholar]
- 12.Zabell AP, Schroff AD, Jr, Bain BE, Van Etten RL, Wiest O, Stauffacher CV. J Biol Chem. 2006;281:6520. doi: 10.1074/jbc.M506285200. [DOI] [PubMed] [Google Scholar]
- 13.Zhang M, Stauffacher CV, Lin D, Van Etten RL. J Biol Chem. 1998;273:21714. doi: 10.1074/jbc.273.34.21714. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Stauffacher CV, Van Etten RL. Biochemistry. 2000;39:1234. doi: 10.1021/bi991515+. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Tabernero L, Zhang M, Harms E, Van Etten RL, Stauffacher CV. Biochemistry. 2000;39:1903. doi: 10.1021/bi991348d. [DOI] [PubMed] [Google Scholar]
- 16.Weitgenant JA, Katsuyama I, Bigi MA, Corden SJ, Markiewicz JT, Zabell APR, Homan KT, Wiest O, Stauffacher CV, Helquist P. Heterocycles. 2006;70:599. [Google Scholar]
- 17.Soellner MB, Rawls KA, Grundner C, Alber T, Ellman JA. J Am Chem Soc. 2007;129:9613. doi: 10.1021/ja0727520. [DOI] [PubMed] [Google Scholar]
- 18.Pospisil P, Wang K, Al Aowad AF, Iyer LK, Adelstein SJ, Kassis AI. Cancer Res. 2007;67:2197. doi: 10.1158/0008-5472.CAN-06-3309. [DOI] [PubMed] [Google Scholar]
- 19.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. J Med Chem. 2002;45:1712. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 20.Feng BY, Shoichet BK. Nat Protoc. 2006;1:550. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Zhou M, Van Etten RL, Stauffacher CV. Biochemistry. 1997;36:15. doi: 10.1021/bi961804n. [DOI] [PubMed] [Google Scholar]
- 22.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asthagiri D, Liu T, Noodleman L, Van Etten RL, Bashford D. J Am Chem Soc. 2004;126:12677. doi: 10.1021/ja048638o. [DOI] [PubMed] [Google Scholar]
- 24.Goodsell DS, Morris GM, Olson AJ. J Mol Recognit. 1996;9:1. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 25.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Journal of Computational Chemistry. 1998;19:1639. [Google Scholar]
- 26.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. J Med Chem. 2004;47:1739. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 27.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. J Med Chem. 2004;47:1750. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 28.De Lano WL. 2002 [Google Scholar]
- 29.Ltd., P. o. V. P. 2004 [Google Scholar]
- 30.Wo YY, McCormack AL, Shabanowitz J, Hunt DF, Davis JP, Mitchell GL, Van Etten RL. J Biol Chem. 1992;267:10856. [PubMed] [Google Scholar]
- 31.Weitgenant JA, Katsuyama I, Bigi MA, Corden SJ, Markiewicz JT, Zabell APR, Homan KT, Wiest O, Stauffacher CV, Helquist P. Heterocycles. 2006;70:599. [Google Scholar]
- 32.Duggleby RG. Comput Biol Med. 1984;14:447. doi: 10.1016/0010-4825(84)90045-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.