Abstract
The last decade brought tremendous progress in the field of schizophrenia genetics. As a result of extensive collaborations and multiple technological advances, we now recognize many types of genetic variants that increase the risk. These include large copy number variants, rare coding inherited and de novο variants, and over 100 loci harboring common risk variants. While the type and contribution to the risk vary among genetic variants, there is concordance in the functions of genes they implicate, such as those whose RNA binds the fragile X-related protein FMRP and members of the activity-regulated cytoskeletal complex involved in learning and memory. Gene expression studies add important information on the biology of the disease and recapitulate the same functional gene groups. Studies of alternative phenotypes help us widen our understanding of the genetic architecture of mental function and dysfunction, how diseases overlap not only with each other but also with non-disease phenotypes. The challenge is to apply this new knowledge to prevention and treatment and help patients. The data generated so far and emerging technologies, including new methods in cell engineering, offer significant promise that in the next decade we will unlock the translational potential of these significant discoveries.
Keywords: Copy number variation, de novo mutations, Gene expression, Transcriptome, Gene editing, Genome-wide association studies, Genetic variation, Psychosis, Endophenotypes
Introduction
Schizophrenia: A Heterogeneous Phenotype
Schizophrenia is a common and serious psychiatric illness [1] affecting 0.5–1% of the population in early adulthood. Despite continuing progress, current treatments continue to have significant side effects and inconsistent efficacy across patients. The disease remains incurable, with the best outcome being the control of symptoms and preservation of sufficient functionality and independence.
Schizophrenia is a disease with remarkable phenotypic heterogeneity. The symptoms are generally divided into three categories [2]. Positive symptoms, with which the general public is most familiar, include hallucinations and delusions of varying content, and are perhaps the most disruptive. Negative symptoms include lack of motivation, anhedonia, and flat affect. Cognitive symptoms such as defects in attention, concentration, working memory, and critical thinking are the most incapacitating, leading to significant disability [3]. Each patient can have a different mix of the three types of symptoms leading to an overall highly heterogeneous phenotype. Affective symptoms may combine with the classic symptoms of schizophrenia leading to schizoaffective disorder, which some genetic researchers categorize with schizophrenia and others with bipolar disorder (BD). Heterogeneity also manifests in the patients' response to medication, frequently resulting in multiple changes in treatment strategy during the course of the illness as patients navigate through ineffective treatments [4]. It is clear that schizophrenic patients would benefit considerably from a robust prediction of their response through individualized medicine. A good understanding of the underlying genetics, the importance of environmental factors and the interaction of the two, along with careful clinical characterization may achieve that in the not so distant future.
Heritability and Environment
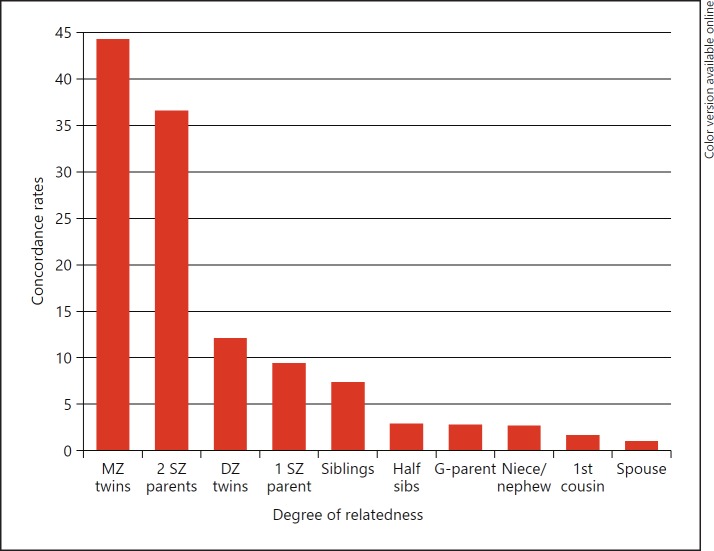
It has long been recognized that schizophrenia runs in families, noted in the first detailed description of the disease by Bleuler in 1911 [5]. Figure 1 shows the concordance rates between different degrees of relatedness (data from McGue and Gottesman [6]) demonstrating the increase from the population prevalence of close to 1% in unrelated spouses to 44% in monozygotic twins. Formally, the heritability of the disease has been calculated at > 80% [6, 7], making genetic makeup the most important factor for developing the disease.
Fig. 1.
Concordance rates for varying degrees of relatedness. MZ, monozygotic; DZ, dizygotic; SZ, schizophrenic; G-parent, grandparent.
In addition to the importance of genetics, the environment also plays a major role in the risk to develop schizophrenia. Environmental factors explain the significant non-heritable fraction of the disease variance; they are also likely to be easier to control and modify than genetic factors. Multiple studies have already implicated a diverse array of environmental factors that increase the risk to develop schizophrenia. These include social factors such as migrant status (odds ratio [OR] = 2.3 [8]) and urban environment (OR = 2.3 [9]), and pre- and perinatal factors such as maternal malnutrition (OR = 2 [10, 11]) and birth month (OR = 1.07 for those born in winter or spring [12]). Finally, there are many studies suggesting increased risk accompanying infections such as toxoplasmosis, rubella, influenza, herpes, and others [13, 14]. It appears from these associations that stress, whether in utero, at birth, or during life, is an important determinant of risk for schizophrenia. These environmental factors are also likely to interact with genetic variation, increasing the risk only in their presence. One such replicated example has been reported for CMV infection and common variants near the CTNNA3 gene [15, 16]. Knowing these relationships will become increasingly important as we learn more about the genetics of the disease, allowing us to achieve individualized prevention and treatment strategies and move precision medicine into psychiatry.
Negative Selection
An interesting, though not unexpected, observation is that patients with schizophrenia have significantly fewer children compared to the general population [17, 18, 19]. In theory, this should be generating an enormous negative selective pressure quickly removing risk alleles from the population; however, the disease maintains a relatively high heritability and prevalence at ∼1%. The reasons for this paradox remain unclear and have sparked much debate and speculation. Among possible explanations are balancing selection favoring genotype diversity; advantage for those who carry the allele but do not get sick; changing environments that expose or protect cryptic variation; or quick replenishment by new mutations, in view of the possibility that disruption of thousands of different genes may be able to lead to disease [20, 21, 22]. The results of genome-wide association studies (GWAS) that we will discuss below support this highly polygenic architecture. In addition, GWAS results are consistent with theoretical predictions [23] that common schizophrenia alleles can only show low odds ratios because of this negative selection. This observation, which has the consequence that GWAS variants explain very little genetic variance, has led some health scientists to challenge the value of GWAS. This, however, is a narrow view of the value of these results. Common variants that survive selection might have small effects on risk, but pharmacological interventions can be designed to have a larger effect on the gene regulation, its product or the related pathway, providing major benefits.
Early Days: Linkage, Candidate Genes, and Lack of Replication
The polygenic nature of schizophrenia has been suspected and debated for a long time [24, 25, 26]. Hoping that at least some families might segregate a single disease-causing variant, or that the overall number of such variants is limited, numerous linkage studies have tested both parametric and non-parametric approaches. Starting as early as 1972, Elston et al. [27] reported possible linkage of schizophrenia with specific blood groups, and many other linkage studies followed. Unfortunately, most were met with disappointment, almost always showing weak results and often failing to replicate one another. The same was true for the first association studies that focused on candidate genes or followed up previous linkage results. At the time, we did not appreciate the large number of risk variants underlying schizophrenia and the small contribution these variants have on the risk. The studies of the era were vastly underpowered and often produced no or false positive results. Only now that we have succeeded in identifying true schizophrenia risk variants have we come to appreciate the serious limitations of earlier work. Very few of the early gene findings remain under investigation today, and those that do are not because of robust evidence for a role in the disease, but rather because of continuing interest in their function revealed by the work initially triggered by the associations.
In this review, we will focus on genetic variation showing robust associations with schizophrenia, including high-penetrance rare variants and low penetrance common variants. We will then describe transcriptomics work, an alternative approach to the genetics of the disease, and the use of alternative phenotypes termed endophenotypes, which is widening our understanding of the dimensionality of mental illness. Finally, we will discuss how cutting-edge technologies are opening new directions in the ways we can experimentally model the disease.
High-Penetrance Genetic Variation
Chromosomal Abnormalities and Copy Number Variation
At the same time that linkage and association studies were not providing the desired results, a strong link between a deletion syndrome and schizophrenia was receiving increasing attention. A recurrent deletion in chromosomal band 22q11.2 causing a phenotype called velo-cardio-facial syndrome (VCFS) was noted to be often accompanied by psychosis [28]. In addition, following the reports of this comorbidity, deletion screening of schizophrenia patients showed that some had the 22q deletion but were not diagnosed with VCFS because of their mild features [29, 30]. Most recently, in a large sample of 21,094 cases and 20,227 controls, this deletion was determined to increase the risk of carriers 68-fold and to be present in 0.3% of individuals diagnosed with schizophrenia [31]. The deleted region includes more than 50 genes including the highly cited catecholamine-degrading enzyme gene COMT. The deletion is recurrent and is due to flanking low copy repeats that mediate unequal meiotic crossing over [32]. There are multiple repeats in the 22q11.2 region so the deletion can vary in size, the most common (90%) being ∼3 Mb followed by ∼1.5 Mb deletions (8%) [33, 34]. Interestingly, the reciprocal duplication has been reported to be protective against schizophrenia [35].
While the 22q11.2 deletion was the first to be discussed in schizophrenia, the ever-increasing use of microarrays, whether for CNV detection or for genotyping, provided data that allowed us to recognize more and smaller copy number variants (CNVs). As a result, many additional CNVs have now been reported to be associated with schizophrenia. These are described in detail below and summarized in Table 1, which includes those that reach significance in the largest case/control CNV analysis to date, on 41,321 cases and controls [31] and have been previously reported.
Table 1.
CNVs that reach significance in the largest case/control CNV analysis to date as reported by Marshall et al. [31] and have been previously reported
| Locus (gene) | Risk allele | Cases (n = 21,094) | Controls (n = 20,227) | OR (95% CI) | BH-FDR |
|---|---|---|---|---|---|
| 22q11.21 | Loss | 64 | 1 | 67.7 (9.3–492.8) | 3.54 × 10–15 |
| 16p11.2, proximal | Gain | 70 | 7 | 9.4 (4.2–20.9) | 5.82 × 10–10 |
| 2p16.3 (NRXN1) | Loss | 35 | 3 | 14.4 (4.2–46.9) | 3.52 × 10–7 |
| 15q13.3 | Loss | 28 | 2 | 15.6 (3.7–66.5) | 2.22 × 10–5 |
| 1q21.1 | Loss + gain | 60 | 14 | 3.8 (2.1–6.9) | 0.00011 |
| 3q29 | Loss | 16 | 0 | INF | 0.00024 |
| 16p11.2, distal | Loss | 11 | 1 | 20.6 (2.6–162.2) | 0.0029 |
| 7q11.23 | Gain | 16 | 1 | 16.1 (3.1–125.7) | 0.0048 |
| 22q11.21 | Gain | 3 | 16 | 0.15 (0.04–0.52) | 0.024 |
| 15q11.2 | Loss | 98 | 50 | 1.8 (1.2–2.6) | 0.044 |
| 7p36.3 | Loss + gain | 20 | 6 | 3.5 (1.3–9.0) | 0.046 |
The location, allele of higher risk, number detected in cases and controls, odds ratio (OR) with 95% confidence interval (CI) and the Benjamini-Hochberg false discovery rate (BH-FDR) are shown.
The 16p11.2 region first received attention after an association of its deletion with autism [36]. Later, it was shown that the reciprocal duplication is associated with schizophrenia [37]. Interestingly, the deletion and duplication show opposite effects on intracranial volume, brain size, compartmental measures of gray and white matter, subcortical structures, and the cerebellum [38, 39]. They also show reciprocal effects on head circumference [40] visual evoked potential amplitude [41] and BMI phenotypes [42]. It has been suggested that the major driver of the neuroanatomical phenotypes may be the gene KCTD13 which has been implicated in long-term positioning and dendritic maturation of cerebral cortical neurons [43]. A more distal and smaller region on 16p11.2 has also been implicated in schizophrenia when deleted [44, 45], as well as in developmental delay and obesity [45]. The 2p16.3 deletion was first identified by comparative genome hybridization and disrupts the NRXN1 gene encoding neurexin 1 a presynaptic cell adhesion molecule which interacts with neuroligins to induce synapse formation and maturation [46, 47]. The initial association with schizophrenia was confirmed soon after by a larger SNP array-based study [48]. The deletion at 15q13.3 was reported to cause mental retardation and seizures [49] before being associated with schizophrenia [47, 50], and two childhood-onset schizophrenia cases with duplications at this locus have also been reported [50]. It contains the CHRNA7 gene encoding the A7 nicotinic acetylcholine receptor, previously linked to many psychiatric phenotypes including schizophrenia [51]. The 1q21.1 CNV was first reported by the International Schizophrenia Consortium to increase the risk when deleted [52] and later the reciprocal duplication was also found in excess [31, 53] in schizophrenia patients. Like the 16p11.2 CNV, reciprocal phenotypes have been described for the 1q21.1 CNV. Deletion and duplication cause microcephaly and macrocephaly, respectively, and schizophrenia is only one of many associated neurodevelopmental phenotypes [54]. The 3q29 deletion was first described to cause mental retardation, with slight dysmorphic facial features [55] and in some cases autism [56] and was later linked to schizophrenia [57, 58, 59]. The 7q11.23 duplication is reciprocal to the Williams-Beuren syndrome deletion [60] and in addition to schizophrenia has been associated with autism, language delay, and mental retardation [61]. Finally, the 15q11.2 is also associated with developmental and language delay, mild dysmorphic features, autism, and seizures in addition to schizophrenia.
Many additional, perhaps more rare CNVs or CNVs with smaller odds ratios are likely to be below our current detection threshold and are not on this list. Their existence however is supported by the overall enrichment for CNVs in cases compared to controls, even after excluding those reaching significance [31]. Additionally, it is thought, largely due to the likely negative selection of these high-risk variants, that CNV leading to schizophrenia often occur de novo and can be recurrent due to flanking low copy repeats, which has been directly demonstrated at least for a subset of those reported in Table 1 (22q11.2, 3q29, 15q11.2, 15q13.3, and 16p11.2) [62, 63].
An important observation is that these CNVs include both gains and losses of genetic material, and are often also associated with autism and/or intellectual disability. The associations are often with reciprocal alleles (22q, 16p) but sometimes they are with the same CNV allele. This not only supports the notion of genetic overlap between psychiatric disorders (see below) but also adds a level of complexity showing that for specific loci the allelic effects can be different. Overall, within CNV regions in schizophrenia there is excess of genes involved in axon guidance, nervous system development, genes coding for targets of the RNA-binding protein FMRP (responsible for Fragile X syndrome) and for proteins of the activity-regulated cytoskeletal (ARC) complex involved in learning and memory [31].
A special case of chromosomal rearrangement that has been linked to schizophrenia is the DISC1 locus. A balanced 1q43: 11q14 translocation has been described in a large Scottish pedigree with multiple psychiatric phenotypes including schizophrenia. The pedigree showed significant genetic linkage between the disease and the translocation [64] which disrupted two genes, DISC1 on chromosome 1 and DISC2 on chromosome 11. Of the two, DISC1 is the gene that appears most relevant [65]. A later study also identified a frameshift mutation in DISC1 segregating in an American family with schizophrenia and schizoaffective disorder [66], and there has been a significant volume of research on the gene's function supporting its importance in cortical development [67]. The initial DISC1 report has also been followed by numerous association studies with mixed results and significant controversy regarding its role in schizophrenia [68, 69]. The gene also shows no association with schizophrenia in large GWAS [70], which suggests it contains no common alleles that increase the risk, but does not exclude its involvement in disease through rare highly penetrant variation like the 1q43: 11q14 translocation and the reported frameshift mutations [66]. The strong linkage results, but most importantly the large volume of functional information that has now accumulated and linked the gene to brain development, certainly makes its study worthwhile.
Rare and de novo Variation
In the last decade, new technologies have made sequencing of the entire exome or genome much more affordable, having a significant impact on the strategies used to study complex diseases including schizophrenia. Hypothesizing the existence of rare or de novo variants that have a strong impact on the risk, many investigators have sought them by sequencing case/control cohorts or parent-patient trios. However, despite the recent price reduction, the cost of sequencing remains significant and the size of the sequenced cohorts lags behind those studied in GWAS. This, along with the low frequency of such variants, leads to insufficient power at the gene level. Investigators have worked around this limitation by exploring the burden of likely functional variants across schizophrenia-related gene groups. In addition to the limitations of the “candidate pathway” approach and the uncertainties regarding gene group membership, a number of arbitrary thresholds are typically set to filter the variants by allele frequency and evidence of function. Nevertheless, there have been interesting results that are consistent with the results of both CNV and GWAS studies.
In one of the first exome sequencing studies, Girard et al. [71] analyzed the exomes of 14 schizophrenia probands and their parents and reported an excess of exonic de novo mutations, a result that was confirmed almost simultaneously by an independent study of 52 cases by Xu et al. [72]. Soon after, Xu et al.[73] followed up in 231 cases re-iterating the higher prevalence of gene-disruptive de novo mutations and reporting recurrent mutations in four genes (LAMA2, DPYD, TRRAP, and VPS39). Sequencing 623 schizophrenia trios, Fromer et al. [74] reported increased de novo mutation burden in select groups of genes, namely those encoding proteins closely associated with N-methyl-D-aspartate (NMDA) receptors and proteins that interact with the activity-regulated cytoskeleton-associated protein ARC, reiterating the results of CNV studies. They also reported recurrence for the TAF13 gene. McCarthy et al. [75] studying 57 schizophrenia trios reported de novo mutations in genes involved in chromatin remodeling and previously implicated in autism and intellectual disability, re-iterating the disease overlaps reported for CNVs. Studying both autism and schizophrenia, Takata et al. [76] extended these results to include synonymous variants that overlap brain-derived DNase hypersensitivity sites, genomic sites where chromatin is open, suggesting functional significance. One of the genes they identified, SETD1A, showed two loss-of-function and one synonymous de novo mutation. In a different approach to the question, Gulsuner et al. [77] mapped the mutated genes onto transcriptome profiles measured in normal human brains aged between the 13th week of gestation and adulthood. They found that mutated genes mapped on transcriptional coexpression and protein interaction networks involved in regulation of transcription, cellular transport, signaling, neuronal migration, and synaptic transmission.
In addition to de novo variation, sequencing studies have also explored the contribution of rare, segregating, potentially functional variants with relatively large effects. Need et al. [78] followed a two-step approach, identifying rare variants from sequencing data on 166 individuals with schizophrenia and genotyping these in a larger sample of 2,617 cases and 1,800 controls; none of the variants showed study-wide significance. They concluded that such rare variants are unlikely to be a major risk factor for schizophrenia. Timms et al. [79] sequenced 5 families segregating schizophrenia and reported that each of them had a segregating rare variant in 1 of 3 genes associated with the NMDA receptor. In 2 separate studies, Ruderfer et al. [80] and Rees et al. [81] explored the contribution of recessive and compound heterozygous rare likely functional variants but neither was able to show a significant contribution; however, a number of possible limitations including sample size do not allow definite conclusions from these negative results. Guipponi et al. [82] sequenced 53 individuals with sporadic schizophrenia, and while they reported 18 putative candidate genes, they did not observe overall enrichment of variants in the patients. Using a much larger sample of 2,536 schizophrenia cases and 2,543 controls, Purcell et al. [83] reported a high polygenic burden of very rare disruptive mutations in schizophrenics. While no individual gene test achieved study-wide significance, they showed enrichment in genes related to voltage-gated calcium channels and the ARC-associated proteins, once again supporting the same enrichments observed with CNVs. In an even larger study of 4,946 patients and 6,242 unaffected controls, Genovese et al. [84] reiterated the highly significant excess of ultra-rare gene-disruptive variants, particularly in genes expressed in neurons. The corresponding RNAs included many known to interact with synaptic proteins, so they concluded that these rare genetic variants disrupt synaptic function. Most recently, Leonenko et al. [85] reported on 10,011 schizophrenia cases and 13,791 controls. They found that in schizophrenia, genes intolerant of loss-of-function variation and genes whose RNAs bind FMRP, similar to CNV results, carried an excess of rare alleles with minor allele frequency less than 0.1%. The only individual gene to show study-wide significant enrichment for rare loss-of-function variants is SETD1A, specifically suggesting a role for epigenetic dysregulation in the histone H3K4 methylation in schizophrenia [86].
Overall, the evidence of a role of rare and de novo mutations in schizophrenia is overwhelming, and involves some of the same functional categories with genes implicated by CNVs, although their overall contribution to disease is modest. The limited examples of recurrent hits of the same gene with de novo mutations, suggests that the number of genes involved in schizophrenia is quite large. The penetrance of these de novo variants and the extent to which their expression depends on the rest of the genome and the environment remains unknown, and requires large sample sizes to study. A limitation of current sequencing studies is that although they make a valid point on the involvement of rare or de novo variants, what they show is deviations from the expected number of such variants; they do not yet point to specific variants that increase the risk with enough certainty to warrant investing resources on follow-up. This however is likely to change as sample sizes increase.
Common Low-Penetrance Variants and GWAS
Following a seminal paper from Risch and Merikangas in 1996 [87] showing that in complex disorders GWAS have far superior power than genome-wide linkage studies, it became clear to the scientific community that this is the preferred way to map genes for diseases like schizophrenia. Initially, the major obstacle to performing a GWAS was the cost of genotyping. Once new technologies changed this, another problem became evident: sample size. The effect sizes of risk alleles turned out to be much lower than expected and, with few exceptions, a few hundred or even thousand samples were not enough for sufficient statistical power. The first GWAS for schizophrenia to adequately cover the genome in a relatively large collaborative case/control sample was published in 2008 [88]. Following a two-step analysis to reduce genotyping cost, and including BD patients in an effort to increase power, this study reported a single association around the ZNF804A gene, a gene that has been replicated in subsequent studies. This was followed by a larger study combining data from multiple others to reach ∼13,000 cases and 35,000 controls and reporting three genomic loci including the Human Leukocyte Antigen (HLA) region on chromosome 6 and near the genes TCF4 and NRGN on chromosomes 18 and 11, respectively [89]. At the same time, the International Schizophrenia Consortium also reported on the HLA association as well as significant genetic overlap with BD [90]. After these studies, smaller groups began consolidating collected samples and genotypes of patients and controls into larger consortia. Efforts such as the Molecular Genetics of Schizophrenia collection (clinicaltrials.gov/ct2/show/NCT00006418) and the Genetic Association Information Network (GAIN; www.genome.gov/19518664/genetic-association-information-network-gain/) were developed in order to achieve the statistical power necessary for robust discovery. The biggest collaborative consortium, the Psychiatric Genomics Consortium (PGC), was started in 2007, and according to its website (http://www.med.unc.edu/pgc/pgc-workgroups) its schizophrenia group currently includes over 400 investigators from 40 countries. The PGC published its first GWAS in 2011 identifying five new loci for schizophrenia using a discovery sample of 21,856 Europeans and 29,839 independent subjects for replication [91]. Many other important papers followed until most recently in 2014 they published on 36,989 cases and 113,075 controls [70], reporting 108 significant loci that represent 128 independent association signals, 83 of which had not been previously reported. The authors mapped the variants onto epigenetic marks characteristic of active enhancers in 56 tissues and cell lines. As expected, they found enrichment in brain tissue enhancers (highest in midfrontal and angular gyrus), but also in tissues with important roles in immunity (highest in CD19 and CD20 B cells). The same group also developed an analytical framework to use summary statistics data from this GWAS to identify and rank common gene/functional pathways between schizophrenia, BD, and major depressive disorder (MDD). They reported associations for the histone methylation pathway as well as for immune and neuronal signaling and postsynaptic density [92]. Li et al.[93] have recently added 30 new loci to the PGC results by adding a large Chinese sample of ∼36,000 individuals and combining them with the PGC data in meta-analysis. The PGC has also continued increasing their sample size and at the 2017 World Congress of Psychiatric Genetics, they reported the identification of 248 genome-wide significant loci, confirming the speculation that a large number of genes are likely involved in schizophrenia, and providing the basis for a tremendous amount of future work on the etiology of schizophrenia. It should be noted that while combining samples through consortia has tremendous value, there is also the limitation that it is hard to replicate every finding in an independent comparable sample. However, the persistence of signals constitutes equally strong validation to independent replication. For example, the original association in ZNF804A on 20,142 samples [88] moved from a p value of 1.61 × 10–7 to 7.13 × 10–12 with the 5-fold sample increase to 113,075 samples [70], suggesting it is highly unlikely to be a false positive. Each new sample increase brings new candidate associations forward while validating many of the previously reported.
In addition to identifying multiple reliable associations, the advent of GWAS also has led to a novel approach in the study of the genetics of complex diseases, the development of polygenic risk scores (PRSs). The International Schizophrenia Consortium, a consortium that included many of the original members of the PGC schizophrenia working group, was the first to use PRSs. In a paper reporting an early schizophrenia GWAS that identified the HLA locus as mentioned above, the authors performed an additional analysis where they used the GWAS results as a reference dataset to calculate PRSs on other independent datasets [90]. In a first step, the method selects variants from the GWAS at some significance threshold and assigns to their alleles the observed effect on risk. Then, based on the genotypes of the individuals in the target data set at these loci, it calculates a PRS for each individual. While most of the selected loci are not genome-wide significant, and many are false positives, those will have random effect size and direction; so, final score is mostly driven by true risk loci (for a review on PRSs, see Maher [94]). In the original paper, the author used this method to formally demonstrate a long-suspected genetic overlap between schizophrenia and BD that is also supported by CNV and rare variant data. The polygenic scores also became a tool for other explorations in the genetic architecture of schizophrenia and its relationship to other phenotypes. Hamshere et al. [95] showed an overlap of schizophrenia PRS with attention-deficit hyperactivity disorder (ADHD), while the Cross-Disorder Group of the PGC published on overlaps across five disorders, autism, ADHD, BP, MDD, and schizophrenia showing strongest overlaps between schizophrenia BP and MDD [96]. Others have shown positive correlations of schizophrenia PRS with the risk for posttraumatic stress disorder [97], addiction [98], and cortical thinning in patients [99]. In contrast, a large study from Iceland found that an increased score correlated with higher creativity, using membership in artistic societies and creative profession as a proxy [100]. It is clear both from PRSs and from the results of CNV and rare variant analyses that the roles of some genes cut across multiple clinical diagnoses. It may be that subset variants confer risk for specific diseases while others affect general mental health robustness, while yet others do not increase the risk per se but only the outcome following loss of robustness. A recent study on a large sample (n ≈ 14,000) of BD patients and controls showed that only 22 of 107 leading schizophrenia SNPs reached nominal significance [101]. While this is more than expected by chance, it clearly demonstrated that not all variants are important across diseases. As the reference GWAS and the target samples become larger, PRSs will gain power that may allow us to understand how behavioral and other phenotypes relate to each other at the level of the gene. For a recent review of the application of PRS on schizophrenia-related phenotypes, see Mistry et al. [102].
Having identified over 100 robust association signals for schizophrenia is a tremendous achievement and demonstrates the power of collaboration in science. However, the true benefits of these discoveries will only be realized once we begin to understand the disease mechanisms that underlie these associations. There are a number of obstacles in this path forward. First, most associated variants disrupt regulatory sequences as demonstrated by the lack of coding variation in linkage disequilibrium (LD). In contrast to coding sequences, we know very little about the rules governing regulatory sequences and the many association signals per locus due to LD; this makes it difficult to identify the biologically relevant variant. It is also often not clear which gene(s) and which isoform(s) are regulated by such variants and under what conditions or at what time during development the regulation is occurring. In the GTEx database for example (commonfund.nih.gov/gtex, www.gtexportal.org/home/), many of the GWAS variants are not identified as eQTLs in the included tissues, while many others are eQTLs for multiple genes across many tissues. Further, eQTLs that only affect splicing or are only active at specific times/conditions may be near invisible. Another obstacle is that the effect of GWAS variants on the risk is quite small, with most carriers of risk alleles being healthy. It is therefore unlikely to observe a phenotype even if one could imitate the exact same biological effect in a model organism. Applying more extreme disruptions such as gene knockouts might give a phenotype but reduces the credibility of the conclusions.
Despite the obstacles, new disease biology is already emerging from the GWAS results. Beyond the group enrichments and network analyses described above, many studies have begun to link variants to specific genes as eQTLs and experimentally follow them up to understand the biological consequences of these variants. Examples include signals near ZNF804A [103, 104], TCF4 [105], and CACNA1C [106, 107]. The most detailed and well publicized such study has been that of Sekar et al. [108] in the HLA region. The authors investigated the strongest signal from the PGC GWAS and found that the associated SNPs were proxies for the genomic structure of a nearby gene, C4A, which along with C4B show structurally diverse alleles. These alleles correspond to differences in the expression levels of the genes, which in turn lead to differences in synaptic pruning as the authors demonstrated by modeling in mice. A connection between schizophrenia and synaptic pruning was first made hypothetically 35 years ago [109] and the involvement of the complement has also been reported [110]. The work of Sekar et al.[108], took advantage of the GWAS results to provide some of the strongest support for this hypothesis to date.
With 108 loci to investigate and more than twice that expected as the PGC continues to expand their sample size, there is a lot of work to be done. The vision of biology-based psychiatry is becoming a reality. As we understand the basis of each genetic association, their common elements, the differences between them, and their interplay with the environment, we will likely soon make leaps in prevention, treatment, and management tailored to the individual patient.
Insights from Transcriptome Studies
The study of the transcriptome has only been possible in the last two decades with the advent of microarray technologies [111], which have been replaced more recently by transcriptome sequencing. There have been numerous studies attempting to characterize the schizophrenia transcriptome, with the first study on postmortem brain tissue of schizophrenics appearing in 2000 [112] and reporting abnormalities in presynaptic function. Subsequent studies have shown fairly consistent results incriminating mitochondrial function and energy metabolism [113, 114, 115, 116, 117], oligodendrocyte function [117, 118, 119, 120, 121], immunity-related genes [117, 122, 123, 124, 125, 126, 127, 128] and GABA neurotransmission [117, 129]. A study by Cohen et al.[130] implicated alternative splicing in schizophrenia by showing differential expression of particular exons and 3′ untranslated regions, a result that was supported by an independent study from Oldmeadow et al. [131] and later another independent observation by Takata et al.[132], who found enrichment of splicing QTLs among schizophrenia-associated loci. Also studying schizophrenia-associated loci, Birnbaum et al. [133] showed that they were enriched for genes transcribed during fetal life, supporting a developmental origin of the disease. The same finding was independently reported a year later by Ohi et al. [134]. Jaffe et al. [135] mapped DNA methylation across development and found 2,104 CpGs differing between schizophrenia patients and controls. These were enriched for brain development and neuronal differentiation genes, and were often located at GWAS schizophrenia risk loci. Ellis et al. [136] studying both autism and schizophrenia brains found significant excess of shared sets of downregulated genes between them, adding to the evidence of etiological overlaps between psychiatric disorder. Finally, using data from the CommonMind Consortium (commonmind.org) Fromer et al.[137] showed that ∼20% of schizophrenia loci have variants that alter gene expression. In cases where a single gene was involved (FURIN, TSNARE1, CNTN4), they showed that altering its expression changed neurodevelopment in zebrafish. Using gene coexpression network analysis, they showed support for gene networks involved in neurobiological functions that had already been suggested by other studies like the ARC protein complex, targets of FMRP, neuronal markers, postsynaptic density proteins, and NMDA receptors. Most recently, Gandal et al. [138] applied transcriptomic analysis to further explore the overlap between psychiatric disorders. They identified patterns of both shared and distinct gene expression perturbations and similar to reported polygenic overlaps, they found strong overlaps with BD autism and MDD in order of overlap strength and reported specific expression patterns.
Three other studies have used the transcriptome as a readout to characterize the biological consequences of schizophrenia-associated genes. Chen et al.[103] studied the ZNF804A gene by analyzing the effects of a gene knockdown in neurons derived from human induced pluripotent stem cells and reported enrichment of downregulated genes involved in interferon signaling. Pham et al. [139], following up on their own linkage, association, and functional data [140, 141, 142], modified an isoform-specific promoter of the DPYSL2 gene by CRISPR/Cas9 and found a complementary effect to transcriptomic changes induced by antipsychotics, enrichment in immune system process genes, as well as a significant overlap with the results of Chen et al. [103]. This last point is of interest as it connects ZNF804A and DPYSL2, two genes that have no other known functional connection other than their associations with schizophrenia. Finally, Hill et al.[105] knocked down the schizophrenia-associated gene TCF4 and reported that this resulted in RNA-level changes of genes involved in the cell cycle and the proliferation of human cortical progenitor cells.
Across different study designs exploring CNVs, rare variants, de novo variants, common variants, or the disease transcriptome, it is clear that certain gene groups and pathways appear repeatedly. Table 2 lists the groups that are supported by more than one independent type of study.
Table 2.
Gene categories supported by more than one independent approach
| Study subject | FMRP | ARC | NMDA receptors | Synapse | Immunity | CNS development |
|---|---|---|---|---|---|---|
| CNVs | ✓ | ✓ | ✓ | |||
| De novo variants | ✓ | |||||
| Rare variants | ✓ | ✓ | ✓ | ✓ | ||
| Common variants (GWAS) | ✓ | ✓ | ||||
| Transcriptome | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
See text for details.
Intermediate Phenotypes/Endophenotypes
Endophenotypes are a concept initially described by Gottesman and Shields [24] as internal phenotypes discoverable by a “biochemical test or microscopic examination.” They are phenotypes that are not immediately observable, but can be related to a disease because they are present in patients and sometimes their non-affected relatives as a presumed consequence of higher penetrance of the endophenotype compared to the disease phenotype. The initial idea behind their study was that endophenotypes may be the result of a subset of the many disease genes and may have higher expressivity, making it easier to identify these genes. This approach has been successfully used in diseases other than schizophrenia. A prominent example is the identification of genes for long QT syndrome, an endophenotype for syncope, ventricle arrhythmias, and sudden death [143, 144]. Other terms that have been used in place of endophenotype include “intermediate phenotype,” “biological marker,” “subclinical trait,” and “vulnerability marker” [145]. The concept is of particular interest for psychiatric diseases for the additional reason that it may provide an objectively measurable phenotype. It is closely related to the Research Domain Criteria (RDoC) in psychiatric research, an NIMH project to create a research framework for genomics and neuroscience aimed to eventually inform classification schemes [146]. Some of the major endophenotypes that have been studied for schizophrenia are sensory motor gating, eye-tracking dysfunction, working memory, and executive cognition [3].
There have been many studies looking for linkage and associations between schizophrenia endophenotypes and common genetic variation. The largest effort to investigate endophenotypes and identify linkage with schizophrenia has been the Consortium on the Genetics of Schizophrenia (COGS), a 7-site study funded by the National Institute of Mental Health. They have reported on the significant impairment of P50 inhibition [147], prepulse inhibition [149, 149], verbal declarative memory [150, 151], and working memory in schizophrenic patients and relatives compared to controls, and they identified smoking status as a moderator in the latter [152]. Positive but weaker results were also reported for the antisaccade task performance [153] and reduced auditory P300 amplitude [154]. A linkage analysis of their 12 phenotypes identified a LOD score of 4.0 on chromosome 3p14 for the antisaccade task but no other genome-wide significant result [155]. Heritability analysis in the COGS-1 family sample has shown comparable levels between endophenotypes and psychotic disorders [156]. More recently, the group has reported on new endophenotypes from measures derived from the original endophenotype tests finding nine to be significantly heritable and discriminate between schizophrenia patients and controls [157].
As GWAS are currently the state-of-the-art and provide much more reliable results, we will not report on the multiple candidate gene association studies. Unfortunately, GWAS for schizophrenia endophenotypes have generally been limited in sample size. One of the first GWAS examined 11 cognitive phenotypes in 750 subjects but did not find genome-wide significant associations [158]. Hatzimanolis et al.[159], in a GWAS on multiple endophenotypes in young male adults that also did not show genome-wide significant results, reported that schizophrenia polygenic risk influences neurocognitive performance, a result recapitulated by a study in childhood [160]. Roussos et al.[161] also showed that in addition to reduced cognition, increased PRS for schizophrenia was associated with reduced PPI validating this additional endophenotype. More recently, a meta-analysis by the Cognitive Genomics Consortium (COGENT) reported on a sample of 35,298 healthy individuals of European ancestry [162]. They found polygenic correlations between cognitive performance, educational attainment, several psychiatric disorders, birth length/weight, smoking behavior and the personality trait of openness. Importantly, they also reported on some specific loci significantly associated with general cognitive function. Finally, schizophrenia-derived polygenic scores have also been correlated with cortical gyrification, an additional potential schizophrenia endophenotype [163].
While endophenotypes were initially considered a way to facilitate disease gene discovery, the results suggest that their genetics are similarly complex. However, these results also highlight the value of polygenic scores in the study of the genetics of schizophrenia and the underlying polygenic architecture. With larger samples, we might be able to better understand their relationships to schizophrenia and how it relates to the underlying genetics. This multifaceted approach to disease may prove to be key in achieving individualized medicine and extending precision medicine into psychiatry.
Closing Remarks
The progress in the genetics of schizophrenia in the last decade has been remarkable. We used to have not a single known genetic risk variant or gene, and failure to replicate prior reports was the norm. That was a difficult period for psychiatric genetics investigators that still has consequences to the way some view our field. Today, we know of numerous robustly supported genetic variants, perhaps more than we hoped for, as well as CNVs, and we have strong evidence for the involvement of rare and de novo variants with stronger effects on the risk. To allow this progress, large consortia have undertaken the recruitment of cohorts of several thousands of subjects, whose genetic analysis has been made possible by technological advancements. As this important work continues, it is now time to make sense out of these hundreds of risk factors and see how they can aid the fight against disease.
Creating animal models for schizophrenia based on the results of GWAS is challenging. The diagnosis in humans is made by interview, which requires verbal interaction, and there is no objectively measurable phenotype such as a laboratory test. The GWAS variants are mostly non-coding, which limits our ability to accurately recapitulate them in other animals. In addition, even if the effect of the GWAS variants could be accurately mimicked, their resulting increase in risk is so small that such a mutant animal would be unlikely to show a phenotype. On the other hand, rare variants of larger effect have a level of uncertainty, as most studies report enrichments across many genes rather than definite involvement of individual variants. Studies in mice have employed knockout or knock-in strategies for a variety of genes with varying degrees of supporting evidence, and have examined schizophrenia-related behavioral domains and phenotypes similar to the endophenotypes described above. Discussing them is beyond the scope of this review, but there are many other excellent reviews of mouse models, the insights they have provided, and the associated challenges [164, 165, 166]. More recently, new approaches to disease models are beginning to show significant promise for schizophrenia research. The ability to manipulate human cells to induce pluripotency [167, 168, 169] and differentiation [170, 171] and the achievement of accurate and efficient genome editing [172, 173, 174] are advancements that may provide a powerful means to study the biology underlying the risk. The major advantage is that they allow us to work on a human genomic background and even to create human pluripotent cells that differ only at specific risk variants. These can be differentiated into the cell type of choice for phenotyping, or even used to generate and study brain organoids [175]. The applications one can imagine are only limited by what is ethical to do in a lab. The small effect sizes may still be a problem, but others and we have shown that when one works with modified or edited cells, small effect variants can lead to robust cellular phenotypes [103, 105, 139, 176]. These technologies are new, and there is still much space for improvement, including editing efficiency [177], uniform and specific differentiation [178, 179], the time needed for maturation, and the faithful recapitulation of cortical circuits by organoids [180]. Nevertheless, the genetic findings are there and the tools for us to understand them are rapidly evolving, giving hope for significant breakthroughs within the next decade.
Despite the successes and promise for the future, obstacles in the biological interpretation of GWAS signals in schizophrenia remain. It is significant to know of the genes and variants leading to risk, but given the low odds ratios, it is difficult to measure and resolve their downstream consequences that can lead to disease. While there is no longer doubt these variants, either themselves of those genotypically correlated to them, increase the risk, they are certainly not deterministic and cannot be referred to as causal. Nevertheless the number of such variants is large and increasing. Cellular models and genome editing may reveal aspects of their weak effects on neuronal homeostasis, and similarities between them might lead to the perturbed gene networks. Such discoveries are likely not far down the road and could provide direct targets for intervention.
The agnostic nature of this type of genetics research does not lend itself to evident narratives on the etiology of the disease, which is reflected in this review. At the same time though, it does not rely on existing knowledge allowing surprising discoveries and breakthroughs. It might be too early to make up the entire story from the current discoveries but, as opposed to just a decade ago, we are certainly on the way there.
Disclosure Statement
The author has no conflicts of interest to disclose.
Acknowledgements
The author would like to thank Kyra Feuer for reviewing the manuscript and providing important feedback for its improvement. The author is supported in part by R56MH113215 (PI) P50MH094268 (project 1 PI) and R01 MH106522 (PI: McCallion).
References
- 1.Jablensky A, Kirkbride JB, Jones PB. Schizophrenia: The epidemiological horizon. In: Weinberger DR, Harrison PJ, editors. Wiley-Blackwell. Schizophrenia: London; 2011. pp. pp 185–225. [Google Scholar]
- 2.American Psychiatric Association . Arlington: American Psychiatric Association; 2013. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- 3.Green MF. Impact of cognitive and social cognitive impairment on functional outcomes in patients with schizophrenia. J Clin Psychiatry. 2016;77((suppl 2)):8–11. doi: 10.4088/JCP.14074su1c.02. [DOI] [PubMed] [Google Scholar]
- 4.Lally J, MacCabe JH. Antipsychotic medication in schizophrenia: a review. Br Med Bull. 2015;114:169–179. doi: 10.1093/bmb/ldv017. [DOI] [PubMed] [Google Scholar]
- 5.Bleuler E. Giessen: Psychosozial-Verlag; 1911. Dementia Paecox oder die Gruppe der Schizophrenien. [Google Scholar]
- 6.McGue M, Gottesman II. The genetic epidemiology of schizophrenia and the design of linkage studies. Eur Arch Psychiatry Clin Neurosci. 1991;240:174–181. doi: 10.1007/BF02190760. [DOI] [PubMed] [Google Scholar]
- 7.Cannon TD, Kaprio J, Lönnqvist J, Huttunen M, Koskenvuo M, The genetic epidemiology of schizophrenia in a Finnish twin cohort A population-based modeling study. Arch Gen Psychiatry. 1998;55:67–74. doi: 10.1001/archpsyc.55.1.67. [DOI] [PubMed] [Google Scholar]
- 8.Pedersen CB, Cantor-Graae E. Age at migration and risk of schizophrenia among immigrants in Denmark: a 25-year incidence study. Am J Psychiatry. 2012;169:1117–1118. doi: 10.1176/appi.ajp.2012.12050614. [DOI] [PubMed] [Google Scholar]
- 9.Vassos E, Pedersen CB, Murray RM, Collier DA, Lewis CM. Meta-analysis of the association of urbanicity with schizophrenia. Schizophr Bull. 2012;38:1118–1123. doi: 10.1093/schbul/sbs096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.St Clair D, Xu M, Wang P, Yu Y, Fang Y, Zhang F, Zheng X, Gu N, Feng G, Sham P, He L. Rates of adult schizophrenia following prenatal exposure to the Chinese famine of 1959–1961. JAMA. 2005;294:557–562. doi: 10.1001/jama.294.5.557. [DOI] [PubMed] [Google Scholar]
- 11.Susser E, Neugebauer R, Hoek HW, Brown AS, Lin S, Labovitz D, Gorman JM. Schizophrenia after prenatal famine. Further evidence. Arch Gen Psychiatry. 1996;53:25–31. doi: 10.1001/archpsyc.1996.01830010027005. [DOI] [PubMed] [Google Scholar]
- 12.Davies G, Welham J, Chant D, Torrey EF, McGrath J. A systematic review and meta-analysis of Northern hemisphere season of birth studies in schizophrenia. Schizophr Bull. 2003;29:587–593. doi: 10.1093/oxfordjournals.schbul.a007030. [DOI] [PubMed] [Google Scholar]
- 13.Yolken RH, Torrey EF. Are some cases of psychosis caused by microbial agents? A review of the evidence. Mol Psychiatry. 2008;13:470–479. doi: 10.1038/mp.2008.5. [DOI] [PubMed] [Google Scholar]
- 14.Niebuhr DW, Millikan AM, Yolken R, Li Y, Weber NS. Results from a hypothesis generating case-control study: herpes family viruses and schizophrenia among military personnel. Schizophr Bull. 2008;34:1182–1188. doi: 10.1093/schbul/sbm139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Børglum AD, Demontis D, Grove J, Pallesen J, Hollegaard MV, Pedersen CB, Hedemand A, Mattheisen M; Group investigators10, Uitterlinden A, Nyegaard M, Ørntoft T, Wiuf C, Didriksen M, Nordentoft M, Nöthen MM, Rietschel M, Ophoff RA, Cichon S, Yolken RH, Hougaard DM, Mortensen PB, Mors O. Genome-wide study of association and interaction with maternal cytomegalovirus infection suggests new schizophrenia loci. Mol Psychiatry. 2014;19:325–333. doi: 10.1038/mp.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avramopoulos D, Pearce BD, McGrath J, Wolyniec P, Wang R, Eckart N, Hatzimanolis A, Goes FS, Nestadt G, Mulle JG, Coneely K, Hopkins M, Ruczinski I, Yolken RH, Pulver AE. Infection and inflammation in schizophrenia and bipolar disorder: a genome wide study for interactions with genetic variation. PLoS One. 2015;10 doi: 10.1371/journal.pone.0116696. e0116696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bundy H, Stahl D, MacCabe JH. A systematic review and meta-analysis of the fertility of patients with schizophrenia and their unaffected relatives. Acta Psychiatr Scand. 2011;123:98–106. doi: 10.1111/j.1600-0447.2010.01623.x. [DOI] [PubMed] [Google Scholar]
- 18.Mansour H, Kandil K, Wood J, Fathi W, Elassy M, Ibrahim I, Salah H, Yassin A, Elsayed H, Tobar S, El-Boraie H, Eissa A, Elhadidy M, Ibrahim NE, El-Bahaei W, Nimgaonkar VL. Reduced fertility and fecundity among patients with bipolar I disorder and schizophrenia in Egypt. Psychiatry Investig. 2011;8:214–220. doi: 10.4306/pi.2011.8.3.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haukka J, Suvisaari J, Lönnqvist J. Fertility of patients with schizophrenia, their siblings, and the general population: a cohort study from 1950 to 1959 in Finland. Am J Psychiatry. 2003;160:460–463. doi: 10.1176/appi.ajp.160.3.460. [DOI] [PubMed] [Google Scholar]
- 20.van Dongen J, Boomsma DI. The evolutionary paradox and the missing heritability of schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:122–136. doi: 10.1002/ajmg.b.32135. [DOI] [PubMed] [Google Scholar]
- 21.Keller MC, Miller G. Resolving the paradox of common, harmful, heritable mental disorders: which evolutionary genetic models work best? Behav Brain Sci. 2006;29 doi: 10.1017/S0140525X06009095. 385–404–452. [DOI] [PubMed] [Google Scholar]
- 22.Uher R. The role of genetic variation in the causation of mental illness: an evolution-informed framework. Mol Psychiatry. 2009;14:1072–1082. doi: 10.1038/mp.2009.85. [DOI] [PubMed] [Google Scholar]
- 23.Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, Lander ES. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottesman II, Shields J. Genetic theorizing and schizophrenia. Br J Psychiatry J Ment Sci. 1973;122:15–30. doi: 10.1192/bjp.122.1.15. [DOI] [PubMed] [Google Scholar]
- 25.Essen-Möller E. Evidence for polygenic inheritance in schizophrenia? Acta Psychiatr Scand. 1977;55:202–207. doi: 10.1111/j.1600-0447.1977.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 26.Odegaard O. Comments on the genetics issue. Schizophr Bull. 1977;3:345–347. doi: 10.1093/schbul/3.3.345. [DOI] [PubMed] [Google Scholar]
- 27.Elston RC, Kringlen E, Namboodiri KK. Possible linkage relationships between certain blood groups and schizophrenia or other psychoses. Behav Genet. 1973;3:101–106. doi: 10.1007/BF01067650. [DOI] [PubMed] [Google Scholar]
- 28.Chow EW, Bassett AS, Weksberg R. Velo-cardio-facial syndrome and psychotic disorders: implications for psychiatric genetics. Am J Med Genet. 1994;54:107–112. doi: 10.1002/ajmg.1320540205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindsay EA, Morris MA, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, Shprintzen R, Antonarakis SE, Baldini A, Pulver AE. Schizophrenia and chromosomal deletions within 22q11.2. Am J Hum Genet. 1995;56:1502–1503. [PMC free article] [PubMed] [Google Scholar]
- 30.Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci USA. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- 33.Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, Wadey R, Patanjali SR, Weissman SM, Anyane-Yeboa K, Warburton D, Scambler P, Shprintzen R, Kucherlapati R, Morrow BE. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am J Hum Genet. 1997;61:620–629. doi: 10.1086/515508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanov D, Kirov G, Norton N, Williams HJ, Williams NM, Nikolov I, Tzwetkova R, Stambolova SM, Murphy KC, Toncheva D, Thapar A, O'Donovan MC, Owen MJ. Chromosome 22q11 deletions, velo-cardio-facial syndrome and early-onset psychosis. Molecular genetic study. Br J Psychiatry J Ment Sci. 2003;183:409–413. doi: 10.1192/bjp.183.5.409. [DOI] [PubMed] [Google Scholar]
- 35.Rees E, Kirov G, Sanders A, Walters JTR, Chambert KD, Shi J, Szatkiewicz J, O'Dush laine C, Richards AL, Green EK, et al. Evidence that duplications of 22q11.2 protect against schizophrenia. Mol Psychiatry. 2014;19:37–40. doi: 10.1038/mp.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MAR, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu B-L, Daly MJ, Autism Consortium Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 37.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qureshi AY, Mueller S, Snyder AZ, Mukherjee P, Berman JI, Roberts TPL, Nagarajan SS, Spiro JE, Chung WK, Sherr EH, Buckner RL, Simons VIP Consortium Opposing brain differences in 16p11.2 deletion and duplication carriers. J Neurosci. 2014;34:11199–11211. doi: 10.1523/JNEUROSCI.1366-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang YS, Owen JP, Pojman NJ, Thieu T, Bukshpun P, Wakahiro MLJ, Marco EJ, Berman JI, Spiro JE, Chung WK, Buckner RL, Roberts TPL, Nagarajan SS, Sherr EH, Mukherjee P. Reciprocal white matter alterations due to 16p11.2 chromosomal deletions versus duplications. Hum Brain Mapp. 2016;37:2833–2848. doi: 10.1002/hbm.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinawi M, Liu P, Kang S-HL, Shen J, Belmont JW, Scott DA, Probst FJ, Craigen WJ, Graham BH, Pursley A, Clark G, Lee J, Proud M, Stocco A, Rodriguez DL, Kozel BA, Sparagana S, Roeder ER, McGrew SG, Kurczynski TW, Allison LJ, Amato S, Savage S, Patel A, Stankiewicz P, Beaudet AL, Cheung SW, Lupski JR. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LeBlanc JJ, Nelson CA. Deletion and duplication of 16p11.2 are associated with opposing effects on visual evoked potential amplitude. Mol Autism. 2016;7:30. doi: 10.1186/s13229-016-0095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, Kutalik Z, Martinet D, Shen Y, Valsesia A, Beckmann ND, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gladwyn-Ng I, Huang L, Ngo L, Li SS, Qu Z, Vanyai HK, Cullen HD, Davis JM, Heng JI-T. Bacurd1/Kctd13 and Bacurd2/Tnfaip1 are interacting partners to Rnd proteins which influence the long-term positioning and dendritic maturation of cerebral cortical neurons. Neural Dev. 2016;11:7. doi: 10.1186/s13064-016-0062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guha S, Rees E, Darvasi A, Ivanov D, Ikeda M, Bergen SE, Magnusson PK, Cormican P, Morris D, Gill M, et al. Implication of a rare deletion at distal 16p11.2 in schizophrenia. JAMA Psychiatry. 2013;70:253–260. doi: 10.1001/2013.jamapsychiatry.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barge-Schaapveld DQCM, Maas SM, Polstra A, Knegt LC, Hennekam RCM. The atypical 16p11.2 deletion: a not so atypical microdeletion syndrome? Am J Med Genet A. 2011;155A:1066–1072. doi: 10.1002/ajmg.a.33991. [DOI] [PubMed] [Google Scholar]
- 46.Cao X, Tabuchi K. Functions of synapse adhesion molecules neurexin/neuroligins and neurodevelopmental disorders. Neurosci Res. 2017;116:3–9. doi: 10.1016/j.neures.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 47.Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, O'Donovan MC, Erdogan F, Owen MJ, Ropers H-H, Ullmann R. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 48.Rujescu D, Ingason A, Cichon S, Pietiläinen OPH, Barnes MR, Toulopoulou T, Picchioni M, Vassos E, Ettinger U, Bramon E, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De Gregori M, Ciccone R, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stefansson H, Rujescu D, Cichon S, Pietiläinen OPH, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sinkus ML, Graw S, Freedman R, Ross RG, Lester HA, Leonard S. The human CHRNA7 and CHRFAM7A genes: a review of the genetics, regulation, and function. Neuropharmacology. 2015;96:274–288. doi: 10.1016/j.neuropharm.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dolcetti A, Silversides CK, Marshall CR, Lionel AC, Stavropoulos DJ, Scherer SW, Bassett AS. 1q21.1 Microduplication expression in adults. Genet Med. 2013;15:282–289. doi: 10.1038/gim.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willatt L, Cox J, Barber J, Cabanas ED, Collins A, Donnai D, FitzPatrick DR, Maher E, Martin H, Parnau J, Pindar L, Ramsay J, Shaw-Smith C, Sistermans EA, Tettenborn M, Trump D, de Vries BBA, Walker K, Raymond FL. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am J Hum Genet. 2005;77:154–160. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nava C, Keren B, Mignot C, Rastetter A, Chantot-Bastaraud S, Faudet A, Fonteneau E, Amiet C, Laurent C, Jacquette A, Whalen S, Afenjar A, Périsse D, Doummar D, Dorison N, Leboyer M, Siffroi J-P, Cohen D, Brice A, Héron D, Depienne C. Prospective diagnostic analysis of copy number variants using SNP microarrays in individuals with autism spectrum disorders. Eur J Hum Genet. 2014;22:71–78. doi: 10.1038/ejhg.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulle JG, Dodd AF, McGrath JA, Wolyniec PS, Mitchell AA, Shetty AC, Sobreira NL, Valle D, Rudd MK, Satten G, Cutler DJ, Pulver AE, Warren ST. Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet. 2010;87:229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grozeva D, Conrad DF, Barnes CP, Hurles M, Owen MJ, O'Donovan MC, Craddock N, Kirov G, WTCCC Independent estimation of the frequency of rare CNVs in the UK population confirms their role in schizophrenia. Schizophr Res. 2012;135:1–7. doi: 10.1016/j.schres.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, Zhang N, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Kendler KS, Freedman R, Dudbridge F, Pe'er I, Hakonarson H, Bergen SE, Fanous AH, Holmans PA, Gejman PV. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nickerson E, Greenberg F, Keating MT, McCaskill C, Shaffer LG. Deletions of the elastin gene at 7q11.23 occur in approximately 90% of patients with Williams syndrome. Am J Hum Genet. 1995;56:1156–1161. [PMC free article] [PubMed] [Google Scholar]
- 61.Depienne C, Heron D, Betancur C, Benyahia B, Trouillard O, Bouteiller D, Verloes A, LeGuern E, Leboyer M, Brice A. Autism, language delay and mental retardation in a patient with 7q11 duplication. J Med Genet. 2007;44:452–458. doi: 10.1136/jmg.2006.047092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 63.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, van de Lagemaat LN, Bayés A, Fernandez E, Olason PI, Böttcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SGN, Purcell S, Sklar P, O'Donovan MC, Owen MJ. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosden C, Evans HJ. Association within a family of a balanced autosomal translocation with major mental illness. Lancet Lond Engl. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 65.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 66.Sachs NA, Sawa A, Holmes SE, Ross CA, DeLisi LE, Margolis RL. A frameshift mutation in disrupted in schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Mol Psychiatry. 2005;10:758–764. doi: 10.1038/sj.mp.4001667. [DOI] [PubMed] [Google Scholar]
- 67.Narayan S, Nakajima K, Sawa A. DISC1: a key lead in studying cortical development and associated brain disorders. Neuroscientist. 2013;19:451–464. doi: 10.1177/1073858412470168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sullivan PF. Questions about DISC1 as a genetic risk factor for schizophrenia. Mol Psychiatry. 2013;18:1050–1052. doi: 10.1038/mp.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Porteous DJ, Thomson PA, Millar JK, Evans KL, Hennah W, Soares DC, McCarthy S, McCombie WR, Clapcote SJ, Korth C, Brandon NJ, Sawa A, Kamiya A, Roder JC, Lawrie SM, McIntosh AM, St Clair D, Blackwood DH. DISC1 as a genetic risk factor for schizophrenia and related major mental illness: response to Sullivan. Mol Psychiatry. 2014;19:141–143. doi: 10.1038/mp.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, Thibodeau P, Bachand I, Bao JYJ, Tong AHY, Lin C-H, Millet B, Jaafari N, Joober R, Dion PA, Lok S, Krebs M-O, Rouleau GA. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 72.Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, Levy S, Gogos JA, Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, Mistry M, Pavlidis P, Solomon R, Ghiban E, Antoniou E, Kelleher E, O'Brien C, Donohoe G, Gill M, Morris DW, McCombie WR, Corvin A. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takata A, Ionita-Laza I, Gogos JA, Xu B, Karayiorgou M. De novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron. 2016;89:940–947. doi: 10.1016/j.neuron.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H; Consortium on the Genetics of Schizophrenia (COGS), PAARTNERS Study Group, Nimgaonkar VL, Go RCP, Savage RM, Swerdlow NR, Gur RE, Braff DL, King M-C, McClellan JM. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Need AC, McEvoy JP, Gennarelli M, Heinzen EL, Ge D, Maia JM, Shianna KV, He M, Cirulli ET, Gumbs CE, Zhao Q, Campbell CR, Hong L, Rosenquist P, Putkonen A, Hallikainen T, Repo-Tiihonen E, Tiihonen J, Levy DL, Meltzer HY, Goldstein DB. Exome sequencing followed by large-scale genotyping suggests a limited role for moderately rare risk factors of strong effect in schizophrenia. Am J Hum Genet. 2012;91:303–312. doi: 10.1016/j.ajhg.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Timms AE, Dorschner MO, Wechsler J, Choi KY, Kirkwood R, Girirajan S, Baker C, Eichler EE, Korvatska O, Roche KW, Horwitz MS, Tsuang DW. Support for the N-methyl-D-aspartate receptor hypofunction hypothesis of schizophrenia from exome sequencing in multiplex families. JAMA Psychiatry. 2013;70:582–590. doi: 10.1001/jamapsychiatry.2013.1195. [DOI] [PubMed] [Google Scholar]
- 80.Ruderfer DM, Lim ET, Genovese G, Moran JL, Hultman CM, Sullivan PF, McCarroll SA, Holmans P, Sklar P, Purcell SM. No evidence for rare recessive and compound heterozygous disruptive variants in schizophrenia. Eur J Hum Genet. 2015;23:555–557. doi: 10.1038/ejhg.2014.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rees E, Kirov G, Walters JT, Richards AL, Howrigan D, Kavanagh DH, Pocklington AJ, Fromer M, Ruderfer DM, Georgieva L, et al. Analysis of exome sequence in 604 trios for recessive genotypes in schizophrenia. Transl Psychiatry. 2015;5 doi: 10.1038/tp.2015.99. e607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guipponi M, Santoni FA, Setola V, Gehrig C, Rotharmel M, Cuenca M, Guillin O, Dikeos D, Georgantopoulos G, Papadimitriou G, Curtis L, Méary A, Schürhoff F, Jamain S, Avramopoulos D, Leboyer M, Rujescu D, Pulver A, Campion D, Siderovski DP, Antonarakis SE. Exome sequencing in 53 sporadic cases of schizophrenia identifies 18 putative candidate genes. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112745. e112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O'Dushlaine C, Chambert K, Bergen SE, Kähler A, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landén M, Moran JL, Purcell SM, Sklar P, Sullivan PF, Hultman CM, McCarroll SA. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19:1433–1441. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leonenko G, Richards AL, Walters JT, Pocklington A, Chambert K, Al Eissa MM, Sharp SI, O'Brien NL, Curtis D, Bass NJ, McQuillin A, Hultman C, Moran JL, McCarroll SA, Sklar P, Neale BM, Holmans PA, Owen MJ, Sullivan PF, O'Donovan MC. Mutation intolerant genes and targets of FMRP are enriched for nonsynonymous alleles in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2017;174:724–731. doi: 10.1002/ajmg.b.32560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J, Suvisaari J, Chheda H, Blackwood D, Breen G, et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci. 2016;19:571–577. doi: 10.1038/nn.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 88.O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 89.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietiläinen OPH, Mors O, Mortensen PB, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.International Schizophrenia Consortium; Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Z, Chen J, Yu H, He L, Xu Y, Zhang D, Yi Q, Li C, Li X, Shen J, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49:1576–1583. doi: 10.1038/ng.3973. [DOI] [PubMed] [Google Scholar]
- 94.Maher BS. Polygenic scores in epidemiology: risk prediction, etiology, and clinical utility. Curr Epidemiol Rep. 2015;2:239–244. doi: 10.1007/s40471-015-0055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hamshere ML, Stergiakouli E, Langley K, Martin J, Holmans P, Kent L, Owen MJ, Gill M, Thapar A, O'Donovan M, Craddock N. Shared polygenic contribution between childhood attention-deficit hyperactivity disorder and adult schizophrenia. Br J Psychiatry J Ment Sci. 2013;203:107–111. doi: 10.1192/bjp.bp.112.117432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cross-Disorder Group of the Psychiatric Genomics Consortium; Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, Mowry BJ, Thapar A, Goddard ME, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sumner JA, Duncan L, Ratanatharathorn A, Roberts AL, Koenen KC. PTSD has shared polygenic contributions with bipolar disorder and schizophrenia in women. Psychol Med. 2016;46:669–671. doi: 10.1017/S0033291715002135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reginsson GW, Ingason A, Euesden J, Bjornsdottir G, Olafsson S, Sigurdsson E, Oskarsson H, Tyrfingsson T, Runarsdottir V, Hansdottir I, Steinberg S, Stefansson H, Gudbjartsson DF, Thorgeirsson TE, Stefansson K. Polygenic risk scores for schizophrenia and bipolar disorder associate with addiction. Addict Biol. 2018;23:485–492. doi: 10.1111/adb.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neilson E, Bois C, Gibson J, Duff B, Watson A, Roberts N, Brandon NJ, Dunlop J, Hall J, McIntosh AM, Whalley HC, Lawrie SM. Effects of environmental risks and polygenic loading for schizophrenia on cortical thickness. Schizophr Res. 2017;184:128–136. doi: 10.1016/j.schres.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 100.Power RA, Steinberg S, Bjornsdottir G, Rietveld CA, Abdellaoui A, Nivard MM, Johannesson M, Galesloot TE, Hottenga JJ, Willemsen G, Cesarini D, Benjamin DJ, Magnusson PKE, Ullén F, Tiemeier H, Hofman A, van Rooij FJA, Walters GB, Sigurdsson E, Thorgeirsson TE, Ingason A, Helgason A, Kong A, Kiemeney LA, Koellinger P, Boomsma DI, Gudbjartsson D, Stefansson H, Stefansson K. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci. 2015;18:953–955. doi: 10.1038/nn.4040. [DOI] [PubMed] [Google Scholar]
- 101.Forstner AJ, Hecker J, Hofmann A, Maaser A, Reinbold CS, Mühleisen TW, Leber M, Strohmaier J, Degenhardt F, Treutlein J, et al. Identification of shared risk loci and pathways for bipolar disorder and schizophrenia. PLoS One. 2017;12 doi: 10.1371/journal.pone.0171595. e0171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mistry S, Harrison JR, Smith DJ, Escott-Price V, Zammit S. The use of polygenic risk scores to identify phenotypes associated with genetic risk of schizophrenia: systematic review. Schizophr Res. doi: 10.1016/j.schres.2017.10.037. DOI: 10.1016/j.schres.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 103.Chen J, Lin M, Hrabovsky A, Pedrosa E, Dean J, Jain S, Lachman HM. ZNF804A transcriptional networks in differentiating neurons derived from induced pluripotent stem cells of human origin. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124597. e0124597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hill MJ, Jeffries AR, Dobson RJB, Price J, Bray NJ. Knockdown of the psychosis susceptibility gene ZNF804A alters expression of genes involved in cell adhesion. Hum Mol Genet. 2012;21:1018–1024. doi: 10.1093/hmg/ddr532. [DOI] [PubMed] [Google Scholar]
- 105.Hill MJ, Killick R, Navarrete K, Maruszak A, McLaughlin GM, Williams BP, Bray NJ. Knockdown of the schizophrenia susceptibility gene TCF4 alters gene expression and proliferation of progenitor cells from the developing human neocortex. J Psychiatry Neurosci Jpn. 2017;42:181–188. doi: 10.1503/jpn.160073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eckart N, Song Q, Yang R, Wang R, Zhu H, McCallion AS, Avramopoulos D. Functional characterization of schizophrenia-associated variation in CACNA1C. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157086. e0157086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roussos P, Mitchell AC, Voloudakis G, Fullard JF, Pothula VM, Tsang J, Stahl EA, Georgakopoulos A, Ruderfer DM, Charney A, Okada Y, Siminovitch KA, Worthington J, Padyukov L, Klareskog L, Gregersen PK, Plenge RM, Raychaudhuri S, Fromer M, Purcell SM, Brennand KJ, Robakis NK, Schadt EE, Akbarian S, Sklar P. A role for noncoding variation in schizophrenia. Cell Rep. 2014;9:1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, Rose SA, Handsaker RE; Schizophrenia Working Group of the Psychiatric Genomics Consortium, Daly MJ, Carroll MC, Stevens B, McCarroll SA. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Feinberg I. Schizophrenia: caused by a fault in programmed synaptic elimination during adolescence? J Psychiatr Res. 1982;17:319–334. doi: 10.1016/0022-3956(82)90038-3. [DOI] [PubMed] [Google Scholar]
- 110.Mayilyan KR, Weinberger DR, Sim RB. The complement system in schizophrenia. Drug News Perspect. 2008;21:200–210. doi: 10.1358/dnp.2008.21.4.1213349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jordan B. Historical background and anticipated developments. Ann NY Acad Sci. 2002;975:24–32. doi: 10.1111/j.1749-6632.2002.tb05938.x. [DOI] [PubMed] [Google Scholar]
- 112.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 113.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT-J, Griffin JL, Wayland M, Freeman T, Dudbridge F, Lilley KS, Karp NA, Hester S, Tkachev D, Mimmack ML, Yolken RH, Webster MJ, Torrey EF, Bahn S. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. doi: 10.1038/sj.mp.4001511. 643. [DOI] [PubMed] [Google Scholar]
- 115.Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, Young TA, Bullard J, Yokoe H, Webster MJ, Knable MB, Brockman JA. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 116.Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- 117.Roussos P, Katsel P, Davis KL, Siever LJ, Haroutunian V. A system-level transcriptomic analysis of schizophrenia using postmortem brain tissue samples. Arch Gen Psychiatry. 2012;69:1205–1213. doi: 10.1001/archgenpsychiatry.2012.704. [DOI] [PubMed] [Google Scholar]
- 118.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mitkus SN, Hyde TM, Vakkalanka R, Kolachana B, Weinberger DR, Kleinman JE, Lipska BK. Expression of oligodendrocyte-associated genes in dorsolateral prefrontal cortex of patients with schizophrenia. Schizophr Res. 2008;98:129–138. doi: 10.1016/j.schres.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martins-de-Souza D. Proteome and transcriptome analysis suggests oligodendrocyte dysfunction in schizophrenia. J Psychiatr Res. 2010;44:149–156. doi: 10.1016/j.jpsychires.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 121.Yu H, Bi W, Liu C, Zhao Y, Zhang D, Yue W. A hypothesis-driven pathway analysis reveals myelin-related pathways that contribute to the risk of schizophrenia and bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2014;51:140–145. doi: 10.1016/j.pnpbp.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 122.Arion D, Unger T, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanders AR, Göring HHH, Duan J, Drigalenko EI, Moy W, Freda J, He D, Shi J; MGS, Gejman PV. Transcriptome study of differential expression in schizophrenia. Hum Mol Genet. 2013;22:5001–5014. doi: 10.1093/hmg/ddt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sanders AR, Drigalenko EI, Duan J, Moy W, Freda J, Göring HHH, Gejman PV. Transcriptome sequencing study implicates immune-related genes differentially expressed in schizophrenia: new data and a meta-analysis. Transl Psychiatry. 2017;7 doi: 10.1038/tp.2017.47. e1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sainz J, Mata I, Barrera J, Perez-Iglesias R, Varela I, Arranz MJ, Rodriguez MC, Crespo-Facorro B. Inflammatory and immune response genes have significantly altered expression in schizophrenia. Mol Psychiatry. 2013;18:1056–1057. doi: 10.1038/mp.2012.165. [DOI] [PubMed] [Google Scholar]
- 126.Mistry M, Gillis J, Pavlidis P. Meta-analysis of gene coexpression networks in the post-mortem prefrontal cortex of patients with schizophrenia and unaffected controls. BMC Neurosci. 2013;14:105. doi: 10.1186/1471-2202-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hess JL, Tylee DS, Barve R, de Jong S, Ophoff RA, Kumarasinghe N, Tooney P, Schall U, Gardiner E, Beveridge NJ, Scott RJ, Yasawardene S, Perera A, Mendis J, Carr V, Kelly B, Cairns M, Neurobehavioural Genetics Unit, Tsuang MT, Glatt SJ. Transcriptome-wide mega-analyses reveal joint dysregulation of immunologic genes and transcription regulators in brain and blood in schizophrenia. Schizophr Res. 2016;176:114–124. doi: 10.1016/j.schres.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim S, Hwang Y, Lee D, Webster MJ. Transcriptome sequencing of the choroid plexus in schizophrenia. Transl Psychiatry. 2016;6 doi: 10.1038/tp.2016.229. e964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hashimoto T, Arion D, Unger T, Maldonado-Avilés JG, Morris HM, Volk DW, Mirnics K, Lewis DA. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cohen OS, Mccoy SY, Middleton FA, Bialosuknia S, Zhang-James Y, Liu L, Tsuang MT, Faraone SV, Glatt SJ. Transcriptomic analysis of postmortem brain identifies dysregulated splicing events in novel candidate genes for schizophrenia. Schizophr Res. 2012;142:188–199. doi: 10.1016/j.schres.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oldmeadow C, Mossman D, Evans T-J, Holliday EG, Tooney PA, Cairns MJ, Wu J, Carr V, Attia JR, Scott RJ. Combined analysis of exon splicing and genome wide polymorphism data predict schizophrenia risk loci. J Psychiatr Res. 2014;52:44–49. doi: 10.1016/j.jpsychires.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 132.Takata A, Matsumoto N, Kato T. Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat Commun. 2017;8:14519. doi: 10.1038/ncomms14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Birnbaum R, Jaffe AE, Chen Q, Hyde TM, Kleinman JE, Weinberger DR. Investigation of the prenatal expression patterns of 108 schizophrenia-associated genetic loci. Biol Psychiatry. 2015;77:e43–e451. doi: 10.1016/j.biopsych.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 134.Ohi K, Shimada T, Nitta Y, Kihara H, Okubo H, Uehara T, Kawasaki Y. Specific gene expression patterns of 108 schizophrenia-associated loci in cortex. Schizophr Res. 2016;174:35–38. doi: 10.1016/j.schres.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 135.Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR, Kleinman JE. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci. 2016;19:40–47. doi: 10.1038/nn.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ellis SE, Panitch R, West AB, Arking DE. Transcriptome analysis of cortical tissue reveals shared sets of downregulated genes in autism and schizophrenia. Transl Psychiatry. 2016;6 doi: 10.1038/tp.2016.87. e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, Schork AJ, Appadurai V, Buil A, Werge TM, Liu C, White KP; CommonMind Consortium, PsychENCODE Consortium, iPSYCH-BROAD Working Group, Horvath S, Geschwind DH. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–697. doi: 10.1176/appi.focus.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pham X, Song G, Lao S, Goff L, Zhu H, Valle D, Avramopoulos D. The DPYSL2 gene connects mTOR and schizophrenia. Transl Psychiatry. 2016;6 doi: 10.1038/tp.2016.204. e933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fallin MD, Lasseter VK, Liu Y, Avramopoulos D, McGrath J, Wolyniec PS, Nestadt G, Liang K-Y, Chen P-L, Valle D, Pulver AE. Linkage and association on 8p21.2-p21.1 in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2011;156:188–197. doi: 10.1002/ajmg.b.31154. [DOI] [PubMed] [Google Scholar]
- 141.Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, Steel G, Nestadt G, Liang K-Y, Huganir RL, Valle D, Pulver AE. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77:918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu Y, Pham X, Zhang L, Burzynski G, McGaughey DM, He S, McGrath JA, Wolyniec P, Fallin MD, Pierce MS, McCallion AS, Pulver AE, Avramopoulos D, Valle D. Functional variants in DPYSL2 sequence increase schizophrenia risk and suggest a link to mTOR signaling. G3 (Bethesda) 2014;5:61–72. doi: 10.1534/g3.114.015636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Keating M, Atkinson D, Dunn C, Timothy K, Vincent GM, Leppert M. Linkage of a cardiac arrhythmia, the long QT syndrome, and the Harvey ras-1 gene. Science. 1991;252:704–706. doi: 10.1126/science.1673802. [DOI] [PubMed] [Google Scholar]
- 144.Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, Sotoodehnia N, Rossin EJ, Morley M, Wang X, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46:826–836. doi: 10.1038/ng.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 146.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 147.Olincy A, Braff DL, Adler LE, Cadenhead KS, Calkins ME, Dobie DJ, Green MF, Greenwood TA, Gur RE, Gur RC, Light GA, Mintz J, Nuechterlein KH, Radant AD, Schork NJ, Seidman LJ, Siever LJ, Silverman JM, Stone WS, Swerdlow NR, Tsuang DW, Tsuang MT, Turetsky BI, Wagner BD, Freedman R. Inhibition of the P50 cerebral evoked response to repeated auditory stimuli: results from the Consortium on Genetics of Schizophrenia. Schizophr Res. 2010;119:175–182. doi: 10.1016/j.schres.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Swerdlow NR, Light GA, Sprock J, Calkins ME, Green MF, Greenwood TA, Gur RE, Gur RC, Lazzeroni LC, Nuechterlein KH, Radant AD, Ray A, Seidman LJ, Siever LJ, Silverman JM, Stone WS, Sugar CA, Tsuang DW, Tsuang MT, Turetsky BI, Braff DL. Deficient prepulse inhibition in schizophrenia detected by the multi-site COGS. Schizophr Res. 2014;152:503–512. doi: 10.1016/j.schres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Swerdlow NR, Light GA, Thomas ML, Sprock J, Calkins ME, Green MF, Greenwood TA, Gur RE, Gur RC, Lazzeroni LC, Nuechterlein KH, Radant AD, Seidman LJ, Siever LJ, Silverman JM, Stone WS, Sugar CA, Tsuang DW, Tsuang MT, Turetsky BI, Braff DL. Deficient prepulse inhibition in schizophrenia in a multi-site cohort: Internal replication and extension. Schizophr Res. doi: 10.1016/j.schres.2017.05.013. DOI: 10.1016/j.schres.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stone WS, Giuliano AJ, Tsuang MT, Braff DL, Cadenhead KS, Calkins ME, Dobie DJ, Faraone SV, Freedman R, Green MF, Greenwood TA, Gur RE, Gur RC, Light GA, Mintz J, Nuechterlein KH, Olincy A, Radant AD, Roe AH, Schork NJ, Siever LJ, Silverman JM, Swerdlow NR, Thomas AR, Tsuang DW, Turetsky BI, Seidman LJ. Group and site differences on the California Verbal Learning Test in persons with schizophrenia and their first-degree relatives: findings from the Consortium on the Genetics of Schizophrenia (COGS) Schizophr Res. 2011;128:102–110. doi: 10.1016/j.schres.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stone WS, Mesholam-Gately RJ, Braff DL, Calkins ME, Freedman R, Green MF, Greenwood TA, Gur RE, Gur RC, Lazzeroni LC, Light GA, Nuechterlein KH, Olincy A, Radant AD, Siever LJ, Silverman JM, Sprock J, Sugar CA, Swerdlow NR, Tsuang DW, Tsuang MT, Turetsky BI, Seidman LJ. California Verbal Learning Test-II performance in schizophrenia as a function of ascertainment strategy: comparing the first and second phases of the Consortium on the Genetics of Schizophrenia (COGS) Schizophr Res. 2015;163:32–37. doi: 10.1016/j.schres.2014.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee J, Green MF, Calkins ME, Greenwood TA, Gur RE, Gur RC, Lazzeroni LC, Light GA, Nuechterlein KH, Radant AD, Seidman LJ, Siever LJ, Silverman JM, Sprock J, Stone WS, Sugar CA, Swerdlow NR, Tsuang DW, Tsuang MT, Turetsky BI, Braff DL. Verbal working memory in schizophrenia from the Consortium on the Genetics of Schizophrenia (COGS) study: the moderating role of smoking status and antipsychotic medications. Schizophr Res. 2015;163:24–31. doi: 10.1016/j.schres.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Radant AD, Dobie DJ, Calkins ME, Olincy A, Braff DL, Cadenhead KS, Freedman R, Green MF, Greenwood TA, Gur RE, Gur RC, Light GA, Meichle SP, Millard SP, Mintz J, Nuechterlein KH, Schork NJ, Seidman LJ, Siever LJ, Silverman JM, Stone WS, Swerdlow NR, Tsuang MT, Turetsky BI, Tsuang DW. Antisaccade performance in schizophrenia patients, their first-degree biological relatives, and community comparison subjects: data from the COGS study. Psychophysiology. 2010;47:846–856. doi: 10.1111/j.1469-8986.2010.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Turetsky BI, Dress EM, Braff DL, Calkins ME, Green MF, Greenwood TA, Gur RE, Gur RC, Lazzeroni LC, Nuechterlein KH, Radant AD, Seidman LJ, Siever LJ, Silverman JM, Sprock J, Stone WS, Sugar CA, Swerdlow NR, Tsuang DW, Light G. The utility of P300 as a schizophrenia endophenotype and predictive biomarker: clinical and socio-demographic modulators in COGS-2. Schizophr Res. 2015;163:53–62. doi: 10.1016/j.schres.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Greenwood TA, Swerdlow NR, Gur RE, Cadenhead KS, Calkins ME, Dobie DJ, Freedman R, Green MF, Gur RC, Lazzeroni LC, Nuechterlein KH, Olincy A, Radant AD, Ray A, Schork NJ, Seidman LJ, Siever LJ, Silverman JM, Stone WS, Sugar CA, Tsuang DW, Tsuang MT, Turetsky BI, Light GA, Braff DL. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry. 2013;170:521–532. doi: 10.1176/appi.ajp.2012.12020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Light G, Greenwood TA, Swerdlow NR, Calkins ME, Freedman R, Green MF, Gur RE, Gur RC, Lazzeroni LC, Nuechterlein KH, Olincy A, Radant AD, Seidman LJ, Siever LJ, Silverman JM, Sprock J, Stone WS, Sugar CA, Tsuang DW, Tsuang MT, Turetsky BI, Braff DL. Comparison of the heritability of schizophrenia and endophenotypes in the COGS-1 family study. Schizophr Bull. 2014;40:1404–1411. doi: 10.1093/schbul/sbu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Greenwood TA, Lazzeroni LC, Calkins ME, Freedman R, Green MF, Gur RE, Gur RC, Light GA, Nuechterlein KH, Olincy A, Radant AD, Seidman LJ, Siever LJ, Silverman JM, Stone WS, Sugar CA, Swerdlow NR, Tsuang DW, Tsuang MT, Turetsky BI, Braff DL. Genetic assessment of additional endophenotypes from the Consortium on the Genetics of Schizophrenia Family Study. Schizophr Res. 2016;170:30–40. doi: 10.1016/j.schres.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KL, Hunt P, Ge D, Heinzen EL, Maia JM, Shianna KV, Weale ME, Cherkas LF, Clement G, Spector TD, Gibson G, Goldstein DB. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Hum Mol Genet. 2009;18:4650–4661. doi: 10.1093/hmg/ddp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hatzimanolis A, Bhatnagar P, Moes A, Wang R, Roussos P, Bitsios P, Stefanis CN, Pulver AE, Arking DE, Smyrnis N, Stefanis NC, Avramopoulos D. Common genetic variation and schizophrenia polygenic risk influence neurocognitive performance in young adulthood. Am J Med Genet B Neuropsychiatr Genet. 2015;168B:392–401. doi: 10.1002/ajmg.b.32323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hubbard L, Tansey KE, Rai D, Jones P, Ripke S, Chambert KD, Moran JL, McCarroll SA, Linden DEJ, Owen MJ, O'Donovan MC, Walters JTR, Zammit S. Evidence of common genetic overlap between schizophrenia and cognition. Schizophr Bull. 2016;42:832–842. doi: 10.1093/schbul/sbv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Roussos P, Giakoumaki SG, Zouraraki C, Fullard JF, Karagiorga V-E, Tsapakis E-M, Petraki Z, Siever LJ, Lencz T, Malhotra A, Bitsios P. The relationship of common risk variants and polygenic risk for schizophrenia to sensorimotor gating. Biol Psychiatry. 2016;79:988–996. doi: 10.1016/j.biopsych.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 162.Trampush JW, Yang MLZ, Yu J, Knowles E, Davies G, Liewald DC, Starr JM, Djurovic S, Melle I, Sundet K, et al. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol Psychiatry. 2017;22:336–345. doi: 10.1038/mp.2016.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu B, Zhang X, Cui Y, Qin W, Tao Y, Li J, Yu C, Jiang T. Polygenic risk for schizophrenia influences cortical gyrification in 2 independent general populations. Schizophr Bull. 2017;43:673–680. doi: 10.1093/schbul/sbw051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kvajo M, McKellar H, Gogos JA. Avoiding mouse traps in schizophrenia genetics: lessons and promises from current and emerging mouse models. Neuroscience. 2012;211:136–164. doi: 10.1016/j.neuroscience.2011.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kannan G, Sawa A, Pletnikov MV. Mouse models of gene-environment interactions in schizophrenia. Neurobiol Dis. 2013;57:5–11. doi: 10.1016/j.nbd.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pletnikov MV. Inducible and conditional transgenic mouse models of schizophrenia. Prog Brain Res. 2009;179:35–47. doi: 10.1016/S0079-6123(09)17905-0. [DOI] [PubMed] [Google Scholar]
- 167.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 168.Park I-H, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 169.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Südhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2 doi: 10.7554/eLife.00471. e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Muzio L, Consalez GG. Modeling human brain development with cerebral organoids. Stem Cell Res Ther. 2013;4:154. doi: 10.1186/scrt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang P, Lin M, Pedrosa E, Hrabovsky A, Zhang Z, Guo W, Lachman HM, Zheng D. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in neurodevelopment. Mol Autism. 2015;6:55. doi: 10.1186/s13229-015-0048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Waddington SN, Privolizzi R, Karda R, O'Neill HC. A broad overview and review of CRISPR-Cas technology and stem cells. Curr Stem Cell Rep. 2016;2:9–20. doi: 10.1007/s40778-016-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Panchision DM. Meeting report: using stem cells for biological and therapeutics discovery in mental illness, April 2012. Stem Cells Transl Med. 2013;2:217–222. doi: 10.5966/sctm.2012-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Panchision DM. Concise review: progress and challenges in using human stem cells for biological and therapeutics discovery: neuropsychiatric disorders. Stem Cells. 2016;34:523–536. doi: 10.1002/stem.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci. 2017;18:573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]