Abstract
There is evidence for systemic metabolic impairment in Alzheimer’s disease (AD), and type 2 diabetes (T2D) increases AD risk. Although studies analyzing blood metabolomics signatures have shown differences between cognitively healthy (CH) and AD subjects, these signatures have not been compared with individuals with T2D. We utilized untargeted analysis platforms (primary metabolism and complex lipids) to characterize the serum metabolome of 126 overnight-fasted elderly subjects classified into four groups based upon AD status (CH or AD) and T2D status [nondiabetic (ND) or T2D]. Cognitive diagnosis groups were a priori weighted equally with T2D subjects. We hypothesized that AD subjects would display a metabolic profile similar to cognitively normal elderly individuals with T2D. However, partial least squares-discriminant analysis (PLS-DA) modeling resulted in poor classification across the four groups (<50% classification accuracy of test subjects). Binary classification of AD vs. CH was poor, but binary classification of T2D vs. ND was good, providing >79.5% and >76.9% classification accuracy for held-out samples using primary metabolism and complex lipids, respectively. When modeling was limited to CH subjects, T2D discrimination improved for the primary metabolism platform (>89.5%) and remained accurate for complex lipids (>73% accuracy). Greater abundances of glucose, fatty acids (C20:2), and phosphatidylcholines and lower abundances of glycine, maleimide, octanol, and tryptophan, cholesterol esters, phosphatidylcholines, and sphingomyelins were identified in CH subjects with T2D relative to those without T2D. In contrast, T2D was not accurately discriminated within AD subjects. Results herein suggest that AD may obscure the typical metabolic phenotype of T2D.
Keywords: Alzheimer’s disease, diabetes, metabolomics
INTRODUCTION
Substantial evidence indicates that type 2 diabetes (T2D) is a risk factor for Alzheimer’s disease (AD) (3, 8, 20, 46, 47, 73, 74). Although AD is the most common neurodegenerative disease in the elderly, it currently cannot be slowed, cured, or prevented (2). We and others have shown that insulin resistance and prediabetes are linked to accelerated clinical progression, cognitive decline, and brain atrophy early in the AD process (42, 64). However, it is unclear whether glucose, alterations in related metabolic hormones such as insulin or incretins, or other related pathways and metabolites such as lipids contribute to the observed associations between diabetes status and AD.
Recent plasma metabolomics studies have found differences between AD patients and cognitively normal subjects (49, 50), but the mechanistic underpinnings of the relationships between metabolites and AD remain unclear. Amyloid deposition in the brain is a primary pathological feature of AD development (5). Metabolites in the form of lipid rafts play a key role in the processing of amyloid precursor protein, and alterations in lipid raft composition can affect the amount of pathological amyloid beta (Aβ) protein that is generated (11). In addition, there is evidence from rodent models that the gene associated with greater risk for AD, apolipoprotein ε4 (APOE4), may elicit changes in membrane phospholipids that are related to cognitive deficits (77). Importantly, in human subjects, specific plasma and serum metabolomics signatures have been associated with AD risk, progression, brain atrophy, and neuropathology (33, 34, 53, 60), and are suggested to discern mild cognitive impairment from AD (71). Broad groups of metabolites are altered during different disease stages, such as preclinical vs. symptomatic AD (60).
Given these relationships, it is possible that other factors related to AD risk, such as insulin resistance and T2D, contribute to metabolic abnormalities observed with AD. Metabolomics technologies have recently elucidated disturbances in metabolic pathways not traditionally associated with insulin resistance and T2D (e.g., branched-chain amino acids) (12, 14, 65–67). However, to our knowledge, no studies have attempted to parse out the independent effects of T2D and AD using metabolomics approaches. T2D is known to affect metabolic pathways involving both glucose and lipid metabolism (52). Although T2D increases AD risk (8, 20, 46, 47, 74), not all AD subjects have T2D, and the relationship between metabolic pathways and AD is likely complex. Other AD risk factors, including APOE4 (61) and vascular dysfunction (24), may affect metabolic pathways such as lipid metabolism (44, 48, 51, 68), independent of hyperglycemia and T2D (40). Thus we postulated that the serum metabolomics phenotype would be similarly altered among cognitively normal T2D subjects and AD subjects regardless of T2D status, but distinct from cognitively normal subjects without T2D. Furthermore, the impact of T2D on global metabolism in the elderly, regardless of cognitive health, remains to be fully elucidated. Therefore, in this study we utilized multiple metabolomics technologies (untargeted metabolomics of primary metabolism and complex lipids) to comprehensively examine the serum metabolome of cognitively normal and AD subjects with and without T2D.
MATERIALS AND METHODS
This study was approved by the University of Kansas Medical Center’s Institutional Review Board. The project was performed in accordance with the Declaration of Helsinki, and all study participants provided informed consent according to institutional guidelines. All participants in the study were recruited from the University of Kansas Alzheimer’s Disease Center (KU ADC) clinical cohort.
Sample Collection and Categorization
We divided 126 subjects into four groups based on cognitive and metabolic classifications. All subjects were cognitively evaluated by a clinician for stratification by cognitive diagnosis. The cognitive evaluation included a Clinical Dementia Rating (CDR) and neuropsychometric testing. Diagnosis of no dementia (cognitively healthy) or probable Alzheimer’s disease (AD) was subsequently confirmed at a consensus diagnosis conference. All cognitively normal subjects had a CDR of 0 and no clinically significant neuropsychometric deficits. AD subjects all had a CDR of 0.5 or higher, and a diagnosis of either mild cognitive impairment with AD as the primary etiology or a diagnosis of dementia with AD as the primary etiology. Individuals were considered as T2D if they met the World Health Organization (WHO) fasting glucose criterion (fasting glucose > 126 mg/dl) and/or had a current clinical diagnosis of T2D. All subjects classified as T2D had a clinical diagnosis of T2D except for two subjects, who met WHO fasting glucose criteria for T2D diagnosis. Metabolically healthy subjects had no current T2D diagnosis or past history of T2D and a fasting glucose value in the normal range according to WHO criteria.
Individuals reported to the KU Clinical and Translational Science Unit after an overnight fast. Vital signs and anthropometric measures were documented. Body mass was measured using a digital scale accurate to 0.1 kg (Seca Platform Scale, model 707), and body composition was assessed by dual-energy X-ray absorptiometry (DXA, Lunar Prodigy, version 11.2068). Blood was drawn (typically from the antecubital vein) into red top vacutainer tubes containing clot activator and centrifuged at 1,200 g for 12 min. Serum was carefully removed and frozen into multiple aliquots. Glucose and lactate were measured using a YSI 2300 Glucose and Lactate analyzer, whereas insulin (Genway), amylin (Millipore), and C-peptide (ALPCO) were assessed using ELISA methods. Insulin resistance was estimated using HOMA-IR.
Metabolomics Analyses
Metabolomics analyses were conducted by the West Coast Metabolomics Center (WCMC) at the University of California, Davis. Serum samples were shipped on dry ice from KUMC to WCMC. In-depth details of all protocols have been previously published (6, 15) and are summarized below.
Primary metabolism.
Serum samples were thawed and 15 µl aliquoted and then extracted using 1 ml of degassed acetonitrile:isopropanol:water (3:3:2; vol/vol/vol) at −20°C, centrifuged, decanted, and then dried. Membrane lipids and triglycerides were removed by adding 500 µl acetonitrile:water (1:1; vol/vol) and then evaporating to dryness. A set of internal standards [13 C8–C30 fatty acid methyl esters (FAMEs)] were added as a set of retention markers across chromatograms. These FAMEs were used to control for overall detector sensitivity, column performance, and injector performance. Technical details of FAMEs have been described in detail elsewhere (13). Following the addition of FAMEs, samples were derivatized by adding 10 µl of methoxyamine hydrochloride in pyridine and 90 µl MSTFA for trimethylsilylation of acidic protons. Samples were injected into an Agilent 6890 gas chromatograph and separated using a 30-m-long, 0.25-mm-ID Rtx5Sil- column at a constant flow of 1 ml/min while ramping the oven temperature from 50°C to 330°C with 22 min total run time. Mass spectrometry was assessed by a Leco Pegasus IV time-of-flight mass spectrometer with a 280°C transfer line temperature, electron ionization at −70 eV, and an ion source temperature of 250°C. Mass spectrometer operated between m/z 85 and 500 at 17 spectra/s. Raw spectra data were exported to dedicated WCMC servers and further processed by the WCMC BinBase database (14, 16). The BinBase database matches sample mass spectrum information and retention index against spectra from the Fiehn laboratory mass spectral library of over 1,200 in-house standards and the NIST05 commercial library. Metabolites were only reported if present in >25% of all of the samples and true peak detection in at least 50% of a given experimental condition (54). Reliably measured peaks found in some samples, but not of the quality matching others, were replaced by searching the raw data for the highest signal within 2 s of the target retention time minus the lowest signal within 5 s of the target retention time. Individual metabolites were normalized by the sum of identified metabolite quantifier ion peak heights present in each sample. Normalized quantifier ion peak heights were then used for all statistical analyses.
Complex lipids.
Serum samples (20 µl) were first extracted via the Matyash protocol using methyl tert-butyl ether (MTBE) (29). Extracts (30 µl) were transferred to two separate vials with microinserts for ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS) analysis. Samples (3 µl) were injected at 65°C and separated using a Waters Acquity UPLC charged-surface hybrid C18 column (100 mm × 2.1 mm) with a particle size of 1.9 µm and a flow rate of 0.6 ml/min. Mass spectrometry was conducted for positively charged ions (PC, lysoPC, PE, and PS) with an Agilent 6530 QTOF MS (resolution: 10,000) and for negatively charged ions (free fatty acids and phosphatidylinositols) with an Agilent 6550 QTOF MS (resolution: 20,000). Both mass spectrometers operated at full scan range m/z 65–1,700. Peak identification was assessed primarily with an internal retention time-corrected mz/rt library as well as MS/MS spectra for selected annotation using MS-Dial. Briefly, compounds in the LipidBlast library are initially selected within a mass window of MS1 0.01 Da and MS2 0.05 Da. MS-Dial will then annotate MS/MS spectra if one of the LipidBlast library spectra is similar based on an accurate mass score, dot product score, and reverse dot score, which is followed by manual checking of the MS/MS spectra against the library reference spectra (23). A minimum of three fragments must match along with fragment height similarity, which is reflected in the dot product and reverse dot product. In-depth details have previously been published (62). Peak alignment was conducted by MassProfilerProfessional. Final data are provided as quantifier ion peak heights and normalized to the sum of all peak heights for all identified metabolites for each sample. Additional details can be found at Metabolomics Workbench (“The Metabolomics Workbench, http://www.metabolomicsworkbench.org/”) under protocol number 163 (http://www.metabolomicsworkbench.org/protocols/protocoldetails.php?file_id=163).
Statistical Analyses
Participant characteristics and cognitive outcomes (Table 1) were assessed using one-way ANOVA. Analyses were controlled for age and sex. The least significant difference (LSD) post hoc test was used to distinguish differences between groups. For categorical variables (i.e., APOE, sex) the chi square test was used. Nonnormally distributed variables were log-transformed before statistical analyses, although raw physiological values are presented in the table.
Table 1.
Participant characteristics
| Outcome | ND/CH (n = 39) | ND/AD (n = 45) | T2D/CH (n = 23) | T2D/AD (n = 19) | P |
|---|---|---|---|---|---|
| Age, yr | 73.0 (7.4) | 74.9 (7.8) | 72.5 (6.9) | 73.9 (7.9) | 0.579 |
| Sex, n (% male) | 14 (36%) | 21 (47%) | 9 (39%) | 12 (63%) | 0.240 |
| Education, yr | 17.0 (3.1) | 15.8 (3.1) | 16.3 (2.9) | 14.9 (2.8) | 0.076 |
| APOE ε4, n (%) | 9 (23%)a | 28 (62%)b | 5 (22%)a | 11 (61%)b | <0.001 |
| T2D medication, n (%) | 0 (0%)a | 0 (0%)a | 16 (70%)b | 9 (47%)b | <0.001 |
| AD duration, yr | N/A | 4.9 (3.0) | N/A | 4.7 (2.9) | 0.945 |
| Glucose, mg/dl | 90.5 (9.4)a | 90.9 (9.9)a | 125.7 (34.1) | 110.7 (26.8) | <0.01 |
| Insulin, µU/ml | 7.7 (7.3)a | 7.4 (6.1)a | 11.1 (9.8)a,b | 11.4 (5.9)b | 0.014 |
| Amylin, pM | 9.9 (12.0) | 18.5 (52.1) | 13.5 (17.8) | 15.8 (21.1) | 0.683 |
| HOMA-IR | 1.78 (1.9)a | 1.71 (1.5)a | 3.48 (3.6)b | 3.34 (2.5)b | <0.01 |
| Weight, kg | 77.6 (12.8)a,c | 74.1 (19.9)a | 88.9 (15.0)b | 82.1 (10.3)b,c | 0.002 |
| BMI | 27.8 (4.0)a,b | 26.4 (5.7)a | 31.9 (4.3)b,c | 29.4 (5.1)b,c | <0.001 |
| MMSE | 29.4 (0.9)a | 23.1 (4.4)b | 29.4 (1.0)a | 22.9 (6.8)b | <0.001 |
| CDR-SB | 0 (0)a | 4.9 (3.2)b | 0 (0)a | 5.1 (3.6)b | <0.001 |
| Logical Memory II | 13.1 (4.1)a | 1.6 (2.6)b | 12.2 (3.5)a | 3.2 (3.3)b | <0.001 |
| SRT Free Recall | 30.4 (5.0)a | 9.2 (7.2)c | 28.5 (5.1)a | 14.4 (8.6)b | <0.001 |
| Fat mass, kg | 29.1 (8.7)a | 26.3 (11.7)a | 36.9 (9.2) | 29.9 (7.0)a | 0.001 |
| Lean mass, kg | 43.9 (8.2) | 44.3 (10.7) | 47.9 (9.1) | 48.6 (9.6) | 0.250 |
Values are means (SD) unless otherwise noted. Analytes were measured in serum from overnight-fasted subjects. ND, nondiabetic; AD, Alzheimer’s disease; CH, cognitively healthy; T2D, type 2 diabetes; APOE, apolipoprotein E; HOMA-IR, homeostasis model assessment of insulin resistance; BMI, body mass index; MMSE, mini-mental state examination; SRT, selective reminding test (units recalled); CDR-SB, Clinical Dementia Rating-Sum of Boxes. P values are for 1-way ANOVA, controlling for age and sex unless otherwise noted. P values for age and sex were not adjusted. Means without a common letter differ with the use of LSD post hoc test.
Preprocessing of modeling data.
Analyses of metabolomics data were conducted in R version 3.3.2. Primary metabolites and complex lipids data were assessed separately for multivariate analyses, but followed the same preprocessing workflow. Relative standard deviations (RSD) were assessed on technical replicates, and metabolites with RSD > 40% were removed from the analysis. Metabolites were then screened for missing data and removed if missing in >10% of all samples. Data were then screened for outliers with an iterative Grubbs’ test for outliers at α ≤ 0.01. Metabolites were further removed from analyses if >10% of metabolite data were removed after screening for outliers. In total, 22.1% and 2.9% of all metabolites from complex lipids and primary metabolites analyses, respectively, were removed after these screening procedures. Missing data were imputed using the K-nearest neighbor algorithm from the impute package. Imputations represented 0.54% and 0.50% of complex lipids and primary metabolites, respectively, used in multivariate analyses.
Statistical modeling of metabolomics data.
Partial least squares-discriminant analysis (PLS-DA) was used to discriminate classifiers due to the high dimensionality and collinear nature of metabolomics data. Before modeling, two-thirds of the data were randomly divided into a “model training” set and used only to develop PLS-DA models and identify discriminant metabolites. The remaining one-third of the data were considered as a validation set and not used in any aspects of model development. Training data were log transformed and scaled to unit variance before PLS-DA modeling. The optimal number of PLS-DA latent variables was identified by the greatest predicted classification rate in the validation set. Metabolites that discriminated classifiers were identified by variable importance measurements (VIM). VIM are rankings based on weighted sums of the absolute regression coefficients and are calculated separately for each classifier. Final sets of discriminant metabolites were metabolites that were above the 90th percentile of the mean VIM distribution. VIM-selected metabolites were used to fit PLS-DA models to identify if feature-selected metabolites could improve classification of subjects held out in the validation data set. Model performance measurements included overall classification rate, sensitivity, and specificity. Visualization of classifier discrimination was shown in PLS-DA scores and loadings plots. Group differences of feature selected metabolites from two-class models were then assessed by Mann-Whitney U-test using all data. P values from Mann-Whitney U-tests were adjusted for multiple comparisons by using Benjamini and Hochberg’s method at factor level [e.g., main effects were assessed with Mann-Whitney U-tests (4)]. All statistical tests were considered significant at a false discovery rate (FDR) ≤ 0.05.
RESULTS
Participants
This analysis included a total of 126 individuals divided into four groups: nondiabetic/cognitively healthy (ND/CH; n = 39), type 2 diabetic/cognitively healthy (T2D/CH; n = 23), nondiabetic/Alzheimer’s disease (ND/AD; n = 45), and Type 2 diabetic/Alzheimer’s disease (T2D/AD; n = 19). As presented in Table 1, these groups were not significantly different in age, sex, years of education, or AD duration. As expected, APOE ε4 carriers were overrepresented in both groups of AD subjects (62% ND/AD and 61% T2D/AD) compared with both groups of CH subjects (23% ND/CH and 22% T2D/CH). T2D/CH subjects had higher body weight compared with both nondiabetic groups (P < 0.01). Although T2D/CH subjects also had higher body weight compared with T2D/AD subjects (P < 0.05), the T2D/AD group had higher body weight compared with AD subjects without diabetes (P < 0.05). BMI was also higher in T2D/CH subjects vs. both ND/CH (P = 0.002) and ND/AD groups (P < 0.001), but was not different from T2D/AD subjects. T2D/AD subjects had a higher BMI compared with ND/AD subjects (P = 0.02).
Metabolic and Cognitive Characteristics
By definition, T2D/CH and T2D/AD subjects had higher fasting glucose compared with both nondiabetic groups (P < 0.001 for both comparisons; Table 1). Of note, T2D/CH subjects had higher fasting glucose compared with T2D/AD subjects (P < 0.05). Insulin levels were higher in the T2D/AD group compared with both nondiabetic groups (P = 0.01 vs. ND/CH, P = 0.02 vs. nondiabetic/AD). In contrast, there was no difference in fasting insulin between T2D/CH individuals and other groups. The HOMA-IR index of insulin resistance was higher in T2D/CH (P < 0.005) and T2D/AD (P < 0.02) subjects compared with both nondiabetic groups. No significant differences in serum amylin or C-peptide were detected between groups.
As expected, both ND/AD and T2D/AD groups performed more poorly than both cognitively normal groups on the Mini Mental State Examination (MMSE), Delayed Logical Memory test (DLM), and Selective Reminding Test (SRT; Table 1, P < 0.001 all comparisons). AD groups were not different from each other on MMSE or LMII score. However, T2D/AD had better performance than the ND/AD subjects on the SRT memory exam (total free recall; P = 0.009).
Metabolomics Characteristics
A total of 1,080 and 277 serum metabolites were detected from the untargeted metabolomics complex lipids and primary metabolism assays, respectively. In the complex lipid assay, 256 metabolites were either fully structurally identified or had structural features that sufficiently identified the chemical class of the metabolite. In the primary assay, 117 metabolites were fully structurally annotated. These metabolites were considered as “known” metabolites. The remaining metabolites were considered as “unknown” metabolites and were annotated with a unique ID.
Poor classification when modeling all diagnosis groups.
To investigate whether the AD groups exhibited a similar metabolomics fingerprint as T2D subjects, PLS-DA models including participants from all four experimental groups (ND/CH, ND/AD, T2D/CH, and T2D/AD) were fitted for each metabolomics platform separately. These models resulted in poor classification of participants held out for validation purposes across all platforms (<50% total accuracy) when fit with either all metabolites or only selected discriminatory metabolites (Table 2). These findings suggest that global serum metabolite differences do not resolve groups from one another when all four groups are included in the multivariate statistical model.
Table 2.
Partial least squares-discriminant analysis (PLS-DA) model performance for 4-class modela
| Training Set (n = 87) |
Test Set (n = 39) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full model |
Reduced model |
Full model |
Reduced model |
|||||||||
| CV Accb | LVc | pd | CV Acc | LV | p | Factor level | Acce | Sensf | Specf | Acc | Sensf | Specf |
| Complex lipid assay | ||||||||||||
| 40.1% | 1 | 1080 | 59.3% | 6 | 118 | 46.2% | 41.0% | |||||
| ND/CH | 0.33 | 0.70 | 0.67 | 0.59 | ||||||||
| T2D/CH | 0.57 | 0.90 | 0.0 | 1.0 | ||||||||
| ND/AD | 0.57 | 0.64 | 0.57 | 0.52 | ||||||||
| T2D/AD | 0.33 | 0.97 | 0.0 | 1.0 | ||||||||
| Primary metabolism assay | ||||||||||||
| 46.8 | 4 | 277 | 71.7% | 5 | 30 | 46.2% | 41.0% | |||||
| ND/CH | 0.42 | 0.81 | 0.67 | 0.59 | ||||||||
| T2D/CH | 0.57 | 0.88 | 0.0 | 1.0 | ||||||||
| ND/AD | 0.57 | 0.72 | 0.57 | 0.52 | ||||||||
| T2D/AD | 0.17 | 0.85 | 0.0 | 1.0 | ||||||||
CV, cross-validation; Acc, accuracy; LV, latent variable; p, parameters; Sens, sensitivity; Spec, specificity; ND, nondiabetic; AD, Alzheimer’s disease; CH, cognitively healthy; T2D, type 2 diabetes.
4-Class PLS-DA model includes classifiers for ND/CH, T2D/CH, ND/AD, and T2D/AD. Training and test sets were split 66% and 34%, respectively, within groups (i.e., distribution of within groups was equal in test and training sets). Full model fit with all known and unknown metabolites. Reduced models fit with known and unknown metabolites with variable importance measurements that were >90th percentile of the mean VIM distribution.
Highest CV prediction accuracy from ninefold cross validation.
Represents the number of LVs to reach highest CV accuracy.
Number of metabolites used to fit model.
Overall prediction accuracy of test set.
Sensitivity and specificity calculated for each group by comparing each group against all remaining levels, i.e., one level vs. all remaining samples.
T2D alone can be discriminated in all participants.
Binary group comparisons can offer a more sensitive approach to detecting discriminating variables. Thus we next focused on the main effects of T2D in all participants. Models were designed to classify T2D status (nondiabetic vs. T2D) in all subjects regardless of cognitive diagnosis. Results from each metabolomics platform are given below.
Primary metabolism (Fig. 1A).
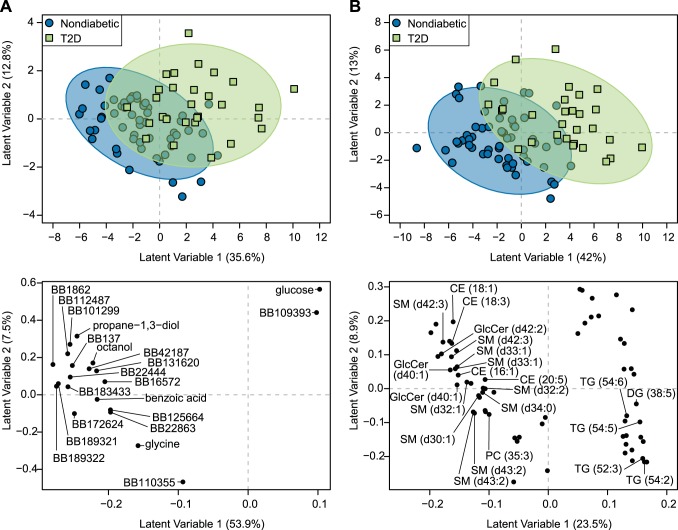
Fig. 1.
Type 2 diabetes (T2D) associates with a unique serum metabolomics signature in overnight-fasted elderly subjects. Partial least squares-discriminant analysis (PLS-DA) accurately discriminated elderly men and women with (n = 42) or without (n = 84) T2D using serum analyses of untargeted metabolomics of primary metabolism (A) and complex lipids (B). All participants were utilized in PLS-DA model development regardless of their dementia status. PLS-DA scores plots of models developed with training subset are displayed in the top panels. PLS-DA scores representing individuals participating in the study are displayed as blue circles (metabolic healthy) and green squares (T2D). Confidence regions of metabolic healthy and T2D groups are presented as 95% confidence ellipses based on Hotelling’s T2 statistic. PLS-DA loadings plots are displayed in the bottom panel of the graphic. PLS-DA loadings representing discriminant metabolites are displayed as black circles. Known and unknown metabolites are labeled in the loadings plot from the primary analysis (A); only known metabolites are labeled in the loadings plot from the complex lipid analysis for the sake of brevity (B). Unknown primary metabolites are labeled with their BinBase (BB) ID. All data were log transformed and scaled to unit variance before analysis.
PLS-DA models using data obtained from the primary metabolism platform resulted in >79.5% classification accuracy in held out data, while models fitted with metabolites from model-based feature selection maintained this classification accuracy (Table 3). Discrimination of T2D in nondiabetic participants is mostly apparent along latent variable 1 in the PLS-DA scores plot from the model fit with feature-selected metabolites (Fig. 1A, top panel). Metabolites associated with T2D subjects (e.g., greater in participants with T2D relative to nondiabetic participants) included glucose and a nonannotated BB109393 metabolite. Propane-1,3-diol, octanol, benzoic acid, glycine, and several other nonannotated metabolites discriminated T2D and were significantly lower in subjects with T2D compared with those that were nondiabetic (Fig. 1A, bottom panel, Table 4).
Table 3.
Partial least squares-discriminant analysis (PLS-DA) model performance for 2-class models fit with untargeted metabolomics of primary metabolisma
| Training Set |
Test Set |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full model (p = 277) |
Reduced model |
Full model |
Reduced model |
||||||||||
| Groupb | n | CV Accc | LVd | CV Acc | LV | pe | n | Accf | Sens | Spec | Acc | Sens | Spec |
| Classifiers: CH vs. AD | |||||||||||||
| All | 87 | 62.8% | 4 | 72.7% | 1 | 17 | 39 | 66.7% | 0.75 | 0.58 | 51.8% | 0.50 | 0.53 |
| ND | 58 | 66.4% | 1 | 75.6% | 1 | 18 | 26 | 50.0% | 0.64 | 0.33 | 46.2% | 0.57 | 0.33 |
| T2D | 29 | 55.6% | 2 | 87.0% | 3 | 22 | 13 | 61.5% | 0.50 | 0.71 | 69.2% | 0.67 | 0.71 |
| Classifiers: ND vs. T2D | |||||||||||||
| All | 87 | 75.0% | 4 | 80.8% | 2 | 21 | 39 | 79.5% | 0.85 | 0.69 | 82.1% | 0.88 | 0.69 |
| CH | 43 | 81.7% | 2 | 85.6% | 2 | 19 | 19 | 89.5% | 0.83 | 1.00 | 100% | 1.00 | 1.00 |
| AD | 44 | 78.9% | 1 | 86.5% | 1 | 22 | 20 | 60.0% | 0.71 | 0.33 | 65.0% | 0.71 | 0.50 |
CV, cross-validation; Acc, accuracy; LV, latent variable; p, parameters; Sens, sensitivity; Spec, specificity; ND, nondiabetic; AD, Alzheimer’s disease; CH, cognitively healthy; T2D, type 2 diabetes.
2-Class PLS-DA model. Training and test sets were split 66% and 34%, respectively, using all data and within groups (i.e., distribution of within groups was equal in test and training sets). Training/test splits for nested models were based on initial split using all data, i.e., training and test data were consistent between all models described in the study. Full models were fit with all known and unknown metabolites. Reduced models fit with known and unknown metabolites with variable importance measurements that were >90th percentile of the mean VIM distribution.
Experimental group used to fit PLS-DA models.
Highest CV prediction accuracy from ninefold cross validation.
Number LVs to reach highest CV accuracy.
Number of metabolites used to fit model.
Overall prediction accuracy of test set.
Table 4.
Metabolites that discriminate type 2 diabetes in overnight-fasted cognitively healthy elderly subjects
| Retention Index | Nondiabetic | Type 2 Diabetes | Pc | |
|---|---|---|---|---|
| Primary metabolitesa | ||||
| Benzoic acid | 339214 | 9.53 (9.4,9.6) | 9.39 (9.2,9.5) | <0.001 |
| Glucose | 659798 | 13.0 (12.9,13.1) | 13.23 (13,13.4) | <0.001 |
| Glycine | 368707 | 10.86 (10.8,11.1) | 10.69 (10.6,10.8) | <0.001 |
| Octanol | 247010 | 7.07 (6.9,7.2) | 6.9 (6.8,7.1) | <0.001 |
| Propane-1,3-diol | 214380 | 7.87 (7.7,8) | 7.66 (7.5,7.8) | <0.001 |
| Mode; Retention time, min (m/z) | ||||
| Complex lipid metabolitesb | ||||
| CE (16:1) | POS; 10.51 (640.6028_645.5884) | 10.76 (10.4,11) | 10.36 (9.9,10.6) | <0.001 |
| CE (18:1) | POS; 11.09 (673.5914_668.6349 | 12.07 (12,12.2) | 11.93 (11.8,12) | 0.003 |
| CE (18:3) | POS; 10.19 (669.5574_664.602) | 11.14 (10.7,11.4) | 10.82 (10.5,11.2) | 0.008 |
| CE (20:5) | POS; 9.93 (693.5601_664.602) | 11.59 (11.1,12) | 10.98 (10.5,11.5) | 0.004 |
| DG (38:5) | POS; 6.43 (665.5088) | 9.34 (9.2,9.7) | 9.71 (9.3,10.1) | 0.008 |
| GlcCer (d40:1) | POS; 7.15 (806.6472) | 8.97 (8.7,9.2) | 8.72 (7.8,9) | 0.009 |
| GlcCer (d40:1) | NEG; 7.13 (818.6282_842.6719) | 9.65 (9.3,9.8) | 9.37 (9,9.7) | 0.005 |
| GlcCer (d42:2) | NEG; 7.10 (844.6445) | 9.25 (9,9.5) | 8.96 (8.8,9.2) | 0.003 |
| PC (35:3) | POS; 4.99 (770.5698) | 10.79 (10.5,11) | 10.6 (10.4,10.9) | 0.005 |
| SM (d30:1) | POS; 3.71 (647.5117) | 9.26 (9,9.7) | 8.99 (8.8,9.3) | 0.003 |
| SM (d32:1) | POS; 4.26 (675.5448_697.5250) | 12.51 (12.4,12.8) | 12.35 (12.1,12.6) | 0.005 |
| SM (d32:2) | POS; 3.8 (673.5277) | 10.04 (9.7,10.3) | 9.69 (9.5,10) | 0.004 |
| SM (d33:1) | POS; 4.56 (689.5602_711.5410) | 11.91 (11.8,12.1) | 11.75 (11.5,11.9) | 0.015 |
| SM (d34:0) | POS; 5.10 (727.575_705.5921) | 11.55 (11.4,11.8) | 11.38 (11.1,11.7) | 0.016 |
| SM (d42:3) | POS; 6.32 (811.6664_833.6517) | 13.42 (13.3,13.6) | 13.35 (13.1,13.5) | 0.023 |
| SM (d43:2) | POS; 7.12 (827.6993) | 10.23 (9.9,10.5) | 9.99 (9.8,10.3) | 0.032 |
| SM (d33:1) | NEG; 4.53 (747.5658) | 9.85 (9.7,10) | 9.73 (9.6,9.9) | 0.016 |
| SM (d42:3) | NEG; 6.29 (869.675) | 10.52 (10.4,10.6) | 10.39 (10.3,10.6) | 0.025 |
| SM (d43:2) | NEG; 7.09 (885.7059) | 8.44 (7.3,8.6) | 7.48 (7.1,8.5) | 0.017 |
| TG (52:3) | POS; 10.67 (879.7481_874.793_857.7594) | 15.65 (15.5,15.8) | 15.73 (15.6,15.9) | 0.032 |
| TG (54:2) | POS; 11.41 (909.7946_904.8413) | 13.43 (13.2,13.8) | 13.68 (13.4,14.1) | 0.025 |
| TG (54:5) | POS; 10.5 (903.7443) | 12.3 (12.1,12.6) | 12.59 (12.3,13) | 0.008 |
| TG (54:6) | POS; 10.16 (901.7267_896.7675) | 12.95 (12.7,13.3) | 13.42 (12.8,13.6) | 0.008 |
Data are medians (25th quartile, 75th quartile) of log-transformed quantifier ion peak height. Presented metabolites were identified as the top 95th percentile of partial least squares-discriminant analysis-variable importance in projection (PLS-DA-VIP) distributions.
Primary metabolites determined by GC-QTOF-MS.
Complex lipid metabolites determined by UPLC-QTOF-MS.
P values are Benjamini and Hochberg adjusted P values derived from Mann Whitney U tests on log transformed quantifier ion peak heights.
Complex lipids (Fig. 1B).
The complex lipid platform yielded ≥76.9% classification accuracy in PLS-DA models using all metabolites or those that were fit with only feature selected metabolites (Table 5), and primarily discriminated T2D along latent variable 1 (Fig. 1B, top panel). Subjects with T2D were characterized by higher abundances of several triglycerides and lower abundances of certain sphingomyelins, glucosyl-ceramides, and cholesterol esters (Fig. 1B, bottom panel, Table 4).
Table 5.
Partial least squares-discriminant analysis (PLS-DA) model performance for 2-class models fit with untargeted metabolomics of complex lipidsa
| Training Set |
Test Set |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full model (p = 1080) |
Reduced model |
Full model |
Reduced model |
||||||||||
| Groupb | n | CV Accc | LVd | CV Acc | LV | pe | n | Accf | Sens | Spec | Acc | Sens | Spec |
| Classifiers: CH vs. AD | |||||||||||||
| All | 87 | 56.0% | 2 | 74.5% | 2 | 70 | 39 | 53.9% | 0.55 | 0.53 | 53.9% | 0.55 | 0.53 |
| ND | 58 | 61.9% | 4 | 92.1% | 4 | 80 | 26 | 61.5% | 0.71 | 0.50 | 61.5% | 0.57 | 0.67 |
| T2D | 29 | 63.9% | 2 | 86.1% | 2 | 76 | 13 | 61.5% | 0.33 | 0.86 | 61.5% | 0.50 | 0.71 |
| Classifiers: ND vs. T2D | |||||||||||||
| All | 87 | 73.3% | 4 | 86.1% | 2 | 65 | 39 | 79.5% | 0.81 | 0.77 | 76.9% | 0.85 | 0.62 |
| CH | 43 | 63.3% | 3 | 97.8% | 4 | 79 | 19 | 73.7% | 0.83 | 0.57 | 73.7% | 0.75 | 0.71 |
| AD | 44 | 73.3% | 4 | 90.9% | 4 | 65 | 20 | 75.0% | 0.86 | 0.50 | 60.0% | 0.86 | 0.00 |
CV, cross-validation; Acc, accuracy; LV, latent variable; p, parameters; Sens, sensitivity; Spec, specificity; ND, nondiabetic; AD, Alzheimer’s disease; CH, cognitively healthy; T2D, type 2 diabetes.
2-Class PLS-DA model. Training and test sets were split 66% and 34%, respectively, using all data and within groups (i.e., distribution of within groups was equal in test and training sets). Training/test splits for nested models were based on initial split using all data, i.e., training and test data were consistent between all models described in the study. Full models were fit with all known and unknown metabolites. Reduced models fit with known and unknown metabolites with variable importance measurements that were >90th percentile of the mean VIM distribution.
Experimental group used to fit PLS-DA models.
Highest CV prediction accuracy from ninefold cross validation.
Number of LVs to reach highest CV accuracy.
Number of metabolites used to fit model.
Overall prediction accuracy of test set.
The serum metabolome can discriminate T2D in cognitively healthy elderly.
To investigate whether T2D-associated metabolite variables differ within cognitively healthy subjects, we modeled the main effect of T2D separately in this group. In cognitively healthy participants, PLS-DA models accurately discriminated T2D status using the primary and complex lipid platforms (Fig. 2, A and B, respectively) as described below.
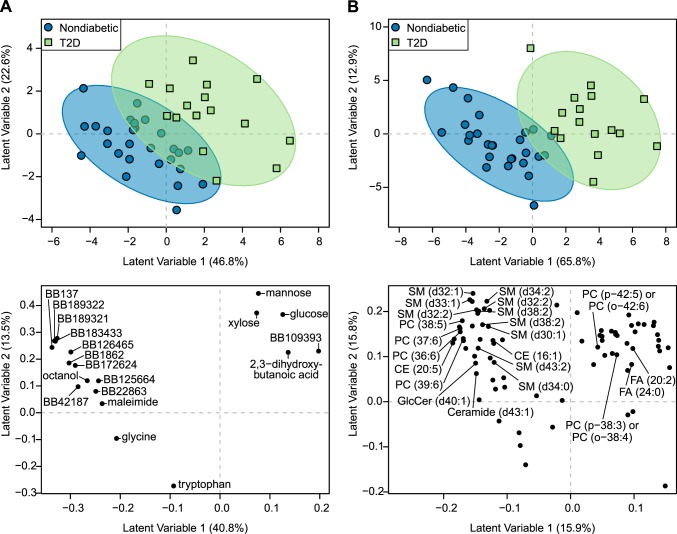
Fig. 2.
Type 2 diabetes (T2D) associates with a unique serum metabolomics signature in overnight-fasted elderly subjects without dementia. Partial least squares-discriminant analysis (PLS-DA) accurately discriminated elderly subjects with (n = 23) or without (n = 39) T2D using serum analyses of untargeted metabolomics of primary metabolism (A) and complex lipids (B). Only participants without dementia were used for PLS-DA model development. PLS-DA scores plots of models developed with training subset are displayed in the top panels. PLS-DA scores representing individuals participating in the study are displayed as blue circles (metabolic healthy) and green squares (T2D). Confidence regions of metabolic healthy and T2D groups are presented as 95% confidence ellipses based on Hotelling’s T2 statistic. PLS-DA loadings plots are displayed in the bottom panel of the graphic. PLS-DA loadings representing discriminant metabolites are displayed as black circles. Known and unknown metabolites are labeled in the loadings plot from the primary analysis (A); only known metabolites are labeled in the loadings plot from the complex lipid analysis for the sake of brevity (B). Unknown primary metabolites are labeled with their BinBase (BB) ID. All data were log transformed and scaled to unit variance before analysis.
Primary metabolism.
PLS-DA models fit with metabolomics of primary metabolites correctly classified ≥89.4% of held out subjects using the primary metabolite data when only cognitively healthy subjects were included (Table 3). Discrimination of patients with or without T2D was, again, explained along PLS-DA latent variable 1 (Fig. 2A, top panel). Serum glucose, glycine, octanol, and several unknown metabolites were selected as important discriminant variables for T2D vs. nondiabetes in cognitively healthy subjects (Fig. 2A, bottom panel, and Fig. 3A): i.e., glucose and BB109393 were significantly higher in participants diagnosed with T2D, and glycine and octanol significantly higher in nondiabetic relative to T2D participants. Mannose, xylose, and 2,3-dihydroxybutanoic acid were also selected as important discriminators of T2D status, but only mannose and xylose were statistically greater in T2D participants relative to those without when assessed by Mann-Whitney U-tests. Maleimide and tryptophan were also identified as discriminatory metabolites and were significantly higher in nondiabetics.
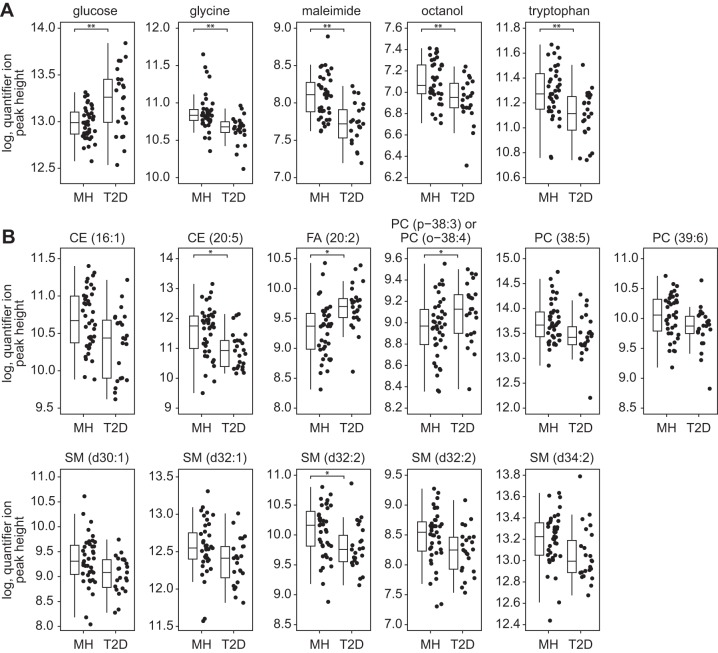
Fig. 3.
Unique serum metabolomics signature components differentiate type 2 diabetes (T2D) status in overnight-fasted elderly subjects without dementia. Serum metabolites derived from untargeted metabolomics of primary metabolism (A) and complex lipids (B) are shown for elderly participants with (n = 23) or without (n = 39) T2D. Data are displayed as boxplots and are accompanied with the data distribution. Upper and lower hinges of boxplots are data within the interquartile range (IQR) and are values between the 1st and 3rd quartile. The median value is represented by a solid line. Whiskers include data based on (IQR × 1.5) + upper hinge and (IQR × 1.5) – lower hinge. Units are log-transformed quantifier ion peak height. Displayed metabolites were identified in PLS-DA analyses. Group differences were assessed by Mann Whitney U-test. Metabolites with ** or * denote FDR < 0.01 and < 0.05, respectively.
Complex lipids.
The classification accuracy was 73.7% in PLS-DA models fit from complex lipid data in cognitively healthy patients (Table 5). Here, there is almost full discrimination of the 95% confidence ellipses for T2D and nondiabetic subjects along latent variable 1, suggesting that this PLS-DA variable explains T2D in these data (Fig. 2B, top panel). Two free fatty acids (C20:2 and C24:0) and two phosphatidylcholines (PC38:3p/PC38:4o and PC42:5p/PC42:6o) were important discriminant metabolites for cognitively healthy participants with T2D. Several sphingomyelins, phosphatidylcholines, and cholesterol esters were also discriminating metabolites between groups, in addition to a single ceramide and glucosyl-ceramide. Of these, CE16:1, CE20:5, PC38:5, PC39:6, SM30:1d, SM32:1d, SM32:2d, SM32:2d, and SM34:2d were significantly lower in cognitively healthy subjects with T2D (Fig. 2B, bottom panel, and Fig. 3B).
The serum metabolome cannot accurately discriminate T2D in AD subjects.
In contrast to the results described above for cognitively healthy individuals, PLS-DA models classifying T2D in participants with AD did not result in accurate classification of T2D status for either platform (75% using primary metabolomics, 60% using complex lipids in held-out subjects)(Tables 3 and 5). Although the overall classification accuracy in the primary metabolite platform appeared adequate, it had low specificity (50%) for T2D classifications compared with non-T2D classifications; i.e., all of the T2D subjects were correctly classified, but only 50% of the non-T2D subjects were correctly classified (Table 3). Isolating discriminating metabolites with model-dependent or model-independent methods did not improve the overall accuracy of predicting T2D status in the test set.
AD is poorly classified across all participants.
We next attempted to identify metabolites that discriminate AD in all participants regardless of T2D status. However, regardless of analytical platform, PLS-DA models classifying AD status (cognitively healthy vs. AD) resulted in poor classification of held out subjects across both platforms (all models ≤ 66.7% classification accuracy)(Tables 3 and 5).
APOE ε4 carriers do not produce a robust serum metabolite signature.
We have previously observed lower fasting insulin and HOMA-IR in APOE ε4 carriers in a larger group of subjects from the KU ADC Clinical Cohort, suggesting that APOE could influence insulin sensitivity in both nondemented subjects and those with Alzheimer’s disease (40). Thus we utilized a subset of this cohort to identify whether the serum metabolome could distinguish APOE4 carrier status. PLS-DA modeling with all subjects combined resulted in <60% of held-out subjects correctly classified as carriers or noncarriers across both platforms (Table 6). This was not improved when filtering for discriminating metabolites. PLS-DA models assessing carrier status in nondementia subjects had poor classification rates in held-out samples (≤70%) on both platforms. Overall classification rates from PLS-DA models in AD subjects were ≥70% in the complex lipid and primary assays mainly due to the disproportionate amount of carriers to noncarriers (Table 6). These models tended to predict all held-out subjects as APOE4 carriers, suggesting poor modeling performance. Poor PLS-DA modeling outcomes were also observed when examining the subset of participants with T2D status (Table 6). Altogether, there was not definitive evidence that the serum metabolome is impacted by the presence of APOE ε4 in this population.
Table 6.
Partial least squares-discriminant analysis (PLS-DA) model performance for APOE ε4 fit with untargeted metabolomicsa
| Training Set |
Test Set |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full model |
Reduced model |
Full model |
Reduced model |
||||||||||
| Groupb | n | CV Accc | LVd | CV Acc | LV | pe | n | Accf | Sens | Spec | Acc | Sens | Spec |
| Complex lipid assay | |||||||||||||
| All | 86 | 66.3% | 4 | 73.2% | 4 | 72 | 39 | 61.5% | 0.62 | 0.62 | 61.5% | 0.69 | 0.46 |
| ND | 58 | 50.3% | 2 | 73.0% | 4 | 83 | 26 | 65.4% | 0.71 | 0.56 | 65.4% | 0.71 | 0.56 |
| T2D | 28 | 63.0% | 4 | 88.0% | 2 | 79 | 13 | 61.5% | 0.78 | 0.25 | 69.2% | 0.78 | 0.50 |
| CH | 43 | 50.5% | 2 | 90.9% | 2 | 78 | 19 | 79.0% | 0.93 | 0.25 | 73.7% | 0.80 | 0.50 |
| AD | 43 | 69.1% | 4 | 71.1% | 2 | 70 | 20 | 70.0% | 0.45 | 1.00 | 75.0% | 0.55 | 1.00 |
| Primary metabolism assay | |||||||||||||
| All | 86 | 54.4% | 2 | 76.8% | 3 | 20 | 39 | 51.3% | 0.62 | 0.31 | 41.0% | 0.42 | 0.38 |
| ND | 58 | 60.6% | 1 | 84.4% | 4 | 21 | 26 | 69.2% | 0.53 | 1.00 | 42.3% | 0.47 | 0.33 |
| T2D | 28 | 57.4% | 3 | 87.0% | 2 | 21 | 13 | 53.9% | 0.67 | 0.25 | 61.5% | 0.56 | 0.75 |
| CH | 43 | 74.4% | 1 | 86.5% | 3 | 25 | 19 | 78.9% | 0.93 | 0.25 | 63.2% | 0.73 | 0.25 |
| AD | 43 | 52.6% | 2 | 79.6% | 1 | 19 | 20 | 45.0% | 0.36 | 0.56 | 55.0% | 0.27 | 0.89 |
CV, cross-validation; Acc, accuracy; LV, latent variable; p, parameters; Sens, sensitivity; Spec, specificity; ND, nondiabetic; AD, Alzheimer’s disease; CH, cognitively healthy; T2D, type 2 diabetes.
2-Class PLS-DA models with APOE ε4 carrier vs. noncarriers. Training and test sets were split 66% and 34%, respectively, using all data and within groups (i.e., distribution of within groups was equal in test and training sets). Training and test data were consistent between all models described in the study. Full models were fit with all known and unknown metabolites. Reduced models fit with known and unknown metabolites with variable importance measurements that were >90th percentile of the mean VIM distribution.
Experimental group used to fit PLS-DA models.
Highest CV prediction accuracy from ninefold cross validation.
Number LVs to reach highest CV accuracy.
Number of metabolites used to fit model.
Overall prediction accuracy of test set.
DISCUSSION
T2D is a risk factor for AD (3, 8, 20, 46, 47, 73, 74), and there is evidence for peripheral insulin resistance and increased prevalence of T2D in AD (22, 41, 43). However, other non-T2D risk factors, such as APOE and vascular dysfunction, are also related to lipid dynamics (48, 51, 68), and these risk factors may share common underlying disturbances in metabolomics or lipidomics-related outcomes. Therefore, we hypothesized that we would be able to cleanly discriminate the metabolic profile of cognitively healthy elderly subjects without T2D from those with AD, T2D, or AD and T2D. We also expected that AD subjects (with or without T2D) and cognitively healthy elderly with T2D would share a similar metabolomics phenotype. However, when modeling all four groups together, we were unable to accurately discriminate the metabolic profile of cognitively healthy subjects from the other three groups, nor could we clearly discriminate any of the four groups from one another. This indicated that the combined metabolic disturbances associated with T2D and AD are not readily unmasked in the serum metabolome of elderly subjects. Importantly, follow-up binary group analyses revealed that whereas we could independently classify T2D status in the overall cohort and in cognitively healthy elderly, we could not discriminate T2D status within AD subjects. This suggests that the serum metabolomics profile of T2D is obscured in AD patients.
The absence of a T2D-like effect on the serum metabolome of individuals with AD is noteworthy, given that AD subjects with T2D in this study did exhibit differences in metabolic parameters including higher fasting glucose, HOMA-IR, BMI, and body weight compared with AD subjects without T2D. Still, a T2D-like dysregulation in the insulin signaling pathway occurs in AD brain (26, 38, 39, 57, 59) and peripheral insulin resistance and T2D occur disproportionately in AD subjects (22, 43). The mechanistic reasons for the obscured serum metabolomics T2D phenotype in AD subjects are unclear. Some factors, such as smaller elevations in fasting blood glucose and body weight in AD subjects with T2D compared with cognitively normal subjects withT2D, may also play a role. However, declines in brain glucose metabolism and body weight are well-characterized to occur in AD (10, 18), and the way that T2D modifies these changes is not well-understood. In addition, disparate therapeutic treatment and adherence (including both AD and T2D medication) could potentially alter the circulating metabolome. As mentioned, other investigators, such as Proitsi et al. (50), have previously shown evidence that the circulating metabolome is altered in AD, identifying a group of molecules including cholesterol esters, long-chain triglycerides, and phosphatidylcholines that were associated with clinical diagnosis and/or neuroimaging measures. However, we used a different modeling approach and a priori included equal representation of T2D subjects in our groups. Given the potential role of metabolic impairment in AD our equal inclusion of diabetic subjects between cognitive diagnosis groups may have hampered our ability to replicate findings from prior metabolic profiling studies that were able to discriminate cognitive status in the broader populations (49, 50).
Studies in younger populations, typically examining the overnight-fasted state, have identified disparate circulating metabolomics profiles when comparing individuals with T2D against metabolically healthy and/or prediabetic individuals (12, 14, 32, 66, 67), suggesting a robust and broad effect of diabetes on intermediary metabolism. Several other reports have documented T2D or insulin-resistant phenotypes associated with alterations in amino acids and amino acid products (7, 14, 30, 31, 65), acylcarnitines reflective of incomplete long-chain fatty acid oxidation (1, 28, 35), and nonesterified fatty acids, epoxides (C18 and C20 chain lengths), and N-acylethanolamides (19). However, to our knowledge there are no studies that have assessed the effect of T2D in elderly populations independent of cognitive function.
In this report, we accurately identified a T2D serum metabolomics phenotype with both untargeted platforms in cognitively healthy elderly subjects. Not surprisingly, glucose and other sugars were higher in cognitively healthy participants with T2D, but T2D was also characterized by lower abundances of glycine, tryptophan, octanol, and maleimide. Similar to our findings, lower glycine concentrations have been previously associated with T2D risk and insulin resistance (12, 14, 36, 65). Although the mechanism is unclear, the drop in glycine has been postulated to be due to changes in hepatic gluconeogenesis or conjugation with β-oxidation by-products (21, 45). Since glycine is a building block for glutathione synthesis, this may have a negative effect on antioxidant capacity (55). Metabolism of tryptophan through the kynurenine pathway, the major catabolic pathway for tryptophan in mammals, has been linked to inflammation (9), a condition often observed in T2D, and prior work suggests that the tryptophan-kynurenine pathway is increased in individuals with gestational diabetes (25). It is possible that elderly individuals with T2D exhibit lower serum tryptophan levels due to flux through this pathway, but further work is warranted. Octanol is used in the laboratory to extract lipids from plasma (56), and there is some evidence that it affects the stability of lipoproteins (69), which would have important implications on lipid transport. Maleimide is an unsaturated imide that has not been widely detected in biological samples. Unfortunately, little is known about the role of maleimide in T2D.
Elevated circulating branched-chain amino acids (BCAA) have been widely identified in metabolomics-based studies as markers of insulin dysregulation and/or T2D (12, 58, 72), whereas others have shown lower concentrations in T2D populations relative to healthy control groups (36, 63, 76). This phenomenon is not fully understood, but hypothesized to be related to dysregulation of BCAA related enzymes (e.g., branched-chain keto acid dehydrogenase) or differences in BCAA catabolism pathways, likely related to insulin resistance (27). In contrast to these studies, no BCAAs were discriminant of T2D in either the cognitive healthy or AD subjects and none were significantly altered by T2D. To our knowledge, BCAA metabolism in relation to insulin resistance and T2D has not been investigated in elderly populations as all of the studies mentioned above are in younger populations. Our results would suggest that the links between insulin resistance and circulating BCAAs in the elderly are not as prominent relative to younger individuals. As for lipids, we found lower abundances of several sphingomyelin and phosphatidylcholine species in cognitively healthy elderly participants with T2D, which concurs with observations seen in other studies examining lipid profiles in T2D participants relative to metabolically healthy controls (17, 65). In mice, knockout of sphingomyelin synthase, which catalyzes the formation of sphingomyelins, results in mitochondrial dysfunction and impaired glucose-stimulated insulin secretion (75), providing a physiological basis for the observation of lower sphingomyelin species in T2D subjects. Likewise, in a recent human study, several PCs were shown to be negatively associated with second-phase insulin secretion, including one that we identified here (PC 38:5) (37). Together, our results suggest that the lipidomics fingerprint we detected in our study of cognitively healthy elderly subjects with T2D may provide important biomarkers that are important in the insulin response.
The study has several limitations that may have made it more difficult to detect robust, specific AD-T2D relationships with respect the serum metabolome. Although we were able to assess the serum metabolome from a set of well-phenotyped subjects and characterized metabolites that defined T2D in subjects without dementia, our sample size was determined by the availability of samples in the KU ADC clinical cohort. Thus this study may not have been adequately powered to uncover subtle AD-related metabolic effects, such as differences based on disease stage (i.e., MCI vs. dementia). This is likely exacerbated by heterogeneity of human samples from differences in diet, medication, physical activity, genetics, and many other factors. Although we had information regarding clinical diagnosis of T2D, the duration of T2D and the glycemic control were unknown, which are critical drivers of metabolic findings in T2D subjects. Future work to investigate the long-term vs. short-term effects of T2D and glucose variations (HbA1c) on the metabolome are warranted. Furthermore, we utilized multiple metabolomics platforms, which had the advantage of greater pathway coverage but also greatly increases the variable-to-sample ratio (i.e., small n/large p paradigm). We used PLS-DA, a supervised data reduction technique that is amenable to small sample sizes and colinearity commonly found in metabolomics data. Still, PLS-DA is prone to overfitting and can result in spurious results if not properly validated (70). We utilized a training/test paradigm to gauge the performance of the PLS-DA models and minimize overfitting. At best, we found 70–75% classification rates and suggested that this performance is adequate; however, we acknowledge that there is no current standard criterion for classification rates in held-out samples from classifier algorithms such as PLS-DA. Interestingly, Proitsi et al. (50) also reported 70–75% classification accuracy in held-out samples from Random Forest models discriminating clinical classifiers of AD progression and brain atrophy in a larger set of AD participants and controls. Finally, our findings must be interpreted in the context of our study design, where we a priori weighted the groups equally in terms of T2D subjects. Previous work suggests greater insulin resistance and increased prevalence of impaired fasting glucose and T2D in AD (22, 41, 43), so this may have hampered our ability to replicate findings in which metabolic profiling discriminated cognitive status (49, 50). Nonetheless, our study had several strengths, including good matching of cohorts based upon age, sex, and education, equal distribution of APOE ε4 carriers among nondiabetic and T2D subjects, and careful diagnostic classification based upon disease status and clinical examination. The results include the most comprehensive metabolite coverage reported to date comparing AD and T2D in an elderly population.
In conclusion, we were able to accurately distinguish T2D individuals based on a metabolomics phenotype in cognitively healthy elderly, but were unable to do so in an AD cohort. This suggests that AD may obscure the serum metabolic phenotype of T2D. Contrary to our hypothesis, we did not find evidence that the serum metabolome in AD mimics the T2D metabolome of cognitively healthy elderly. Future work to clarify the complex relationships between peripheral metabolism and AD is warranted.
GRANTS
This project was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences of the National Institutes of Health through Grant Numbers UL1-TR-000001 and UL1-TR-001449, as well as K99-AG-050490 (J. K. Morris), R01-DK-088940 (J. P. Thyfault), R21-AG-056062 (J. P. Thyfault, J. K. Morris), P20-GM-121293 (B. D. Piccolo), P30-AG-035982 (Univ. of Kansas Alzheimer’s Disease Center), U24-DK-097154 (West Coast Metabolomics Center), and Veterans Affairs Merit Review Award 1I01BX002567-01 (J. P. Thyfault).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.K.M., B.D.P., K.S., J.P.T., and S.H.A. conceived and designed research; J.K.M. and B.D.P. performed experiments; J.K.M., B.D.P., and S.H.A. analyzed data; J.K.M., B.D.P., J.P.T., and S.H.A. interpreted results of experiments; J.K.M. and B.D.P. drafted manuscript; J.K.M., B.D.P., K.S., J.P.T., and S.H.A. edited and revised manuscript; J.K.M., B.D.P., K.S., J.P.T., and S.H.A. approved final version of manuscript; B.D.P. prepared figures.
REFERENCES
- 1.Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE, Hwang DH, Newman JW, Garvey WT. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr 139: 1073–1081, 2009. doi: 10.3945/jn.108.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzheimer’s Association 2016 Alzheimer’s disease facts and figures. Alzheimers Dement 12: 459–509, 2016. doi: 10.1016/j.jalz.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Arvanitakis Z, Wilson RS, Bienias JL, Evans DA, Bennett DA. Diabetes mellitus and risk of Alzheimer disease and decline in cognitive function. Arch Neurol 61: 661–666, 2004. doi: 10.1001/archneur.61.5.661. [DOI] [PubMed] [Google Scholar]
- 4.Benjamini H. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser A Stat Soc 57: 289–300, 1995. [Google Scholar]
- 5.Bloom GS. Amyloid-β and tau: the trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol 71: 505–508, 2014. doi: 10.1001/jamaneurol.2013.5847. [DOI] [PubMed] [Google Scholar]
- 6.Cajka T, Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt Chem 61: 192–206, 2014. doi: 10.1016/j.trac.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen T, Ni Y, Ma X, Bao Y, Liu J, Huang F, Hu C, Xie G, Zhao A, Jia W, Jia W. Branched-chain and aromatic amino acid profiles and diabetes risk in Chinese populations. Sci Rep 6: 20594, 2016. doi: 10.1038/srep20594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng D, Noble J, Tang MX, Schupf N, Mayeux R, Luchsinger JA. Type 2 diabetes and late-onset Alzheimer’s disease. Dement Geriatr Cogn Disord 31: 424–430, 2011. doi: 10.1159/000324134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis I, Liu A. What is the tryptophan kynurenine pathway and why is it important to neurotherapeutics? Expert Rev Neurother 15: 719–721, 2015. doi: 10.1586/14737175.2015.1049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drzezga A, Riemenschneider M, Strassner B, Grimmer T, Peller M, Knoll A, Wagenpfeil S, Minoshima S, Schwaiger M, Kurz A. Cerebral glucose metabolism in patients with AD and different APOE genotypes. Neurology 64: 102–107, 2005. doi: 10.1212/01.WNL.0000148478.39691.D3. [DOI] [PubMed] [Google Scholar]
- 11.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol 160: 113–123, 2003. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrannini E, Natali A, Camastra S, Nannipieri M, Mari A, Adam KP, Milburn MV, Kastenmüller G, Adamski J, Tuomi T, Lyssenko V, Groop L, Gall WE. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes 62: 1730–1737, 2013. doi: 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiehn O. Metabolomics by gas chromatography-mass spectrometry: combined targeted and untargeted profiling. Curr Protoc Mol Biol 114: 30.4.1–30.4.32, 2016. doi: 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiehn O, Garvey WT, Newman JW, Lok KH, Hoppel CL, Adams SH. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS One 5: e15234, 2010. doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiehn O, Kind T. Metabolite profiling in blood plasma. Methods Mol Biol 358: 3–17, 2007. doi: 10.1007/978-1-59745-244-1_1. [DOI] [PubMed] [Google Scholar]
- 16.Fiehn O, Wohlgemuth G, Scholz M. Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. Lect Notes Comput Sci 3615: 224–239, 2005. doi: 10.1007/11530084_18. [DOI] [Google Scholar]
- 17.Floegel A, Stefan N, Yu Z, Mühlenbruch K, Drogan D, Joost HG, Fritsche A, Häring HU, Hrabě de Angelis M, Peters A, Roden M, Prehn C, Wang-Sattler R, Illig T, Schulze MB, Adamski J, Boeing H, Pischon T. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 62: 639–648, 2013. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillette-Guyonnet S, Nourhashemi F, Andrieu S, de Glisezinski I, Ousset PJ, Riviere D, Albarede JL, Vellas B. Weight loss in Alzheimer disease. Am J Clin Nutr 71: 637S–642S, 2000. doi: 10.1093/ajcn/71.2.637s. [DOI] [PubMed] [Google Scholar]
- 19.Grapov D, Adams SH, Pedersen TL, Garvey WT, Newman JW. Type 2 diabetes associated changes in the plasma non-esterified fatty acids, oxylipins and endocannabinoids. PLoS One 7: e48852, 2012. doi: 10.1371/journal.pone.0048852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudala K, Bansal D, Schifano F, Bhansali A. Diabetes mellitus and risk of dementia: A meta-analysis of prospective observational studies. J Diabetes Investig 4: 640–650, 2013. doi: 10.1111/jdi.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hetenyi G Jr, Anderson PJ, Raman M, Ferrarotto C. Gluconeogenesis from glycine and serine in fasted normal and diabetic rats. Biochem J 253: 27–32, 1988. doi: 10.1042/bj2530027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janson J, Laedtke T, Parisi JE, O’Brien P, Petersen RC, Butler PC. Increased risk of type 2 diabetes in Alzheimer disease. Diabetes 53: 474–481, 2004. doi: 10.2337/diabetes.53.2.474. [DOI] [PubMed] [Google Scholar]
- 23.Kind T, Liu KH, Lee DY, DeFelice B, Meissen JK, Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods 10: 755–758, 2013. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kivipelto M, Helkala EL, Hänninen T, Laakso MP, Hallikainen M, Alhainen K, Soininen H, Tuomilehto J, Nissinen A. Midlife vascular risk factors and late-life mild cognitive impairment: A population-based study. Neurology 56: 1683–1689, 2001. doi: 10.1212/WNL.56.12.1683. [DOI] [PubMed] [Google Scholar]
- 25.Law KP, Han TL, Mao X, Zhang H. Tryptophan and purine metabolites are consistently upregulated in the urinary metabolome of patients diagnosed with gestational diabetes mellitus throughout pregnancy: A longitudinal metabolomics study of Chinese pregnant women part 2. Clin Chim Acta 468: 126–139, 2017. doi: 10.1016/j.cca.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Liu F, Grundke-Iqbal I, Iqbal K, Gong CX. Deficient brain insulin signalling pathway in Alzheimer’s disease and diabetes. J Pathol 225: 54–62, 2011. doi: 10.1002/path.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol 10: 723–736, 2014. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mai M, Tönjes A, Kovacs P, Stumvoll M, Fiedler GM, Leichtle AB. Serum levels of acylcarnitines are altered in prediabetic conditions. PLoS One 8: e82459, 2013. doi: 10.1371/journal.pone.0082459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 49: 1137–1146, 2008. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormack SE, Shaham O, McCarthy MA, Deik AA, Wang TJ, Gerszten RE, Clish CB, Mootha VK, Grinspoon SK, Fleischman A. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr Obes 8: 52–61, 2013. doi: 10.1111/j.2047-6310.2012.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menge BA, Schrader H, Ritter PR, Ellrichmann M, Uhl W, Schmidt WE, Meier JJ. Selective amino acid deficiency in patients with impaired glucose tolerance and type 2 diabetes. Regul Pept 160: 75–80, 2010. doi: 10.1016/j.regpep.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Menni C, Fauman E, Erte I, Perry JR, Kastenmüller G, Shin SY, Petersen AK, Hyde C, Psatha M, Ward KJ, Yuan W, Milburn M, Palmer CN, Frayling TM, Trimmer J, Bell JT, Gieger C, Mohney RP, Brosnan MJ, Suhre K, Soranzo N, Spector TD. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 62: 4270–4276, 2013. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mielke MM, Bandaru VV, Haughey NJ, Xia J, Fried LP, Yasar S, Albert M, Varma V, Harris G, Schneider EB, Rabins PV, Bandeen-Roche K, Lyketsos CG, Carlson MC. Serum ceramides increase the risk of Alzheimer disease: the Women’s Health and Aging Study II. Neurology 79: 633–641, 2012. doi: 10.1212/WNL.0b013e318264e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mielke MM, Haughey NJ, Bandaru VV, Schech S, Carrick R, Carlson MC, Mori S, Miller MI, Ceritoglu C, Brown T, Albert M, Lyketsos CG. Plasma ceramides are altered in mild cognitive impairment and predict cognitive decline and hippocampal volume loss. Alzheimers Dement 6: 378–385, 2010. doi: 10.1016/j.jalz.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mihalik SJ, Goodpaster BH, Kelley DE, Chace DH, Vockley J, Toledo FG, DeLany JP. Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity (Silver Spring) 18: 1695–1700, 2010. doi: 10.1038/oby.2009.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mihalik SJ, Michaliszyn SF, de las Heras J, Bacha F, Lee S, Chace DH, DeJesus VR, Vockley J, Arslanian SA. Metabolomic profiling of fatty acid and amino acid metabolism in youth with obesity and type 2 diabetes: evidence for enhanced mitochondrial oxidation. Diabetes Care 35: 605–611, 2012. doi: 10.2337/DC11-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molnos S, Wahl S, Haid M, Eekhoff EMW, Pool R, Floegel A, Deelen J, Much D, Prehn C, Breier M, Draisma HH, van Leeuwen N, Simonis-Bik AMC, Jonsson A, Willemsen G, Bernigau W, Wang-Sattler R, Suhre K, Peters A, Thorand B, Herder C, Rathmann W, Roden M, Gieger C, Kramer MHH, van Heemst D, Pedersen HK, Gudmundsdottir V, Schulze MB, Pischon T, de Geus EJC, Boeing H, Boomsma DI, Ziegler AG, Slagboom PE, Hummel S, Beekman M, Grallert H, Brunak S, McCarthy MI, Gupta R, Pearson ER, Adamski J, ter Hart LM. Metabolite ratios as potential biomarkers for type 2 diabetes: a DIRECT study. Diabetologia 61: 117–129, 2018. doi: 10.1007/s00125-017-4436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moloney AM, Griffin RJ, Timmons S, O’Connor R, Ravid R, O’Neill C. Defects in IGF-1 receptor, insulin receptor and IRS-1/2 in Alzheimer’s disease indicate possible resistance to IGF-1 and insulin signalling. Neurobiol Aging 31: 224–243, 2010. doi: 10.1016/j.neurobiolaging.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Morris JK, Burns JM. Insulin: an emerging treatment for Alzheimer’s disease dementia? Curr Neurol Neurosci Rep 12: 520–527, 2012. doi: 10.1007/s11910-012-0297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris JK, Uy RAZ, Vidoni ED, Wilkins HM, Archer AE, Thyfault JP, Miles JM, Burns JM. Effect of APOE ε4 Genotype on Metabolic Biomarkers in Aging and Alzheimer’s Disease. J Alzheimers Dis 58: 1129–1135, 2017. doi: 10.3233/JAD-170148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris JK, Uy RAZ, Vidoni ED, Wilkins HM, Archer AE, Thyfault JP, Miles JM, Burns JM. Effect of APOE e4 genotype on metabolic biomarkers in aging and Alzheimer’s Disease. J Alzheimers Dis 58: 1129–1135, 2017. doi: 10.3233/JAD-170148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris JK, Vidoni ED, Honea RA, Burns JM; Alzheimer’s Disease Neuroimaging Initiative . Impaired glycemia increases disease progression in mild cognitive impairment. Neurobiol Aging 35: 585–589, 2014. doi: 10.1016/j.neurobiolaging.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris JK, Vidoni ED, Mahnken JD, Montgomery RN, Johnson DK, Thyfault JP, Burns JM. Cognitively impaired elderly exhibit insulin resistance and no memory improvement with infused insulin. Neurobiol Aging 39: 19–24, 2016. doi: 10.1016/j.neurobiolaging.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Notkola IL, Sulkava R, Pekkanen J, Erkinjuntti T, Ehnholm C, Kivinen P, Tuomilehto J, Nissinen A. Serum total cholesterol, apolipoprotein E epsilon 4 allele, and Alzheimer’s disease. Neuroepidemiology 17: 14–20, 1998. doi: 10.1159/000026149. [DOI] [PubMed] [Google Scholar]
- 45.Okekunle AP, Li Y, Liu L, Du S, Wu X, Chen Y, Li Y, Qi J, Sun C, Feng R. Abnormal circulating amino acid profiles in multiple metabolic disorders. Diabetes Res Clin Pract 132: 45–58, 2017. doi: 10.1016/j.diabres.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 46.Ott A, Stolk RP, van Harskamp F, Pols HAP, Hofman A, Breteler MMB. Diabetes mellitus and the risk of dementia: The Rotterdam Study. Neurology 53: 1937–1942, 1999. doi: 10.1212/WNL.53.9.1937. [DOI] [PubMed] [Google Scholar]
- 47.Peila R, Rodriguez BL, Launer LJ; Honolulu-Asia Aging Study . Type 2 diabetes, APOE gene, and the risk for dementia and related pathologies: The Honolulu-Asia Aging Study. Diabetes 51: 1256–1262, 2002. doi: 10.2337/diabetes.51.4.1256. [DOI] [PubMed] [Google Scholar]
- 48.Prasinou P, Dafnis I, Giacometti G, Ferreri C, Chroni A, Chatgilialoglu C. Fatty acid-based lipidomics and membrane remodeling induced by apoE3 and apoE4 in human neuroblastoma cells. Biochim Biophys Acta 1859: 1967–1973, 2017. doi: 10.1016/j.bbamem.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 49.Proitsi P, Kim M, Whiley L, Pritchard M, Leung R, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, Vellas B, Sham P, Lovestone S, Powell JF, Dobson RJ, Legido-Quigley C. Plasma lipidomics analysis finds long chain cholesteryl esters to be associated with Alzheimer’s disease. Transl Psychiatry 5: e494, 2015. doi: 10.1038/tp.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Proitsi P, Kim M, Whiley L, Simmons A, Sattlecker M, Velayudhan L, Lupton MK, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, Vellas B, Lovestone S, Powell JF, Dobson RJ, Legido-Quigley C. Association of blood lipids with Alzheimer’s disease: A comprehensive lipidomics analysis. Alzheimers Dement 13: 140–151, 2017. doi: 10.1016/j.jalz.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Rebeck GW. The role of APOE on lipid homeostasis and inflammation in normal brains. J Lipid Res 58: 1493–1499, 2017. doi: 10.1194/jlr.R075408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savage DB, Petersen KF, Shulman GI. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev 87: 507–520, 2007. doi: 10.1152/physrev.00024.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savica R, Murray ME, Persson XM, Kantarci K, Parisi JE, Dickson DW, Petersen RC, Ferman TJ, Boeve BF, Mielke MM. Plasma sphingolipid changes with autopsy-confirmed Lewy Body or Alzheimer’s pathology. Alzheimers Dement (Amst) 3: 43–50, 2016. doi: 10.1016/j.dadm.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scholz M, Fiehn O. SetupX–a public study design database for metabolomic projects. Pac Symp Biocomput 12: 169–180, 2007. [PubMed] [Google Scholar]
- 55.Sekhar RV, McKay SV, Patel SG, Guthikonda AP, Reddy VT, Balasubramanyam A, Jahoor F. Glutathione synthesis is diminished in patients with uncontrolled diabetes and restored by dietary supplementation with cysteine and glycine. Diabetes Care 34: 162–167, 2011. doi: 10.2337/dc10-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sethi PK, Muralidhara S, Bruckner JV, White CA. Measurement of plasma protein and lipoprotein binding of pyrethroids. J Pharmacol Toxicol Methods 70: 106–111, 2014. doi: 10.1016/j.vascn.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Steen E, Terry BM, Rivera EJ, Cannon JL, Neely TR, Tavares R, Xu XJ, Wands JR, de la Monte SM. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease–is this type 3 diabetes? J Alzheimers Dis 7: 63–80, 2005. doi: 10.3233/JAD-2005-7107. [DOI] [PubMed] [Google Scholar]
- 58.Suhre K, Meisinger C, Döring A, Altmaier E, Belcredi P, Gieger C, Chang D, Milburn MV, Gall WE, Weinberger KM, Mewes HW, Hrabé de Angelis M, Wichmann HE, Kronenberg F, Adamski J, Illig T. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One 5: e13953, 2010. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Talbot K, Wang HY, Kazi H, Han LY, Bakshi KP, Stucky A, Fuino RL, Kawaguchi KR, Samoyedny AJ, Wilson RS, Arvanitakis Z, Schneider JA, Wolf BA, Bennett DA, Trojanowski JQ, Arnold SE. Demonstrated brain insulin resistance in Alzheimer’s disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J Clin Invest 122: 1316–1338, 2012. doi: 10.1172/JCI59903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toledo JB, Arnold M, Kastenmuller G, Chang R, Baillie RA, Han X, Thambisetty M, Tenenbaum JD, Suhre K, Thompson JW, John-Williams LS, MahmoudianDehkordi S, Rotroff DM, Jack JR, Motsinger-Reif A, Risacher SL, Blach C, Lucas JE, Massaro T, Louie G, Zhu H, Dallmann G, Klavins K, Koal T, Kim S, Nho K, Shen L, Casanova R, Varma S, Legido-Quigley C, Moseley MA, Zhu K, Henrion MY, van der Lee SJ, Harms AC, Demirkan A, Hankemeier T, van Duijn CM, Trojanowski JQ, Shaw LM, Saykin AJ, Weiner MW, Doraiswamy PM, Kaddurah-Daouk R; Alzheimer’s Disease Neuroimaging Initiative; the Alzheimer Disease Metabolomics Consortium . Metabolic network failures in Alzheimer’s disease-A biochemical road map. Alzheimers Dement 13: 965–984, 2017. doi: 10.1016/j.jalz.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai MS, Tangalos EG, Petersen RC, Smith GE, Schaid DJ, Kokmen E, Ivnik RJ, Thibodeau SN. Apolipoprotein E: risk factor for Alzheimer disease. Am J Hum Genet 54: 643–649, 1994. [PMC free article] [PubMed] [Google Scholar]
- 62.Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods 12: 523–526, 2015. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Doorn M, Vogels J, Tas A, van Hoogdalem EJ, Burggraaf J, Cohen A, van der Greef J. Evaluation of metabolite profiles as biomarkers for the pharmacological effects of thiazolidinediones in Type 2 diabetes mellitus patients and healthy volunteers. Br J Clin Pharmacol 63: 562–574, 2007. doi: 10.1111/j.1365-2125.2006.02816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Velayudhan L, Poppe M, Archer N, Proitsi P, Brown RG, Lovestone S. Risk of developing dementia in people with diabetes and mild cognitive impairment. Br J Psychiatry 196: 36–40, 2010. doi: 10.1192/bjp.bp.109.067942. [DOI] [PubMed] [Google Scholar]
- 65.Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, Heim K, Campillos M, Holzapfel C, Thorand B, Grallert H, Xu T, Bader E, Huth C, Mittelstrass K, Döring A, Meisinger C, Gieger C, Prehn C, Roemisch-Margl W, Carstensen M, Xie L, Yamanaka-Okumura H, Xing G, Ceglarek U, Thiery J, Giani G, Lickert H, Lin X, Li Y, Boeing H, Joost HG, de Angelis MH, Rathmann W, Suhre K, Prokisch H, Peters A, Meitinger T, Roden M, Wichmann HE, Pischon T, Adamski J, Illig T. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol 8: 615, 2012. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O’Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med 17: 448–453, 2011. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, Ghorbani A, O’Sullivan J, Cheng S, Rhee EP, Sinha S, McCabe E, Fox CS, O’Donnell CJ, Ho JE, Florez JC, Magnusson M, Pierce KA, Souza AL, Yu Y, Carter C, Light PE, Melander O, Clish CB, Gerszten RE. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest 123: 4309–4317, 2013. doi: 10.1172/JCI64801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warnakula S, Hsieh J, Adeli K, Hussain MM, Tso P, Proctor SD. New insights into how the intestine can regulate lipid homeostasis and impact vascular disease: frontiers for new pharmaceutical therapies to lower cardiovascular disease risk. Can J Cardiol 27: 183–191, 2011. doi: 10.1016/j.cjca.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 69.Waters LL. The extraction of lipids from human plasma or serum by 2-octanol. Yale J Biol Med 37: 204–210, 1964. [PMC free article] [PubMed] [Google Scholar]
- 70.Westerhuis JA, Hoefsloot HCJ, Smit S, Vis DJ, Smilde AK, van Velzen EJJ, van Duijnhoven JPM, van Dorsten FA. Assessment of PLSDA cross validation. Metabolomics 4: 81–89, 2008. doi: 10.1007/s11306-007-0099-6. [DOI] [Google Scholar]
- 71.Wood PL, Locke VA, Herling P, Passaro A, Vigna GB, Volpato S, Valacchi G, Cervellati C, Zuliani G. Targeted lipidomics distinguishes patient subgroups in mild cognitive impairment (MCI) and late onset Alzheimer’s disease (LOAD). BBA Clin 5: 25–28, 2015. doi: 10.1016/j.bbacli.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu F, Tavintharan S, Sum CF, Woon K, Lim SC, Ong CN. Metabolic signature shift in type 2 diabetes mellitus revealed by mass spectrometry-based metabolomics. J Clin Endocrinol Metab 98: E1060–E1065, 2013. doi: 10.1210/jc.2012-4132. [DOI] [PubMed] [Google Scholar]
- 73.Xu W, Qiu C, Gatz M, Pedersen NL, Johansson B, Fratiglioni L. Mid- and late-life diabetes in relation to the risk of dementia: a population-based twin study. Diabetes 58: 71–77, 2009. doi: 10.2337/db08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu WL, Qiu CX, Wahlin A, Winblad B, Fratiglioni L. Diabetes mellitus and risk of dementia in the Kungsholmen project: a 6-year follow-up study. Neurology 63: 1181–1186, 2004. doi: 10.1212/01.WNL.0000140291.86406.D1. [DOI] [PubMed] [Google Scholar]
- 75.Yano M, Watanabe K, Yamamoto T, Ikeda K, Senokuchi T, Lu M, Kadomatsu T, Tsukano H, Ikawa M, Okabe M, Yamaoka S, Okazaki T, Umehara H, Gotoh T, Song WJ, Node K, Taguchi R, Yamagata K, Oike Y. Mitochondrial dysfunction and increased reactive oxygen species impair insulin secretion in sphingomyelin synthase 1-null mice. J Biol Chem 286: 3992–4002, 2011. doi: 10.1074/jbc.M110.179176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X, Wang Y, Hao F, Zhou X, Han X, Tang H, Ji L. Human serum metabonomic analysis reveals progression axes for glucose intolerance and insulin resistance statuses. J Proteome Res 8: 5188–5195, 2009. doi: 10.1021/pr900524z. [DOI] [PubMed] [Google Scholar]
- 77.Zhu L, Zhong M, Elder GA, Sano M, Holtzman DM, Gandy S, Cardozo C, Haroutunian V, Robakis NK, Cai D. Phospholipid dysregulation contributes to ApoE4-associated cognitive deficits in Alzheimer’s disease pathogenesis. Proc Natl Acad Sci USA 112: 11965–11970, 2015. doi: 10.1073/pnas.1510011112. [DOI] [PMC free article] [PubMed] [Google Scholar]