Abstract
Flavan-3-ols and oligomeric proanthocyanidins (PAs) are the main nutritional polyphenols in green tea (Camellia sinensis), which provide numerous benefits to human health. To date, the regulatory mechanism of flavan-3-ol biosynthesis in green tea remains open to study. Herein, we report the characterization of a C. sinensis tryptophan-aspartic acid repeat protein (CsWD40) that interacts with myeloblastosis (MYB) and basic helix-loop-helix (bHLH) transcription factors (TFs) to regulate the biosynthesis of flavan-3-ols. Full length CsWD40 cDNA was cloned from leaves and was deduced to encode 342 amino acids. An in vitro yeast two-hybrid assay demonstrated that CsWD40 interacted with two bHLH TFs (CsGL3 and CsTT8) and two MYB TFs (CsAN2 and CsMYB5e). The overexpression of CsWD40 in Arabidopsis thaliana transparent testa glabra 1 (ttg1) restored normal trichome and seed coat development. Ectopic expression of CsWD40 alone in tobacco resulted in a significant increase in the anthocyanins of transgenic petals. CsWD40 was then coexpressed with CsMYB5e in tobacco plants to increase levels of both anthocyanins and PAs. Furthermore, gene expression analysis revealed that CsWD40 expression in tea plants could be induced by several abiotic stresses. Taken together, these data provide solid evidence that CsWD40 partners with bHLH and MYB TFs to form ternary WBM complexes to regulate anthocyanin, PA biosynthesis, and trichome development.
Keywords: Camellia sinensis, anthocyanin biosynthesis, CsWD40, flavan-3-ols, MYB–bHLH–WD40 complex
1. Introduction
Flavonoids are widely distributed and ubiquitous secondary metabolites in the plant kingdom [1]. Flavonoid compounds have been confirmed to be involved in various important physiological functions in plants, such as seed germination, protection from ultraviolet radiation, and pathogenic microorganism defense erosion [2,3,4]. More importantly, flavonoids have health benefits to human beings, such as antioxidative, antihypertensive, anti-inflammatory, antiaging, and insulin-sensitizing activities [5,6,7].
In plants, chalcones, flavones, flavonols, flavan-3-ols, anthocyanins, and proanthocyanidins (PAs, also called condensed tannins) are the primary subgroups of flavonoids [1]. Their biosynthetic pathways have been extensively studied over the past three decades [1,8,9]. The main pathway enzymes involved in flavonoid biosynthesis include chalcone synthase (CHS), chalconeisomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), flavonoid 3′,5′-hydroxylase (F3′5′H), flavonol synthase (FLS), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and anthocyanidin reductase (ANR) [1].
In addition, regulation of the flavonoid pathway has been studied in-depth. To date, many transcription factors (TFs) have been identified from different plant species. Three TF families, WD40, basic helix-loop-helix, and MYB (v-myb avian myeloblastosis viral oncogene homolog), have been extensively studied to elucidate the regulation mechanism.
Numerous past studies have demonstrated that these three families form MBW (MYB-bHLH-WD40) ternary complexes to regulate the biosynthesis of anthocyanins and PA in all investigated plants, and the development of root hairs and trichomes in some plants [10,11,12,13,14]. For example, in Arabidopsis, four MBW complexes, PAP1(MYB75)–TT8/GL3–TTG1(WD40), PAP2(MYB90)–TT8/GL3–TTG1, MYB113(PAP3)–TT8/GL3–TTG1, and PAP4(MYB114)–TT8/GL3–TTG1 have been shown to activate the expression of late anthocyanin biosynthetic genes, such as AtDFR, AtANS/LDOX, and UF3GT [10,15,16]. The MBW complex TT2 (MYB123)–TT8/GL3–TTG1 regulated the expression of AtDFR, AtANS/LDOX, AtANR/BANYULS (BAN), glutathione-S-transferase (TT19), and a MATE (multidrug and toxic compound extrusion) transporter-encoding (TT12) involved in the PA biosynthesis [16,17].
In all of the MBW complexes, the WD40 repeat protein is localized in the center of ternary structure [18,19]. Unlike the bHLH and MYB members, WD40 has been shown to functionally enhance complex activation rather than directly participate in the recognition of the target gene promoter [20]. To date, several other WD40 repeat proteins that are orthologs to Arabidopsis TTG1 have been reported in a few of plant species, such as Perilla frutescens, petunia (Petunia hybrida), cotton (Gossypium hirsutum), and maize (Zea mays). It is interesting that the gene encoding WD40 has been shown to have only one copy in the same species [21,22,23].
To date, the biosynthetic pathways of flavan-3-ols and PAs have gained intensive studies for human health benefits. Recently, the genome of green tea has been sequenced from different cultivars. These studies provide important knowledge to enhance tea industries for improved health benefits. However, the regulation mechanism of the flavonoid pathway in green tea remains to be elucidated. Herein, we report the cloning and functional analysis of cDNA encoding a WD40 repeat protein from green tea, namely CsWD40. Transgenic analysis, genetic complementation, and protein–protein interactions were performed to characterize that CsWD40 partner with R2R3-MYB and bHLH to form complexes which regulate the biosynthesis of anthocyanins, flavan-3-ols, and PAs. All data are fundamental to comprehensively elucidate the regulation of the green tea flavonoid pathway.
2. Results
2.1. Molecular Cloning and Sequence Analysis of CsWD40
The full-length cDNA of CsWD40 was obtained from tender leaves of the local cultivar C. sinensis Nongkangzao’ through rapid amplification of cDNA ends polymerase chain reaction (RACE-PCR). The 1029-bp open reading frame (ORF) of CsWD40 codes a protein with 342 amino acid residues. Alignment with CsWD40 proteins from other species revealed that four WD40 repeat domains are highly conserved among all WD40 repeat proteins (Figure S1). CsWD40 shared 79.62% and 77.17% identity with MdTTG1 (GU173813) from Malus domestica and PgTTG1 (HQ199314) from Punicagranatum, respectively (Figure S1). MdTTG1 and PgTTG1 TFs have been reported to regulate anthocyanin biosynthesis in M. domestica and P. granatum plants, respectively [24,25]. Therefore, their orthologous gene CsWD40 was predicted to be a TF that regulates anthocyanin biosynthesis in tea plants.
2.2. CsWD40 Interacts with MYB and bHLH TFs
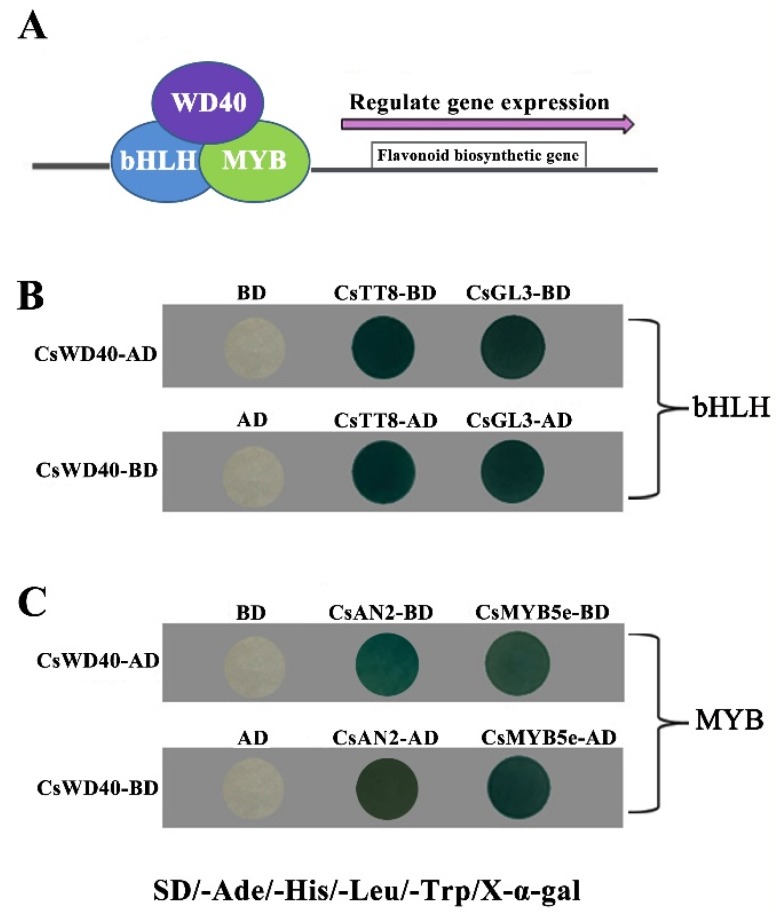
The bHLH type TF genes CsTT8 and CsGL3 in C. sinensis are the orthologs of AtTT8 and AtGL3 in Arabidopsis thaliana, respectively. The MYB type TF genes CsAN2 and CsMYB5e are the orthologs of AtMYB75 in A. thaliana and MtMYB5 in M. truncatula, respectively. AtTT8, AtGL3, and AtTT2 have been demonstrated to form MBW complexes with WD40 proteins; these complexes regulate the phenylpropanoid pathway [20]. Therefore, CsWD40, CsMYB5e, CsTT8, and CsGL3 were selected to determine whether interactions occur among them in the yeast two-hybrid system.
The yeast two-hybrid test results revealed that CsWD40 specifically interacted with the bHLH TFs CsTT8 and CsGL3 (Figure 1B). Moreover, both CsAN2 and CsMYB5e could physically interact with CsWD40 (Figure 1C). CsWD40 exhibited no autoactivation. The results indicated that CsGL3, CsTT8, CsAN2, CsMYB5e, and CsWD40 may be involved in jointly regulating the flavonoid pathway in tea plants.
Figure 1.
Protein interactions between CsWD40 and other transcript factors from tea plants by two-hybrid system. (A) A model pattern of the MYB–bHLH–WD40 ternary complex. (B) Protein–protein interactions between CsWD40 and bHLH transcript factors (CsTT8 and CsGL3) from tea plants by two-hybrid system. (C) Protein–protein interactions between CsWD40 and MYB transcript factors (CsAN2 and CsMYB5e) from tea plants by two-hybrid system.
2.3. CsWD40 Complements the Arabidopsis ttg1 Deficient Phenotype
In A. thaliana, TTG1 not only regulates anthocyanin synthesis but is also involved in trichome organogenesis, seed coat pigment synthesis, root hair development, and regulation and control of negative cotyledon hypocotyl stomatal cell movement [18,19].
To determine whether CsWD40 is a functional ortholog of TTG1 that can restore the deficient phenotypes in the Arabidopsis ttg1 mutant, the ORF of CsWD40, under the control of the 35S promoter, was transformed into the mutant. The surface of the ttg1 mutant leaves appeared smooth and there was no trichome on them. The seed coats lost the pigment store and appeared yellow in comparison with the wild type. The ttg1 mutant lost red pigmentation in the stem of seedlings even in those induced by 6% sucrose. The results showed that overexpression of CsWD40 fully complemented the trichome deficiency and pigmentation phenotypes in the stems of the ttg1 mutant (Figure 2). The seed coat color of the transgenic mutant recovered partially (Figure 2).
Figure 2.
CsWD40 complements the phenotypes of Arabidopsis ttg1 mutant. (A) Leaf trichome occurrence. Bar = 0.5mm (B) Seed coat pigmentation. (C) Trichome seedling.
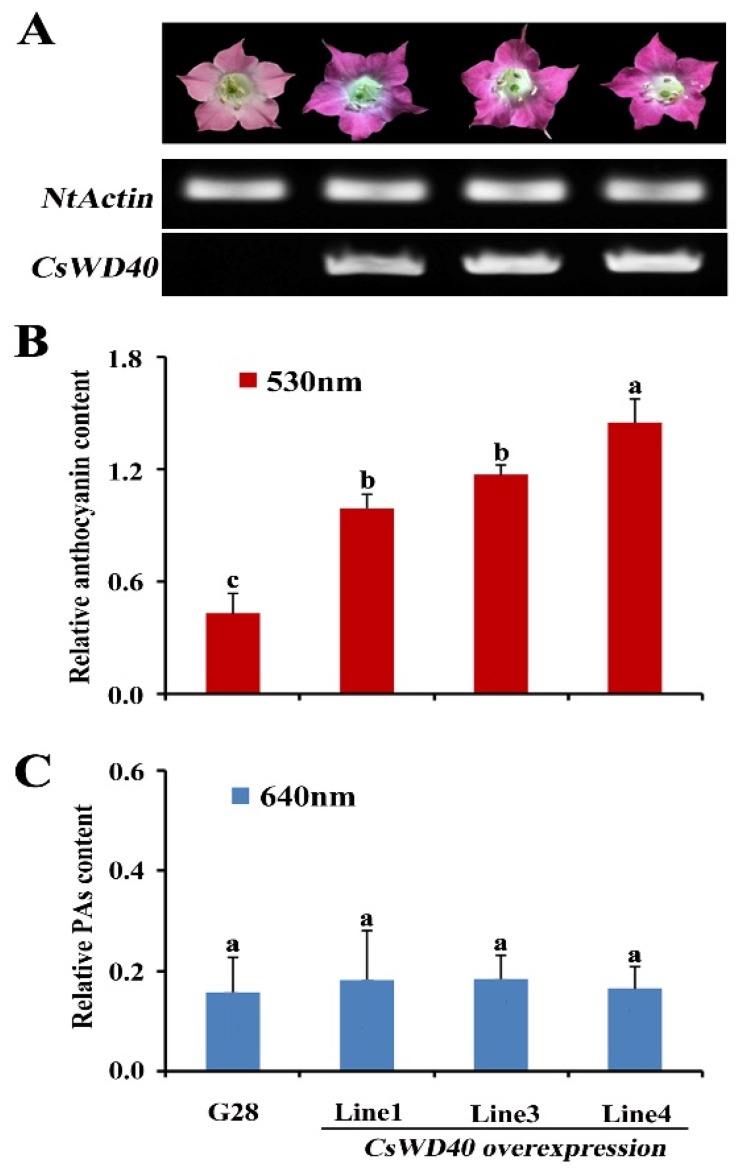
2.4. Anthocyanin and PA Accumulation in the Flowers of CsWD40-Overexpressing Tobacco Plants
To ascertain the putative function of CsWD40, a tobacco gene transformation system was used for ectopic expression experiments. The petals of T1- and T2-generation CsWD40-overexpressing tobacco plants showed a deeper pink color than those of the control (G28) (Figure 3A). The expression level of CsWD40 was positively associated with anthocyanin content (Figure 3A,B). This result indicates that CsWD40 is involved in anthocyanin biosynthesis. Pang reported that MtWD40 in Medicago truncatula participates in PA accumulation [26]. Therefore, in this study, PA content in the flowers of transgenic tobacco plants was determined. No blue color was developed after a reaction with the DMACA (7-DIMETHYLAMINOCOUMARIN-4-ACETICACID) reagent for transgenic and wild-type tobacco plants (Figure 3C), indicating that CsWD40 overexpression did not affect PA biosynthesis in the flowers of transgenic tobacco plants.
Figure 3.
Identification of the CsWD40 function in transgenic tobacco. (A) Analysis of CsWD40 transcription levels in the flowers by semiquantitative PCR. (B) Relative content of anthocyanin in the flowers of CsWD40 overexpressing tobacco. (C) Relative soluble proanthocyanidin content in the flowers of CsWD40 overexpressing tobacco. All data are the means of three biological replicates, and the error bars represent the standard deviation of three replicates. Statistical significance was analyzed using ANOVA software (ANOVA ALL MAC VERSION 2.0, Thomas Hanson, OR, USA). Means followed by the same letter are not significantly different (p > 0.05).
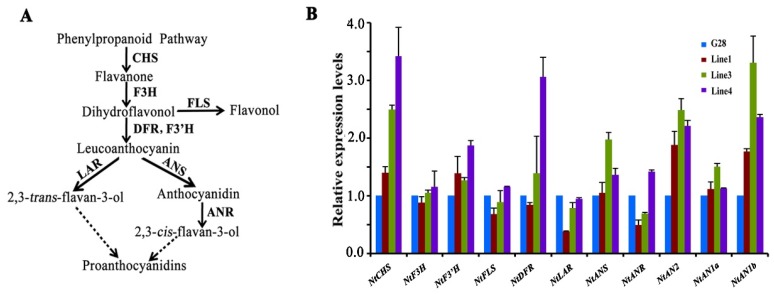
2.5. Analysis of Expression of Genes Involved in Flavonoid Biosynthesis in the Flowers of CsWD40-Overexpressing Tobacco Plants
qRT-PCR was performed to analyze the expression of genes involved in the flavonoid biosynthetic pathway in transgenic tobacco petals. The results showed that the key structural genes, NtCHS, NtF3′H, NtDFR, and NtANS were upregulated significantly (Figure 4B). Furthermore, the transcription factor genes NtAN2 and NtAN1b were expressed highly in all transgenic tobacco petals (Figure 4B). NtAN2 and NtAN1b were clearly activated in anthocyanin biosynthesis in tobacco [27]. These results indicate that CsWD40 acts as a positive participator and upregulates key structural and transcript factor genes involved in the anthocyanin pathway in transgenic tobacco plants.
Figure 4.
Expression of genes involved in flavonoid biosynthesis in the flowers of CsWD40-overexpressing tobacco plants. (A) A schematic diagram of flavonoid biosynthetic pathway. (B) Expression profiles of genes in flavonoid pathway in flowers of transgenic CsWD40 tobacco lines. CHS, chalcone synthase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3-hydroxylase; DFR, dihydroflavonol reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; FLS, flavonol synthase; AN2, N. tabacum Anthocyanin 2; AN1a, N. tabacum Anthocyanin 1a; AN1b, N. tabacum Anthocyanin 1b.
2.6. Anthocyanin and PA Content in CsWD40- and CsMYB5e-Overexpressing Tobacco Plants
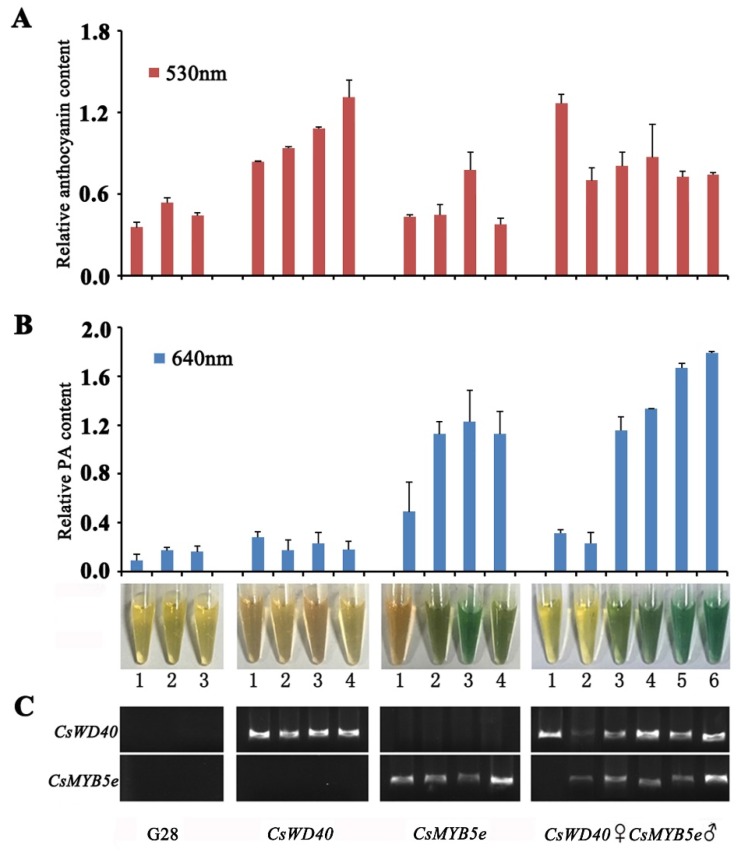
The aforementioned yeast two-hybrid test results suggested that CsWD40 could positively interact with CsMYB5e. To validate the synergistic effects of CsMYB5e and CsWD40 function, we generated CsWD40- and CsMYB5e-overexpressing tobacco plants. We then cross-pollinated CsWD40 and CsMYB5e transgenic tobacco plants in both ♀ × ♂ and ♂ × ♀ directions to produce co-overexpressing transgenic tobacco. Eleven CsWD40♀CsMYB5e♂ and nine CsWD40♂CsMYB5e♀ transgenic plants were verified by RT-PCR. There was no phenotypical difference between the flowers of CsWD40♀CsMYB5e♂ and CsWD40♂CsMYB5e♀ transgenic plants (Figure S2).
CsWD40 expression resulted in significantly increased anthocyanin content in the flowers of transgenic tobacco plants but did not affect PA content (Figure 5A,B). The extract solution of petals from the flowers of CsMYB5e lines turned blue after a reaction with DMACA (Figure 5B), indicating high PA accumulation in CsMYB5e-overexpressing tobacco plants, but slight changes in anthocyanin content in CsMYB5e tobacco plants than in control plants (Figure 5).
Figure 5.
Anthocyanin and proanthocyanidin accumulation variated in petals of co-overexpressing CsWD40 and CsMYB5e transgenic tobacco. (A) The relative content of anthocyanin was calculated on the record of absorbance at 530 nm. (B) The relative content of proanthocyanidin was calculated on the record of absorbance at 640 nm after reaction with DMACA regent. (C) RT–PCR determination of the CsWD40 and CsMYB5e expression levels in CsWD40, CsMYB5e, and CsWD40♀CsMYB5e♂ transgenic tobacco flowers. All the data were present based on three biological and technical repeats.
The data showed that anthocyanin and PA accumulation in CsWD40♀CsMYB5e♂ transgenic lines were markedly higher than those in control plants (Figure 5). In the CsWD40♀CsMYB5e♂ transgenic plants, high CsWD40 expression and low CsMYB5e expression resulted in marked anthocyanin accumulation but slight PA accumulation. The highest PA accumulation was detected in lines with high CsWD40 and CsMYB5e expression. The petals of CsMYB5e and CsWD40 co-overexpressing transgenic tobacco plants exhibited higher levels of anthocyanin than those of lines overexpressing CsMYB5e alone (Figure 5, Figure S2), but anthocyanin levels were lower in the co-overexpressing tobacco plants than in the tobacco plants expressing CsWD40 alone (Figure 5A).
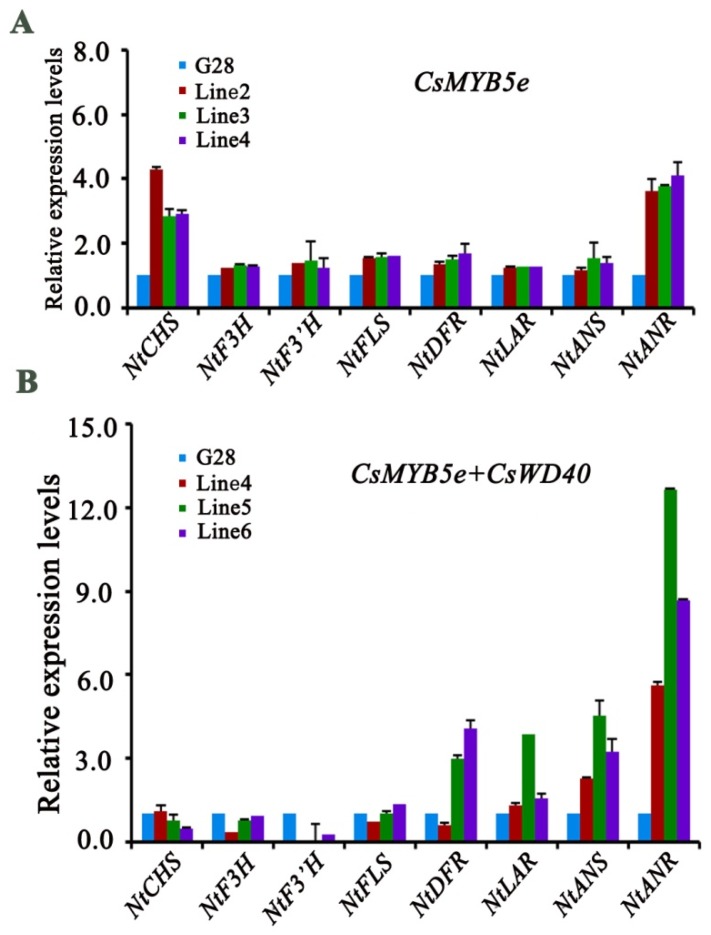
2.7. Expression Analysis of Genes Involved in Flavonoid Biosynthesis in CsWD40 and CsMYB5e Transgenic Tobacco
Gene expression profiling using qRT-PCR analysis was completed to understand the effects of overexpression of CsWD40 and CsMYB5e on flavonoid pathway genes in transgenic tobacco plants. In CsMYB5e alone transgenic flowers, expression levels of NtCHS and NtANR increased more than two-fold (Figure 6A). In CsWD40 (♀) and CsMYB5e (♂) coupled expression transgenic lines, the expression levels NtANS and NtANR increased more than four-fold and five-fold, respectively (Figure 6B). In addition, the expression levels of NtLAR and NtDFR significantly increased. The expression of other genes was either slightly increased or similar between transgenic and wild type flowers.
Figure 6.
(A) Relative expression of flavonoid biosynthetic pathway genes in CsMYB5e overexpression tobacco petals. (B) Relative expression of flavonoid biosynthetic pathway genes in CsWD40 and CsMYB5e overexpressing tobacco petals. CHS, chalcone synthase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3-hydroxylase; DFR, dihydroflavonol reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; FLS, flavonol synthase.
2.8. Expression Patterns of CsWD40 in Tea Leaves under Different Abiotic Stresses
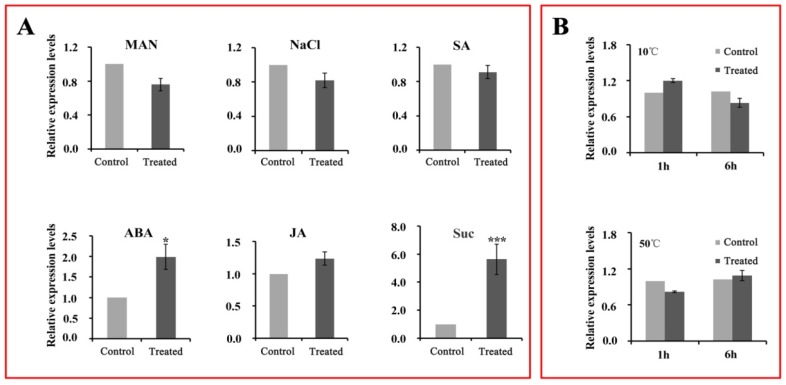
TaWD40D overexpression enhanced the tolerance of Triticum aestivum plants to salt, mannitol, and abscisic acid (ABA) stresses, indicating that WD40 proteins may be involved in the response of plants to environmental stresses [28]. Therefore, we analyzed the expression patterns of CsWD40 induced by different abiotic stresses, including sucrose (Suc), ABA, mannitol (Man), sodium chloride (NaCl), salicylic acid (SA), and jasmonic acid (JA) stresses. The results showed that compared to the control, the expression level of the CsWD40 gene increased nearly 2-fold under ABA stress and approximately 6-fold under sucrose stress (Figure 7A). Under mannitol, NaCl, SA, and JA stresses, the expression level of the CsWD40 gene varied slightly in comparison with the control (Figure 7A). To determine whether CsWD40 expression is related to temperature changes, we compared CsWD40 expression under low (10 °C) and high (50 °C) temperatures (Figure 7B). Our data indicated no significant difference was observed in the expression level of CsWD40 under different temperatures.
Figure 7.
Effects of different abiotic treatments on the expression levels of CsWD40 in tea leaves. (A) The transcript levels of CsWD40 under mannitol (MAN), sodium chloride (NaCl), salicylic acid (SA), abscisic acid (ABA), jasmonic acid (JA), and sucrose (Suc) stresses. (B) The transcript levels of CsWD40 under different temperatures. The asterisks indicate the significant level (n = 3, * p < 0.05, *** p < 0.001) based on a Tukey’s honestly significant difference test.
3. Discussion
Flavonoid compounds are widely distributed and are ubiquitous secondary metabolites, mainly existing in the root, leaf, fruit, and skin of plants ranging from spermatophytes to mosses [9,20]. A complex comprising an R2R3-MYB TF, a bHLH domain protein, and a WD40 repeat protein regulates the production of anthocyanin and PA in seed coats, roots, leaves, and fruit [10,20,26,29]. This complex also controls the formation of root hairs and trichomes on aerial tissues in some, but not all, plants [14].
WD40 repeat proteins are widely present in plants, animals, and unicellular eukaryotes such as fungi and slime molds [30]. WD40 repeat proteins comprise a superfamily of proteins with a β-propeller structure. The core region of WD40 repeat proteins contains 40 amino acid residues, including histidine–glycine and aspartic acid–tryptophane dipeptides. The conserved motifs can be arranged in 4–16 tandems in the same protein. Eight such random repeats are present in the β-subunit of G protein in higher eukaryotic organs.
Plant flavonoid-related WD proteins are grouped into the 4 WD-repeat (4WDR) subfamily of the TTG (Transparent teata glabra) family. The functions of several WD40 repeat proteins have been reported in petunia (Petunia hybrida), Perilla frutescens, cotton (Gossypium hirsutum), and maize (Zea mays) [22,23,31]. In A. thaliana, only one WD40 protein (TTG1), involved in anthocyanin biosynthesis and trichome formation, has been reported [32]. The ortholog (MtWD40-1) of AtTTG1 is necessary for tissue-specific anthocyanin and PA biosynthesis in M. truncatula [33]. Two WD repeat genes (GhTTG1 and GhTTG2) were found in G. hirsutum [23], but only GhTTG1 could restore trichome formation in the Arabidopsis ttg1 mutant and complemented the anthocyanin deficiency in the white-flowered Matthiola incana ttg1 mutant.
A total of 195 candidate WD genes were isolated from the transcriptome data sets of C. sinensis [34]. However, only one WD gene was confirmed to be the ortholog of AtTTG1 in tea plants. As shown in Figure 3, the petals of CsWD40 transgenic tobacco plants accumulated a large amount of anthocyanins, but no observable PA was found in these plants. In the Arabidopsis ttg1 mutant, CsWD40 not only complemented trichome deficiency but also restored seed coat color. The results implied that CsWD40 improved PA production in the seeds of the ttg1 mutant. This result also implied the different roles of CsWD40 in transgenic tobacco and Arabidopsis plants. The cause was probably the difference in the dominant flavonoid biosynthesized in the petals of tobacco plants and the seeds of Arabidopsis plants. Anthocyanin is mainly biosynthesized in the petals of tobacco plants [35], whereas PA is mainly biosynthesized in the seeds of Arabidopsis plants [17].
Besides, not only the structural genes, NtCHS, NtF3′H, NtDFR, and NtANS, but also the transcript factor genes, NtAN2 and NtANb1, were upregulated significantly in CsWD40 transgenic tobacco petals (Figure 5B). A potential reason for this is that CsWD40 improves the stability of some MBW complexes or promotes the formation of some MBW complexes. These complexes may regulate the gene expression of some transcription factors. For example, TT8 expression was found to be directly regulated by TT8 itself through a positive feedback regulatory loop involving redundant MBW complexes [36,37]. TT8 promoter activity is itself was partially regulated by TT1 [37].
The anthocyanin and PA biosynthetic pathways are regulated by different MBW complexes. Different MYB TFs play critical roles in the regulation of the complexes [24]. For example, TT2 (ATMYB123) in subgroup 5 mainly regulates PA biosynthesis in Arabidopsis. PAP1 (ATMYB75), in subgroup 6, significantly promotes anthocyanin accumulation in Arabidopsis [38]. In our study, CsWD40 could interact with CsAN2 (the ortholog of PAP1) and CsMYB5e (the ortholog of TT2) in the yeast two-hybrid system (Figure 1). In our previous study, CsAN2 markedly increased anthocyanin biosynthesis in CsAN2-overexpressing tobacco plants [39]. In CsMYB5e transgenic tobacco plants, the PA pathway was markedly upregulated [40]. Expression levels of both anthocyanin and PA biosynthesis related genes were upregulated to varying degrees in the petals of CsWD40 and CsMYB5e co-overexpressing transgenic tobacco plants. CsWD40 and CsMYB5e co-overexpressing transgenic tobacco plants tended to show less anthocyanin production than transgenic tobacco plants expressing CsWD40 alone. This might be because CsWD40 promoted the regulation effects of CsMYB5e, resulting in higher PA biosynthesis than anthocyan in biosynthesis. In our previous studies, CsANR overexpression reduced anthocyanin production and promoted PA accumulation in the petals of tobacco plants [41]. In the Arabidopsis mutant of BAN (loss of function of ANR), a high level of anthocyanin accumulated in the seed coat. Hybridization and yeast two-hybrid assays further confirmed that CsWD40 played a crucial role in the regulation of anthocyanin and PA biosynthesis through the formation of MBW ternary complexes [42].
In a previous study, the ectopic overexpression of TaWD40D (T. aestivum L.) in Arabidopsis greatly increased the tolerance of the plants to ABA (Abscisic acid), salt, and osmotic stresses during seed germination and seedling development [28]. In this study, the transcript levels of the CsWD40 gene were significantly upregulated under sucrose and ABA stresses, but no response was found to mannitol, NaCl, and SA (Salicylicacid) stresses under different temperatures. Our previous study showed that under ABA and sucrose stresses the most flavonoid was accumulated and the related gene expression increased [43]. This result suggests CsWD40 promotes the accumulation of products of the flavonoid biosynthetic pathway, which led to stress tolerance in tea plants.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Numerous samples from different organs (including young sprout, young stem, and tender root) of the tea plants growing on the grounds of the research station at Anhui Agricultural University were obtained during the growth period in early spring. All samples were immediately frozen in liquid nitrogen and were stored at −80 °C for the present investigation.
Wild-type tobacco “G28” plants were used in genetic transformation studies. Tobacco plants were grown in a controlled environment chamber at a constant temperature of 28 ± 3 °C and a 12/12-h (light/dark) photoperiod with a light intensity of 150–200 μmol·m–2·s–1.
A. thaliana (Ecotype Columbia) ttg1 mutant seeds were purchased from the Arabidopsis Seed Bank (http://www.Arabidopsis.org/). The methods used for seed germination and plant growth for A. thaliana were the same as those used for tobacco [44]. For genetic transformation, A. thaliana plants were grown in a green house. The temperature, light intensity, and photoperiod were 22 ± 2 °C, 50 µmol·m−2·s−1, and 16/8 h (light/dark), respectively.
In order to induce the anthocyanin accumulation in seedlings of Arabidopsis thaliana, Arabidopsis seeds were surface sterilized using 70% alcohol for 1 min followed by 3 times of washing with sterile distilled water, then treated with 1.5% sodium hypochlorite (10% Clorox), (SC Johnson Wax, Racine County, WI, USA) for 8 min followed by 6 times of washing with sterile distilled water. Seeds were germinated on MS medium with 6% sucrose. Seedlings were cultured for five days after germination and their phenotypes were observed under microscope.
4.2. Cloning CsWD40 and CsMYB5e
Total RNA was isolated from plants using the RNAiso-mate for Plant Tissue Kit (Takara, Dalian, China), according to the manufacturer’s protocol. cDNAs were reverse transcribed using the PrimeScript® RT Reagent Kit (Takara, Dalian, China), according to the manufacturer’s protocol. The specific conditions of PCR were as follows: 98 °C for 30 s; followed by 30 cycles of 98 °C for 10 s and 60 °C for 20 s; 72 °C for 30 s; and then a 10-min extension step at 72 °C. The cDNAs of CsGL3, CsTT8, CsAN2, and CsMYB5e were provided by Tong Li and Xiaolan Jiang in our university.
4.3. Extraction and Analysis of Anthocyanins and PAs
To extract anthocyanins and PAs, the samples were ground in liquid nitrogen and extracted with extraction solution (0.5% HCl:80% methanol:19.5% water) by vortexing and then sonicating for 30 min at low temperature (4 °C). The samples were centrifuged at 5000× g for 20 min, and the residues were re-extracted twice. The supernatants were combined and diluted with extraction solution to 2 mL. Absorbance of the aqueous phase was measured at 530 nm. The extraction method for PA is described in the article by Jiang [45].
The total PA content was determined spectrophotometrically at 640 nm after a reaction with the DMACA reagent (0.2% (w/v) DMACA in methanol-3 N HCl), with (−)-epicatechin serving as the standard [33].
4.4. Agrobacterium-Mediated Transformation of Arabidopsis and Tobacco
The ORFs of CsWD40 were cloned into a binary vector pDONR207 by using the Gateway® Cloning System (Invitrogen, Carlsbad, CA, USA). The primers used are listed in Table S1. The vectors were constructed and the genetic transformation of tobacco was performed according to the method of Li et al. [43]. The phenotypes of transgenic plants growing in the controlled environment chamber were recorded to characterize the effect of transgene overexpression on growth.
4.5. Hybridization of Different Transgenic Tobacco Lines
The T1-generation plants of CsWD40 and CsMYB5e transgenic tobacco plants were chosen for research. Transgenic tobacco plants separately overexpressing CsMYB5e and CsWD40 were crossed in both ♀ × ♂ and ♂ × ♀ directions. The pollinated ovaries were collected until the seeds ripened. The next generation of plants was grown for subsequent experiments after PCR verification.
4.6. Abiotic Stress Treatment of Tea Shoots
For abiotic stress treatment, tea shoots sprouting approximately 10 cm were cultivated in 90 mM sucrose, 200 mM mannitol, 50 mM NaCl, 10 µM JA, 100 µM ABA, and 20 mM SA for 12 h. All samples were exposed to treatments at 22 °C, with a light intensity of 150–200 µmol·m−2·s−1. Control shoots were cultivated in deionized water. For low- and high-temperature treatment, the shoots were exposed to treatments at 10 and 50 °C for 1 and 6 h, respectively.
4.7. Yeast Two-Hybrid Assay
The plasmids pGADT7 (Clontech Laboratories, Inc., Terra Bella Ave, Mountain View, CA, USA) and pGBKT7 (Clontech Laboratories, Inc., Terra Bella Ave, Mountain View, CA, USA), which contained the GAL4 activation domain and the GAL4 DNA-binding domain, were used for yeast two-hybrid experiments.
To analyze protein interaction, the ORFs of CsWD40, CsGL3, CsTT8, CsAN2, and CsMYB5e were cloned into the pGADT7 (Clontech Laboratories, Inc., Terra Bella Ave, Mountain View, CA, USA) and pGBKT7 vectors through pfu DNA PCR (Thermo Scientific, Waltham, MA, USA). This cloning was performed using the Matchmaker™ Gold Yeast Two-Hybrid System (Clontech), according to the manufacturer’s instructions described in Clontech Yeast Protocol Handbook. The primers with leading and tailing EcoRI and BamHI restriction enzyme sites were used and are listed in Table S1.
BD-CsWD40, CsGL3, CsTT8, CsAN2, or CsMYB5e and AD-CsWD40, CsGL3, CsTT8, CsAN2, or CsMYB5e were cotransformed into the yeast strain Y2HGold (Clontech) using the PEG/LiAC method described in the Clontech Yeast Protocol Handbook. These transformed colonies were tested on synthetic dropout (SD) medium with X-α-Gal lacking leucine, tryptophan, histidine, and adenine (SD/-Ade/-His/-Leu/-Trp).
4.8. Quantitative Real-Time PCR
RNA was extracted from various tissues and was quantified spectrophotometrically (NANODROP 2000, Thermo Scientific). Reverse transcription of RNA into cDNA was performed using 2 μL of 5× PrimeScript RT Master Mix (Takara) and 500 ng of RNA in a reaction volume of 10 µL.
In addition, cDNA was diluted to 25% (v/v) with deionized water before being used as the template. Quantitative real-time PCR was performed in a reaction volume of 20 µL containing 10 µL of SYBR Green PCR Master Mix (Takara), 1.1 µL of cDNA, and 0.8 µL of forward and reverse primers (10 μM). The PCR cycling parameters used were as follows: 95 °C for 30 s and 40 cycles of 95 °C for 5 s, 30 s at 60 °C, and 30 s at 72 °C, followed by melting curve analysis from 55 to 95 °C. The transcription abundance was normalized to the transcription abundance of the control gene and was calculated from three technical replicates. The gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Accession No. GE651107) in tea plants and ribosomal protein L25 (RPL25, Accession No. L18908) in tobacco were used as control genes in qPCR analysis. Their gene primers were listed in Table S1. The relative expression level was calculated using a previously described method [46].
Acknowledgments
This work was supported by the Natural Science Foundation of China (grant numbers 31570694, 31470689 and 31300577, 31300576), and Anhui Major Demonstration Project for the Leading Talent Team on Tea Chemistry and Health, the Chang-Jiang Scholars and the Innovative Research Team in University (IRT1101).
Abbreviations
| ANR | Anthocyanidinreductase |
| ANS | Anthocyanidin synthase |
| CHS | Chalcone synthase |
| DFR | Dihydroflavonolreductase |
| DMACA | Dimethylaminocinnamaldehyde |
| F3H | Flavanone 3-hydroxylase |
| F3′H | Flavonoid 3-hydroxylase |
| FLS | Flavonol synthase |
| MBW | MYB-bHLH-WD40 |
| Pas | Proanthocyanidins |
| ttg1 | Transparent testa glabra 1 |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/6/1686/s1.
Author Contributions
Y.L. and X.J. conceived and designed the experiments; Y.L. and H.H. performed the experiments and analyzed the data; P.W., W.C., and X.D. contributed reagents/materials/analysis tools; L.G. and T.X. drafted the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreyra M.L.F., Rius S.P., Casati P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012;3:222. doi: 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernández I., Alegre L., Van Breusegem F., Munné-Bosch S. How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 2009;14:125–132. doi: 10.1016/j.tplants.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Nakabayashi R., Yonekurasakakibara K., Urano K., Suzuki M., Yamada Y., Nishizawa T., Matsuda F., Kojima M., Sakakibara H., Shinozaki K., et al. Enhancement of oxidative and drought tolerance in Arabidopsis by over accumulation of antioxidant flavonoids. Plant J. 2014;77:367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tohge T., Fernie A.R. An overview of compounds derived from the shikimate and phenylpropanoid pathways and their medicinal importance. Mini-Rev. Med. Chem. 2016;17:1013–1027. doi: 10.2174/1389557516666160624123425. [DOI] [PubMed] [Google Scholar]
- 6.Khan N., Adhami V.M., Mukhtar H. Apoptosis by dietary agents for prevention and treatment of prostate cancer. Endocr-Relat. Cancer. 2010;17:R39–R52. doi: 10.1677/ERC-09-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross J.A., Kasum C.M. Dietary flavonoids: Bioavailability, metabolic effects, and safety. Annu. Rev. Nutr. 2002;22:19–34. doi: 10.1146/annurev.nutr.22.111401.144957. [DOI] [PubMed] [Google Scholar]
- 8.Holton T.A., Cornish E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pang Y., Peel G.J., Wright E., Wang Z., Dixon R.A. Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol. 2007;145:601–615. doi: 10.1104/pp.107.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L.L., Shi M.Z., Xie D.Y. Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-programmed red cells of Arabidopsis thaliana. Planta. 2012;236:825–837. doi: 10.1007/s00425-012-1674-2. [DOI] [PubMed] [Google Scholar]
- 12.Shi M.Z., Xie D.Y. Engineering of red cells of Arabidopsis thaliana and comparative genome-wide gene expression analysis of red cells versus wild-type cells. Planta. 2011;233:787–805. doi: 10.1007/s00425-010-1335-2. [DOI] [PubMed] [Google Scholar]
- 13.Tohge T., Nishiyama Y., Hirai M.Y., Yano M., Nakajima J., Awazuhara M., Inoue E., Takahashi H., Goodenowe D.B., Kitayama M., et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- 14.Wilkins O., Nahal H., Foong J., Provart N., Campbell M. Expansion and diversification of the populus R2R3-MYB family of transcription factors. Plant Physiol. 2009;149:981–993. doi: 10.1104/pp.108.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi M.Z., Xie D.Y. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat. Biotechnol. 2014;8:47–60. doi: 10.2174/1872208307666131218123538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng Y., Lu S. Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 2017;36:1–34. doi: 10.1080/07352689.2017.1402852. [DOI] [Google Scholar]
- 17.Xu W., Grain D., Bobet S., Le G.J., Thévenin J., Kelemen Z., Lepiniec L., Dubos C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014;202:132–144. doi: 10.1111/nph.12620. [DOI] [PubMed] [Google Scholar]
- 18.Ramsay N.A., Glover B.J. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Lloyd A.M. Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 2000;5:214–219. doi: 10.1016/s1360-1385(00)01597-1. [DOI] [PubMed] [Google Scholar]
- 20.Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2010;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 21.Sompornpailin K., Makita Y., Yamazaki M., Saito K. A WD-repeat-containing putative regulatory protein in anthocyanin biosynthesis in Perilla frutescens. Plant Mol. Biol. 2002;50:485–495. doi: 10.1023/A:1019850921627. [DOI] [PubMed] [Google Scholar]
- 22.De N.V., Quattrocchio F., Mol J., Koes R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997;11:1422–1434. doi: 10.1101/gad.11.11.1422. [DOI] [PubMed] [Google Scholar]
- 23.Humphries J.A., Walker A.R., Timmis J.N., Orford S.J. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 2005;57:67–81. doi: 10.1007/s11103-004-6768-1. [DOI] [PubMed] [Google Scholar]
- 24.An X.H., Tian Y., Chen K.Q., Wang X.F., Hao Y.J. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 2012;169:710–717. doi: 10.1016/j.jplph.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Simhon Z., Judeinstein S., Nadler-Hassar T., Trainin T., Bar-Ya’akov I., Borochov-Neori H., Holland D. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta. 2011;234:865–881. doi: 10.1007/s00425-011-1438-4. [DOI] [PubMed] [Google Scholar]
- 26.Pang Y., Wenger J.P., Saathoff K., Peel G.J., Wen J., Huhman D., Allen S.N., Tang Y., Cheng X., Tadege M. A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 2009;151:1114–1129. doi: 10.1104/pp.109.144022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie X., Zhao J., Hao Y.J., Fang C., Wang Y. The ectopic expression of apple MYB1 and bHLH3 differentially activates anthocyanin biosynthesis in tobacco. Plant Cell Tissue Organ Cult. 2017;131:183–194. doi: 10.1007/s11240-017-1275-7. [DOI] [Google Scholar]
- 28.Kong D., Li M., Dong Z., Ji H., Li X. Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Rep. 2015;34:395–410. doi: 10.1007/s00299-014-1717-1. [DOI] [PubMed] [Google Scholar]
- 29.Lepiniec L., Debeaujon I., Routaboul J.M., Baudry A., Pourcel L., Nesi N., Caboche M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- 30.Nocker S.V., Ludwig P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genom. 2003;4:50. doi: 10.1186/1471-2164-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamazaki M., Makita Y., Springob K., Saito K. Regulatory mechanisms for anthocyanin biosynthesis in chemotypes of Perilla frutescens var. crispa. Biochem. Eng. J. 2003;14:191–197. doi: 10.1016/S1369-703X(02)00222-X. [DOI] [Google Scholar]
- 32.Koornneef M. The complex syndrome of ttg mutanis. Arab. Inf. Serv. 1981;18:45–51. [Google Scholar]
- 33.Pang Y., Peel G.J., Sharma S.B., Tang Y., Dixon R.A. A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc. Natl. Acad. Sci. USA. 2008;105:14210–14215. doi: 10.1073/pnas.0805954105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L., Gao L., Wang H., Chen X., Wang Y., Yang H., Wei C., Wan X., Xia T. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct. Integr. Genom. 2013;13:75–98. doi: 10.1007/s10142-012-0301-4. [DOI] [PubMed] [Google Scholar]
- 35.Xie D.Y., Dixon R.A. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science. 2003;299:396–399. doi: 10.1126/science.1078540. [DOI] [PubMed] [Google Scholar]
- 36.Baudry A., Caboche M., Lepiniec L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006;46:768–779. doi: 10.1111/j.1365-313X.2006.02733.x. [DOI] [PubMed] [Google Scholar]
- 37.Xu W., Grain D., Gourrierec J.L., Harscoët E., Berger A., Jauvion V., Scagnelli A., Berger N., Bidzinski P., Kelemen Z., et al. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 2013;198:59–70. doi: 10.1111/nph.12142. [DOI] [PubMed] [Google Scholar]
- 38.Xie D.Y., Shi M.Z. Differentiation of programmed Arabidopsis cells. Bioengineered. 2012;3:54–59. doi: 10.4161/bbug.3.1.17786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He X., Zhao X., Gao L., Shi X., Dai X., Liu Y., Xia T., Wang Y. Isolation and characterization of key genes that promote flavonoid accumulation in purple-leaf tea (Camellia sinensis L.) Sci. Rep. 2018;8:130. doi: 10.1038/s41598-017-18133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang X., Huang K., Zheng G., Hou H., Wang P., Jiang H., Zhao X., Li M., Zhang S., Liu Y. CsMYB5a and CsMYB5e from Camellia sinensis differentially regulate anthocyanin and proanthocyanidin biosynthesis. Plant Sci. 2018;270:209–220. doi: 10.1016/j.plantsci.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Zhao L., Jiang X.L., Qian Y.M., Wang P.Q., Xie D.Y., Gao L.P., Xia T. Metabolic characterization of the anthocyanidin reductase pathway involved in the biosynthesis of flavan-3-ols in Elite Shuchazao tea (Camellia sinensis) cultivar in the field. Molecules. 2017;22:2241. doi: 10.3390/molecules22122241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogs J., Jaffé F.W., Takos A.M., Walker A.R., Robinson S.P. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007;143:1347–1361. doi: 10.1104/pp.106.093203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qian Y.M., Zhang S.X., Yao S.B., Xia J.X., Liu Y.J., Gao L.P., Xia T. The effects of vitro sucrose on quality components of tea plants (Camellia sinensis) based on transcriptomic and metabolic analysis. BMC Plant Biol. 2018 doi: 10.1186/s12870-018-1335-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M., Li Y., Guo L., Gong N., Pang Y., Jiang W., Liu Y., Jiang X., Zhao L., Wang Y. Functional characterization of tea (Camellia sinensis) MYB4a transcription factor using an integrative approach. Front. Plant Sci. 2017;8:943. doi: 10.3389/fpls.2017.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang X., Liu Y., Wu Y., Tan H., Meng F., Wang Y.S., Li M., Zhao L., Liu L., Qian Y., et al. Analysis of accumulation patterns and preliminary study on the condensation mechanism of proanthocyanidins in the tea plant (Camellia sinensis) Sci. Rep. 2015;5:8742. doi: 10.1038/srep08742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y.S., Xu Y.J., Gao L.P., Yu O., Wang X.Z., He X.J., Jiang X.L., Liu Y.J., Xia T. Functional analysis of flavonoid 3′,5′-hydroxylase from Tea plant (Camellia sinensis): Critical role in the accumulation of catechins. BMC Plant Biol. 2014;14:347. doi: 10.1186/s12870-014-0347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.