Abstract
For sustainable crop cultivation in the face of global warming, it is important to unravel the genetic mechanisms underlying plant adaptation to a warming climate and apply this information to breeding. Thermomorphogenesis and ambient temperature signaling pathways have been well studied in model plants, but little information is available for vegetable crops. Here, we investigated genes responsive to warming conditions from two Brassica rapa inbred lines with different geographic origins: subtropical (Kenshin) and temperate (Chiifu). Genes in Gene Ontology categories “response to heat”, “heat acclimation”, “response to light intensity”, “response to oxidative stress”, and “response to temperature stimulus” were upregulated under warming treatment in both lines, but genes involved in “response to auxin stimulus” were upregulated only in Kenshin under both warming and minor-warming conditions. We identified 16 putative high temperature (HT) adaptation-related genes, including 10 heat-shock response genes, 2 transcription factor genes, 1 splicing factor gene, and 3 others. BrPIF4, BrROF2, and BrMPSR1 are candidate genes that might function in HT adaptation. Auxin response, alternative splicing of BrHSFA2, and heat shock memory appear to be indispensable for HT adaptation in B. rapa. These results lay the foundation for molecular breeding and marker development to improve warming tolerance in B. rapa.
Keywords: warming, BrHSFA2, BrHSP18.2s, transcriptome, alternative splicing, Kenshin
1. Introduction
Global warming poses a serious threat to agriculture, as it threatens crop productivity and food safety worldwide [1,2]. Various strategies have been utilized to facilitate the breeding or engineering of thermotolerant crops, including regulating the expression of heat shock (HS) transcription factor (HSF) and heat shock response (HSR) genes, as well as the use of molecular markers [1,3,4].
In a model plant Arabidopsis thaliana, three types of tolerance responses to heat exposure have been identified: basal thermotolerance, acquired thermotolerance, and warming tolerance [5,6,7]. Plants with basal thermotolerance can survive when grown at 21–22 °C (normal growth condition), exposed to 42–45 °C for 0.5–1 h, and examined after 5–7 days. Plants with acquired thermotolerance can survive when grown under normal conditions, transferred to 36–38 °C (moderate heat stress) for 1.5 h (referred to as “priming”), recovered at 21–22 °C for 2 h, subjected to over 45 °C, and examined after 5–7 days. Plants with warming tolerance (as opposed to heat-stress tolerance) survive when grown under normal conditions, subjected to 12 °C for 2 days, and treated with warming conditions (27 °C) for 3 h. The long-term adaptation of plants to warmer growth conditions is thought to induce developmental reprograming. Identifying and applying warming-related genes in Brassica crop species poses a major challenge for crop breeding for improved tolerance to global warming.
Several marker genes for thermotolerance responses are currently available [8]. Genes encoding an exportin family protein (XPO1A, AT5G17020) and heat shock protein (HSP) 101 (HSP101, AT1G74310) are markers for basal thermotolerance. Acquired thermotolerance by priming is divided into two categories: short-term and long-term acquired thermotolerance. HSP101 expression represents short-term acquired thermotolerance, while long-term acquired thermotolerance is characterized by the expression of several marker genes, including those encoding Rotamase FKBP1/FK506-binding protein 62 (ROF1/FKBP62, AT3G25230), ROF2/FKBP65 (AT5G48570), heat HSF factor A2 (HSFA2, AT2G26150), HSP101, and heat stress-associated 32 kD (Hsa32, AT4G21320). During the long-term acquired thermotolerance response, the expression levels of small HSP genes (sHSPs), HSP70s, ROS genes, and ascorbate peroxidase (APX) also increase [7].
Warming treatment does not trigger the expression of HSR genes, but other genes, such as HSP70 (AT3g12580) [6] and Phytochrome-Interacting Factor4 (PIF4, AT2G43010) [9,10,11], are core components in this process. PIF4 controls morphological acclimation to high temperatures (HT) via auxin [9,12]. Phytochrome B (PhyB) was recently shown to function upstream of PIF4 [13]. Changes in ambient temperatures induce alternative splicing of a large number of genes [14]. Due to increases in global temperatures, the mechanism used by plants to sense small variations in ambient temperatures is becoming an increasing focus of study. It is important to elucidate whether crops that have long been cultivated in regions with different climates, such as Chinese cabbage, have developed similar responses to warming to those found in Arabidopsis.
Two inbred Chinese cabbage lines, Chiifu and Kenshin, have different geographic origins: Chiifu originated in temperate regions, whereas Kenshin originated in subtropical and tropical regions. Kenshin has long been used as a breeding stock to develop heat-tolerant Brassica species [15,16]. In addition, these two inbred lines show different electrolyte leakage rates in response to HT exposure and different expression of many genes [17]. The long history and intensive breeding of these two Chinese cabbage lines make them promising targets for transcriptome analysis after warming treatment to identify warming-related genes in this crop. These genes could then be used to develop molecular markers and to generate climate-change-resilient Brassica crops under global warming conditions. In the current study, we used the Br135K microarray (Version 3) to identify differentially expressed genes (DEGs) upon warming treatment in Chiifu and Kenshin Chinese cabbage, confirmed their expression patterns by qRT-PCR, and further characterized the expression of patterns of several candidate warming-relating genes. The results of this study lay the foundation for breeding Chinese cabbage lines with improved tolerance to warming conditions.
2. Results
2.1. Transcriptome Analysis of Plants under Warming, Minor-Warming, and Low-Temperature (LT) Conditions Using the Br135K Microarray
To identify putative warming-related (or HT adaptation-related) genes, we carried out Br135K microarray analysis of samples from two inbred lines, Chiifu and Kenshin, under three conditions (22 °C, 12 °C, and 12 ⟶ 28 °C) (Figure 1). The experiments were repeated twice; the mean values are summarized in Table S2. The microarray data have been deposited in “NCBI (https://www.ncbi.nlm.nih.gov/)” with [geo] GSE113637. Among the 41,173 genes deposited on the Br135K microarray, 14,222 (35%) showed probe intensity (PI) values <500 in all samples, whereas 26,951 (65%) showed PI values >500 in at least one sample. Of these 26,951 genes, 2104 had no Arabidopsis counterpart, i.e., NA (nonannotated genes). We subjected these 26,951 genes to further analysis because genes with a PI value of 500 (cutoff value) can easily be examined using standard RT-PCR. Responsive genes (i.e., DEGs) were defined as having at least a 2-fold change (cutoff value) in expression between comparative conditions. As shown in Figure 1, three types of comparisons were made: warming (samples treated for 2 days at 12 °C vs. samples after 3 h exposure to 28 °C), minor-warming (samples grown at 22 °C [control] vs. samples after 3 h exposure to 28 °C), and low-temperature (LT) treatment (samples grown at 22 °C vs. samples treated for 2 days at 12 °C).
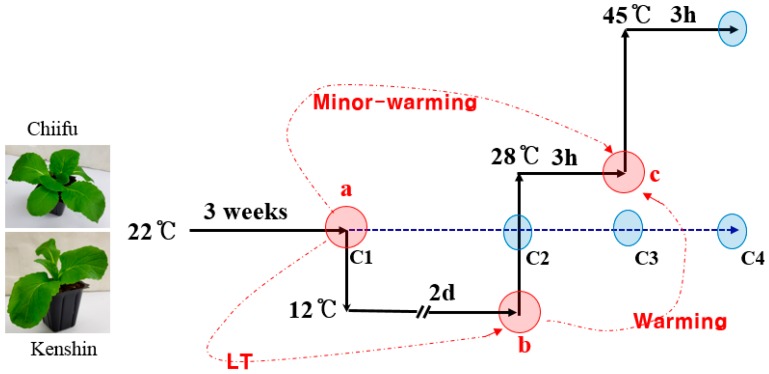
Figure 1.
Temperature treatment and sampling schedule. Collection times are indicated by circles. Red circles (a–c) represent sampling times for the microarray experiments as well as qRT-PCR analysis. Blue circles indicate that the collected samples were only used for qRT-PCR. Shoots from five individual plants were sampled and frozen in liquid nitrogen. Treatments were as follows: (a) (22 °C) to (b) (12 °C); low-temperature (LT) conditions; (a) (22 °C) to (c) (28 °C), minor-warming conditions; and (b) (12 °C) to (c) (28 °C), warming conditions.
2.1.1. Warming-Responsive Genes
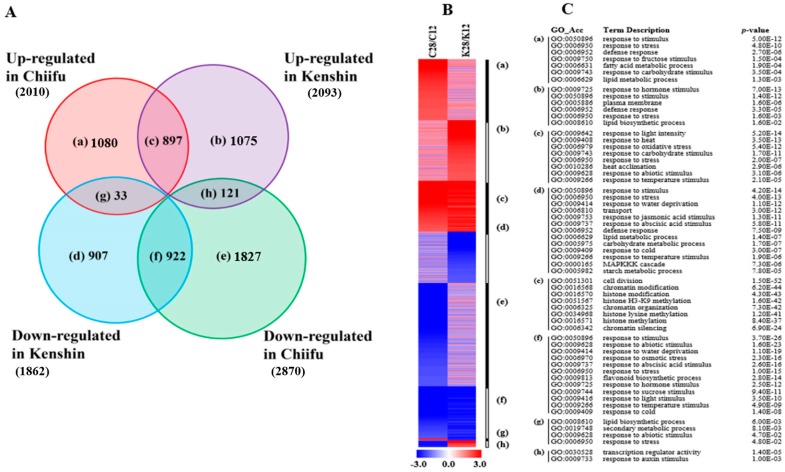
We identified 6862 warming-responsive genes. Similar numbers of these genes were upregulated in Chiifu and Kenshin, but more were downregulated in Chiifu (Figure 2A; Table S3). Genes in the Gene Ontology (GO) biological process categories “response to heat”, “heat acclimation”, “response to light intensity”, “response to oxidative stress”, and “response to temperature stimulus” were enriched among upregulated genes in both lines (group (c) in Figure 2B,C). The categories “response to hormone stimulus”, “plasma membrane”, “defense response”, and “lipid biosynthetic process” were enriched among genes that were specifically upregulated in Kenshin (group (b) in Figure 2B,C). Genes in the categories “response to water deprivation”, “response to osmotic stress”, “response to sucrose stimulus”, and “response to cold” were enriched among downregulated genes in both lines (group (f) in Figure 2B,C). Genes in two categories, “transcription regulatory activity” and “response to auxin stimulus”, were upregulated in Kenshin but downregulated in Chiifu under warming conditions (group (h) in Figure 2B,C).
Figure 2.
Analysis of warming-responsive genes from two contrasting inbred lines, Chiifu and Kenshin. (A) Venn diagram of DEGs with over 2-fold differences in expression; a–h indicate the groups of genes in each category; (B) Heatmap of DEGs in each group based on fold change; (C) Gene Ontology (GO) enrichment analysis of DEGs in each group, with p values obtained using the agriGO tool (http://bioinfo.cau.edu.cn/agriGO/index.php). C and K of Figure 2B indicate Chiifu and Kenshin, respectively.
2.1.2. Minor Warming-Responsive Genes
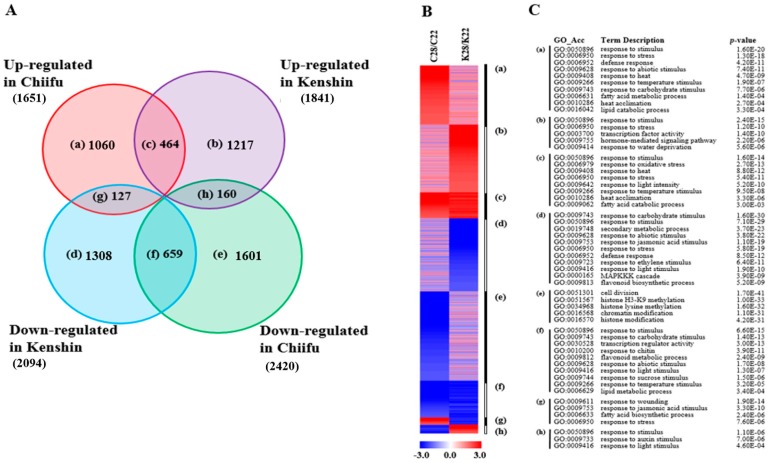
We identified 6596 minor warming-responsive genes; more of these genes were upregulated in Kenshin and downregulated in Chiifu (Figure 3A; Table S4). Upregulated genes in both lines were enriched in similar categories to those of warming-responsive genes, such as “response to heat”, “heat acclimation”, “response to light intensity”, “response to oxidative stress”, and “response to temperature stimulus” (group (c) in Figure 3B,C). Genes upregulated specifically in Kenshin were enriched in the categories “response to stimulus”, “response to stress”, “transcription factor activity”, and “response to water deprivation” (group (b) in Figure 3B,C). Genes in three categories, “response to stimulus”, “response to light stimulus”, and “response to auxin stimulus”, were upregulated in Kenshin but downregulated in Chiifu in response to minor-warming conditions (group (h) in Figure 3B,C). The category “response to auxin stimulus” was enriched among genes upregulated in Kenshin under both warming and minor-warming conditions, suggesting that the auxin response might contribute to heat tolerance in Kenshin.
Figure 3.
Analysis of minor warming-responsive genes from two contrasting inbred lines, Chiifu and Kenshin. (A) Venn diagram of DEGs with over 2-fold differences in expression; a–h indicate the groups of genes in each category; (B) Heatmap of DEGs in each group based on fold change; (C) GO enrichment analysis of DEGs in each group, with p values obtained using the agriGO tool (http://bioinfo.cau.edu.cn/agriGO/index.php). C and K of Figure 3B indicate Chiifu and Kenshin, respectively.
2.1.3. LT-Responsive Genes
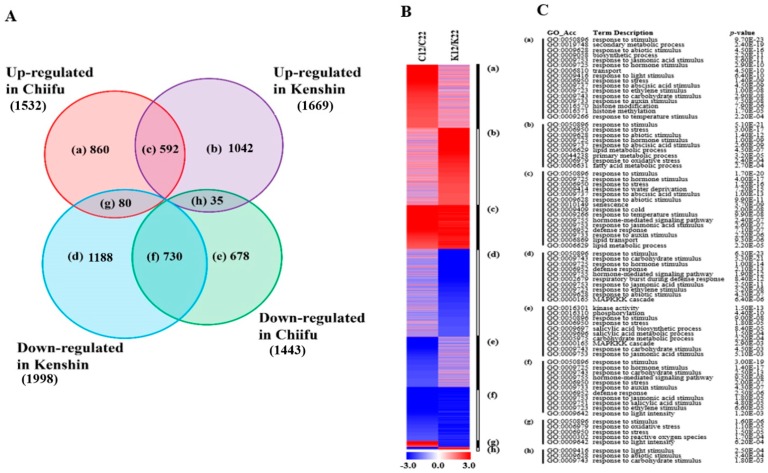
The identification of LT-responsive genes was not the main objective of this study, but associations between cold- and heat-stress signaling through calcium signaling, ROS signaling, and protein degradation have been reported [18,19,20], and changes in DNA methylation were also shown to be involved in heat tolerance [21]. In addition, we wanted to examine the possible association between LT treatment and the warming response. We identified 5205 LT-responsive genes, for which more genes were upregulated in Kenshin and downregulated in Chiifu (Figure 4; Table S5), implying that Kenshin is more sensitive to LT exposure than Chiifu. Many GO categories were identified as enriched among upregulated genes in both lines, including “response to cold”, “response to stress”, and “lipid transport and metabolic process”. Although similar GO categories were enriched among LT-responsive genes and genes in the two categories mentioned above (warming and minor warming), the expression of warming- and minor warming-responsive genes might not be influenced by LT treatment (Table S5).
Figure 4.
Analysis of LT-responsive genes from two contrasting inbred lines, Chiifu and Kenshin. (A) Venn diagram of DEGs with over 2-fold differences in expression; a–h indicate the groups of genes in each category; (B) Heatmap of DEGs in each group based on fold change; (C) GO enrichment analysis of DEGs in each group, with p values obtained using the agriGO tool (http://bioinfo.cau.edu.cn/agriGO/index.php). C and K of Figure 4B indicate Chiifu and Kenshin, respectively.
2.1.4. Genes Upregulated by Both Warming and Minor-Warming Treatment
Among DEGs, we attempted to identify genes that were upregulated by warming, minor warming, and both treatments (Table 1; Tables S6–S14). The numbers of genes upregulated under each condition were the same as those listed in Figure 1, Figure 2 and Figure 3, but 759 and 726 genes were upregulated by both conditions in Chiifu and Kenshin, respectively (Table S6). We reasoned that these genes might be involved in acquired thermotolerance and/or long-term adaptation to HT in both Chiifu and Kenshin. To specifically identify genes that might be involved in long-term adaptation to HT in the subtropical species Kenshin, we extracted specifically expressed genes (SEGs) from our data set (Tables S8–S13). We identified 85 genes that were specifically upregulated over 2-fold by both minor-warming and warming conditions in Chiifu but not in Kenshin (Table S10). In addition, 86 genes were specifically upregulated over 2-fold by both minor-warming and warming conditions in Kenshin but not in Chiifu (Table S13). Among these, 27 genes were nonannotated and unknown genes. Genes commonly upregulated in both Chiifu and Kenshin under both minor-warming and warming conditions included 15 HSPs and chaperone genes (Table S14). We subjected the genes listed in Tables S12–S14 to further GO enrichment analysis (Table 2, Table 3 and Table 4).
Table 1.
Summary of genes upregulated by various treatments. SEGs (specifically expressed genes) represent genes showing an over 2-fold change in expression under the indicated condition but no change or downregulation under the other conditions.
| Inbred Line (Treatment) | Comparison | No. of Genes (Table S5) | SEGs (Tables S7–S12) | Comparison | No. of Genes (Table S6) |
|---|---|---|---|---|---|
| Chiifu (22 °C ⟶ 12 °C ⟶ 28 °C) | 28 °C/22 °C | 1651 | 121 | 12 °C/22 °C | 1532 |
| 28 °C/12 °C | 2010 | 49 | 28 °C/12 °C | 2010 | |
| Both | 759 | 85 | Both | 40 | |
| Kenshin (22 °C ⟶ 12 °C ⟶ 28 °C) | 28 °C/22 °C | 1841 | 193 | 12 °C/22 °C | 1669 |
| 28 °C/12 °C | 2093 | 146 | 28 °C/12 °C | 2093 | |
| Both | 726 | 86 | Both | 59 |
12 °C/22 °C: LT conditions; 28 °C/12 °C: warming conditions; 28 °C/22 °C: minor-warming conditions.
Table 2.
Functional classification of genes specifically expressed in response to HT acclimation and/or adaption conditions. The table was constructed based on Tables S11–S13 using agriGO (http://bioinfo.cau.edu.cn/agriGO/) based on GO information for Arabidopsis homologs. W, warming, MW, minor warming.
| Classification | Kenshin (W) | Kenshin (W + MW) | Kenshin/Chiifu (W + MW) |
|---|---|---|---|
| Heat acclimation | - | 1 | 6 |
| Response to heat | - | - | 13 |
| Response to stress | 8 | 6 | 8 |
| Transcription factor activity | 9 | 9 | 9 |
| Transferase activity | 9 | 5 | 12 |
| Transport | 7 | 6 | 6 |
| Carbohydrate metabolic process | 8 | 4 | 4 |
| Ligase activity | - | 5 | 1 |
| Lipid biosynthetic process | 14 | 4 | 3 |
| Oxidation reduction | - | 1 | 2 |
| Response to auxin stimulus | - | 3 | 1 |
| Response to oxidative stress | - | 3 | 2 |
| Response to salicylic acid stimulus | - | 3 | 3 |
| Chromosome organization | - | - | 5 |
| Response to hormone stimulus | 22 | - | - |
| Intracellular membrane bound organelle | 23 | - | - |
| Signal transduction | 6 | - | - |
| Primary metabolic process | 7 | - | - |
| Catalytic activity | 7 | - | - |
| Plasma membrane | 3 | - | - |
| Ion binding | 5 | - | - |
| Unclassified | 4 | 15 | 34 |
| Unknown protein | 6 | 5 | 14 |
| Not annotated | 8 | 16 | 34 |
| Total | 132 | 86 | 157 |
Table 3.
Genes associated with the adaptation of Kenshin to HT based on microarray analysis. The selection criteria were (1) intrinsic levels of expression in Kenshin at 22 °C over 2-fold higher than those in Chiifu; and (2) expression levels under both warming and minor-warming conditions at least 2-fold higher in Kenshin than in Chiifu. HSR, heat shock response; TF, transcription factor; SF, splicing factor.
| Classification | At_Locus | Gene Description | Br_SEQ_ID | Expression Level (Probe Intensity) | Fold Change | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chiifu (C) | Kenshin (K) | Intrinsic | Warming | Warming | Minor-W | ||||||||
| 22 °C | 12 °C | 28 °C | 22 °C | 12 °C | 28 °C | K22/C22 | K28/K12 | C28/C12 | K28/K22 | ||||
| HSR | AT2G26150 | Heat shock transcription factor A2 (HSFA2) | Bra000557 * | 68 | 223 | 1256 | 256 | 542 | 1786 | 3.8 | 3.3 | 5.6 | 7.0 |
| AT5G62020 | Heat shock transcription factor B2A (HSFB2A) | Bra029292 * | 385 | 374 | 3164 | 875 | 587 | 3814 | 2.3 | 6.5 | 8.5 | 4.4 | |
| AT4G25200 | Mitochondrion-localized small heat shock protein 23.6 (HSP23.6-MITO) | Bra013872 * | 333 | 323 | 1308 | 984 | 265 | 3551 | 3.0 | 13.4 | 4.0 | 3.6 | |
| AT4G10250 | HSP20-like chaperones superfamily protein (HSP22.0) | Bra027999 * | 27 | 71 | 378 | 85 | 37 | 677 | 3.2 | 18.1 | 5.3 | 7.9 | |
| AT1G54050 | HSP20-like chaperones superfamily protein | Bra030910 * | 827 | 945 | 4041 | 2406 | 2649 | 9204 | 2.9 | 3.5 | 4.3 | 3.8 | |
| AT2G29500 | HSP20-like chaperones superfamily protein | Bra018383 * | 1870 | 2774 | 11,166 | 5211 | 2428 | 19,026 | 2.8 | 7.8 | 4.0 | 3.7 | |
| Bra018384 | 2575 | 5095 | 6789 | 5906 | 5362 | 14,692 | 2.3 | 2.7 | 1.3 | 2.5 | |||
| Bra031725 | 1996 | 4409 | 5909 | 6292 | 5275 | 13,450 | 3.2 | 2.5 | 1.3 | 2.1 | |||
| AT5G51440 | HSP20-like chaperones superfamily protein | Bra029174 * | 406 | 376 | 5772 | 1657 | 292 | 7627 | 4.1 | 26.1 | 15.3 | 4.6 | |
| AT5G59720 | Heat shock protein 18.2 (HSP18.2) | Bra002539 * | 73 | 259 | 747 | 206 | 58 | 5114 | 2.8 | 88.5 | 2.9 | 24.8 | |
| AT5G12020 | 17.6 kDa class II heat shock protein (HSP17.6II) | Bra006137 * | 52 | 271 | 2513 | 909 | 310 | 2036 | 17.6 | 6.6 | 9.3 | 2.2 | |
| AT5G48570 | FKBP-type peptidyl-prolyl cis-trans isomerase family protein (ROF2) | Bra037477 * | 309 | 283 | 4967 | 1632 | 352 | 6607 | 5.3 | 18.8 | 17.6 | 4.0 | |
| AT1G72660 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | Bra016043 * | 212 | 185 | 2034 | 967 | 889 | 2723 | 4.6 | 3.1 | 11.0 | 2.8 | |
| AT5G47830 | Unknown protein | Bra020728 * | 523 | 544 | 4910 | 1310 | 1076 | 4854 | 2.5 | 4.5 | 9.0 | 3.7 | |
| AT3G14200 | Chaperone DnaJ-domain superfamily protein | Bra027363 * | 1333 | 2129 | 5365 | 2857 | 2634 | 7477 | 2.1 | 2.8 | 2.5 | 2.6 | |
| TF | AT2G23690 | HTH-type transcriptional regulator | Bra039208 | 1122 | 1383 | 3449 | 2567 | 1072 | 6076 | 2.3 | 5.7 | 2.5 | 2.4 |
| AT5G56840 | MYB-like transcription factor family protein | Bra002790 * | 89 | 154 | 589 | 251 | 144 | 767 | 2.8 | 5.3 | 3.8 | 3.0 | |
| AT5G52600 | MYB domain protein 82 (MYB82) | Bra029113 | 64 | 334 | 132 | 205 | 371 | 1579 | 3.2 | 4.3 | 0.4 | 7.7 | |
| AT2G24645 | Transcriptional factor B3 family protein | Bra032079 | 43 | 85 | 77 | 182 | 244 | 1013 | 4.3 | 4.2 | 0.9 | 5.6 | |
| AT1G70270 | Transcription factor | Bra007905 | 246 | 797 | 368 | 1127 | 952 | 3529 | 4.6 | 3.7 | 0.5 | 3.1 | |
| AT5G15150 | Homeobox 3 (HB3) | Bra023506 | 197 | 389 | 165 | 389 | 372 | 1184 | 2.0 | 3.2 | 0.4 | 3.0 | |
| AT5G66940 | Dof-type zinc finger DNA-binding family protein | Bra012119 | 488 | 1459 | 209 | 1159 | 983 | 2746 | 2.4 | 2.8 | 0.1 | 2.4 | |
| AT3G62090 | Phytochrome interacting factor 3-like 2 (PIL2/PIF6) | Bra007660 * | 218 | 239 | 537 | 426 | 684 | 1883 | 2.0 | 2.8 | 2.2 | 4.4 | |
| AT5G10970 | C2H2 and C2HC zinc fingers superfamily protein | Bra009000 | 303 | 776 | 668 | 823 | 878 | 2180 | 2.7 | 2.5 | 0.9 | 2.6 | |
| AT1G23380 | KNOTTED1-like homeobox gene 6 (KNAT6) | Bra016348 | 140 | 970 | 475 | 273 | 1004 | 2202 | 2.0 | 2.2 | 0.5 | 8.1 | |
| AT4G18610 | Light-dependent short hypocotyl 9 (LSH9) | Bra021000 | 83 | 85 | 77 | 189 | 112 | 605 | 2.3 | 5.4 | 0.9 | 3.2 | |
| AT2G42610 | Light-dependent short hypocotyl 10 (LSH10) | Bra016865 | 422 | 617 | 197 | 1008 | 1013 | 2079 | 2.4 | 2.1 | 0.3 | 2.1 | |
| SF | AT1G65660 | Pre-mRNA splicing Prp18-interacting factor (SMP1) | Bra023741 | 141 | 232 | 1190 | 664 | 589 | 1712 | 4.7 | 2.9 | 5.1 | 2.6 |
| Others | AT4G36430 | Peroxidase superfamily protein | Bra017761 | 125 | 381 | 182 | 543 | 686 | 2278 | 4.4 | 3.3 | 0.5 | 4.2 |
| AT1G16530 | ASYMMETRIC LEAVES 2-like 9 (LBD3/ASL9) | Bra026042 | 171 | 393 | 272 | 522 | 824 | 1729 | 3.1 | 2.1 | 0.7 | 3.3 | |
| Bra026716 | 209 | 325 | 291 | 454 | 534 | 1063 | 2.2 | 2.0 | 0.9 | 2.3 | |||
| AT5G59670 | Leucine-rich repeat protein kinase family protein | Bra020300 | 129 | 93 | 80 | 288 | 147 | 622 | 2.2 | 4.2 | 0.9 | 2.2 | |
| AT4G19530 | Disease resistance protein (TIR-NBS-LRR class) family | Bra027594 | 158 | 206 | 202 | 532 | 536 | 1591 | 3.4 | 3.0 | 1.0 | 3.0 | |
| AT2G32660 | Receptor like protein 22 (RLP22) | Bra021803 | 163 | 72 | 201 | 960 | 963 | 2013 | 5.9 | 2.1 | 2.8 | 2.1 | |
| AT1G51860 | Leucine-rich repeat protein kinase family protein | Bra030411 | 92 | 75 | 41 | 874 | 1051 | 1992 | 9.5 | 1.9 | 0.5 | 2.3 | |
| AT1G53350 | Disease resistance protein (CC-NBS-LRR class) family | Bra037453 * | 185 | 155 | 420 | 539 | 706 | 1425 | 2.9 | 2.0 | 2.7 | 2.6 | |
| AT4G08570 | Heavy metal transport/detoxification superfamily protein | Bra037865 | 37 | 45 | 505 | 255 | 67 | 2044 | 6.9 | 30.5 | 11.1 | 8.0 | |
| AT5G66110 | Heavy metal transport/detoxification superfamily protein (HIPP27) | Bra009662 | 115 | 329 | 449 | 1102 | 1195 | 7086 | 9.5 | 5.9 | 1.4 | 6.4 | |
| AT1G79360 | Organic cation/carnitine transporter 2 (OCT2) | Bra035111 | 53 | 37 | 23 | 135 | 111 | 568 | 2.5 | 5.1 | 0.6 | 4.2 | |
| AT2G04100 | MATE efflux family protein | Bra015133 | 78 | 181 | 255 | 350 | 510 | 1175 | 4.5 | 2.3 | 1.4 | 3.4 | |
| AT2G35460 | Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family | Bra028562 | 215 | 78 | 879 | 575 | 123 | 2595 | 2.7 | 21.1 | 11.3 | 4.5 | |
| AT2G25450 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | Bra021671 | 246 | 68 | 240 | 530 | 644 | 5146 | 2.2 | 8.0 | 3.5 | 9.7 | |
| AT1G28030 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | Bra021552 | 59 | 73 | 121 | 129 | 169 | 1079 | 2.2 | 6.4 | 1.7 | 8.4 | |
| AT2G39310 | Jacalin-related lectin 22 (JAL22) | Bra005053 | 16 | 99 | 45 | 36 | 109 | 826 | 2.3 | 7.6 | 0.5 | 23.1 | |
| AT3G16900 | LURP-one-like protein | Bra021211 | 217 | 205 | 358 | 1035 | 524 | 2674 | 4.8 | 5.1 | 1.7 | 2.6 | |
| AT4G36380 | Cytochrome P450 superfamily protein (ROT3) | Bra011678 * | 52 | 139 | 350 | 395 | 205 | 842 | 7.6 | 4.1 | 2.5 | 2.1 | |
| AT4G24110 | NADP-specific glutamate dehydrogenase | Bra013763 | 92 | 79 | 129 | 204 | 176 | 637 | 2.2 | 3.6 | 1.6 | 3.1 | |
| AT3G51000 | Alpha/beta-Hydrolases superfamily protein | Bra036841 | 69 | 21 | 23 | 676 | 873 | 3074 | 9.8 | 3.5 | 1.1 | 4.5 | |
| AT4G22460 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | Bra013619 | 152 | 251 | 304 | 541 | 555 | 1932 | 3.6 | 3.5 | 1.2 | 3.6 | |
| AT4G13410 | Nucleotide-diphospho-sugar transferases superfamily protein (CSLA15) | Bra008638 | 276 | 385 | 600 | 656 | 523 | 1384 | 2.4 | 2.6 | 1.6 | 2.1 | |
| AT5G40650 | Succinate dehydrogenase 2-2 (SDH2-2) | Bra028469 | 468 | 531 | 545 | 2517 | 2395 | 6064 | 5.4 | 2.5 | 1.0 | 2.4 | |
| AT5G05390 | Laccase 12 (LAC12) | Bra009111 | 279 | 1072 | 1201 | 1118 | 1176 | 2906 | 4.0 | 2.5 | 1.1 | 2.6 | |
| AT3G17820 | Glutamine synthetase 1.3 (GLN1.3) | Bra021276 * | 1197 | 1053 | 2579 | 2798 | 3121 | 7326 | 2.3 | 2.3 | 2.4 | 2.6 | |
| AT5G61260 | Plant calmodulin-binding protein-related | Bra029324 | 256 | 194 | 163 | 811 | 1078 | 2480 | 3.2 | 2.3 | 0.8 | 3.1 | |
| AT5G64870 | SPFH/Band 7/PHB domain-containing membrane-associated protein family | Bra024333 | 270 | 569 | 701 | 559 | 484 | 1113 | 2.1 | 2.3 | 1.2 | 2.0 | |
| AT1G20575 | Nucleotide-diphospho-sugar transferases superfamily protein | Bra025828 | 1293 | 888 | 2710 | 3467 | 3248 | 7310 | 2.7 | 2.3 | 3.1 | 2.1 | |
| AT4G39140 | RING/U-box superfamily protein | Bra025860 | 27 | 27 | 59 | 2422 | 2404 | 5394 | 88.5 | 2.2 | 2.2 | 2.2 | |
| AT3G09260 | Glycosyl hydrolase superfamily protein (BGLU23) | Bra034060 | 278 | 412 | 393 | 1034 | 995 | 2106 | 3.7 | 2.1 | 1.0 | 2.0 | |
| AT3G06550 | O-acetyltransferase family protein | Bra040276 | 250 | 250 | 310 | 492 | 762 | 1570 | 2.0 | 2.1 | 1.2 | 3.2 | |
| AT1G29590 | Eukaryotic initiation factor 4E protein (eIF4E3) | Bra032325 | 299 | 445 | 486 | 1304 | 1365 | 2571 | 4.4 | 1.9 | 1.1 | 2.0 | |
| AT4G19430 | Unknown protein | Bra013396 | 103 | 16 | 42 | 252 | 17 | 726 | 2.5 | 41.6 | 2.7 | 2.9 | |
| NA | NA | Bra012220 | 183 | 112 | 97 | 5190 | 4062 | 11,440 | 28.4 | 2.8 | 0.9 | 2.2 | |
| NA | NA | Bra025861 | 53 | 59 | 147 | 1979 | 1928 | 4522 | 37.3 | 2.3 | 2.5 | 2.3 | |
| NA | NA | Bra010352 | 596 | 604 | 1785 | 4135 | 4187 | 9370 | 6.9 | 2.2 | 3.0 | 2.3 | |
* Gene in Table S13 (genes upregulated in both Chiifu and Kenshin under both minor-warming and warming conditions).
Table 4.
Summary of the expression levels of B. rapa genes shown to be HT responsive in Arabidopsis.
| Marker | At_Locus | Gene Description | Br_SEQ_ID | Expression Level (Probe Intensity) | Fold Change | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chiifu | Kenshin | Chiifu | Kenshin | ||||||||||
| 22 °C | 12 °C | 28 °C | 22 °C | 12 °C | 28 °C | 28/22 °C | 28/12 °C | 28/22 °C | 28/12 °C | ||||
| Basal thermotolerance | AT1G74310 | Heat shock protein 101 (HSP101) | Bra003807 | 1457 | 1411 | 3457 | 1696 | 2238 | 3594 | 2.4 | 2.5 | 2.1 | 1.6 |
| Bra015922 | 2725 | 3674 | 6053 | 5409 | 5601 | 8821 | 2.2 | 1.6 | 1.6 | 1.6 | |||
| AT5G17020 | Exportin 1A (XPO1A) | Bra006382 | 9010 | 8711 | 9368 | 8345 | 9747 | 11,271 | 1.0 | 1.1 | 1.4 | 1.2 | |
| Bra008580 | 10,093 | 10,241 | 10,261 | 9834 | 8047 | 12,608 | 1.0 | 1.0 | 1.3 | 1.6 | |||
| Bra023593 | 6942 | 7056 | 7948 | 6939 | 6511 | 7561 | 1.1 | 1.1 | 1.1 | 1.2 | |||
| Acquired thermotolerance | AT3G25230 | Rotamase FKBP 1 (ROF1/FKBP62) | Bra013224 | 11 | 18 | 42 | 15 | 59 | 40 | 3.8 | 2.4 | 2.7 | 0.7 |
| AT5G48570 | Rotamase FKBP 2 (ROF2/FKBP65) | Bra037477 * | 309 | 283 | 4967 | 1632 | 352 | 6607 | 16.1 | 17.6 | 4.0 | 18.8 | |
| AT2G26150 | Heat shock transcription factor A2 (HSFA2) | Bra000557 * | 68 | 223 | 1256 | 256 | 542 | 1786 | 18.5 | 5.6 | 7.0 | 3.3 | |
| Warming | AT2G18790 | Phytochrome B (PHYB) | Bra001650 | 717 | 770 | 829 | 402 | 1083 | 527 | 1.2 | 1.1 | 1.3 | 0.5 |
| Bra022192 | 13,874 | 10,184 | 7989 | 16,575 | 11,087 | 13,685 | 0.6 | 0.8 | 0.8 | 1.2 | |||
| AT3G12580 | Heat shock protein 70 (HSP70) | Bra001457 | 1560 | 2512 | 8099 | 9758 | 6732 | 13,032 | 5.2 | 3.2 | 1.3 | 1.9 | |
| Bra038734 | 1049 | 1159 | 7871 | 6488 | 4093 | 7597 | 7.5 | 6.8 | 1.2 | 1.9 | |||
| AT2G43010 | Phytochrome interacting factor 4 (PIF4) | Bra000283 * | 19,533 | 12,261 | 15,413 | 13,610 | 9359 | 24,186 | 0.8 | 1.3 | 1.8 | 2.6 | |
| Bra037742 | 3718 | 4145 | 3350 | 1988 | 3218 | 4296 | 0.9 | 0.8 | 2.2 | 1.3 | |||
| Other HSPs | AT2G25140 | Casein lytic proteinase B4 (HSP98.7/CLPB4) | Bra007816 | 645 | 702 | 1353 | 1332 | 1269 | 2425 | 2.1 | 1.9 | 1.8 | 1.9 |
| AT4G16660 | HSP 70 family protein | Bra038496 | 1160 | 1868 | 2286 | 1503 | 2091 | 1505 | 2.0 | 1.2 | 1.0 | 0.7 | |
| AT4G25200 | Mitochondrion-localized small heat shock protein 23.6 (HSP23.6-MITO) | Bra013872 * | 333 | 323 | 1308 | 984 | 265 | 3551 | 3.9 | 4.0 | 3.6 | 13.4 | |
| AT4G10250 | HSP20-like chaperones superfamily protein (HSP22.0-L) | Bra000703 * | 1174 | 1055 | 1638 | 1352 | 2898 | 6069 | 1.4 | 1.6 | 4.5 | 2.1 | |
| Bra027999 * | 27 | 71 | 378 | 85 | 37 | 677 | 14.1 | 5.3 | 7.9 | 18.1 | |||
| AT4G27670 | Heat shock protein 21 (HSP21) | Bra026317 | 45 | 166 | 729 | 310 | 54 | 603 | 16.1 | 4.4 | 1.9 | 11.2 | |
| AT5G47590 | Heat shock protein HSP20/alpha crystallin family | Bra022051 | 1119 | 1560 | 1914 | 1045 | 3295 | 2588 | 1.7 | 1.2 | 2.5 | 0.8 | |
| Bra022079 | 4462 | 6564 | 7455 | 4309 | 9320 | 12,845 | 1.7 | 1.1 | 3.0 | 1.4 | |||
| Bra022083 | 1201 | 1103 | 1815 | 939 | 1902 | 1852 | 1.5 | 1.6 | 2.0 | 1.0 | |||
| Bra022084 | 1042 | 1120 | 1737 | 760 | 1635 | 1540 | 1.7 | 1.6 | 2.0 | 0.9 | |||
| AT1G53540 | HSP20-like chaperones superfamily protein | Bra018216 * | 10,773 | 3454 | 10,773 | 7716 | 3159 | 16,904 | 1.0 | 3.1 | 2.2 | 5.4 | |
| AT1G54050 | HSP20-like chaperones superfamily protein | Bra012949 | 5473 | 4945 | 10796 | 1568 | 1799 | 1843 | 2.0 | 2.2 | 1.2 | 1.0 | |
| Bra030910 * | 827 | 945 | 4041 | 2406 | 2649 | 9204 | 4.9 | 4.3 | 3.8 | 3.5 | |||
| AT2G29500 | HSP20-like chaperones superfamily protein | Bra018383 * | 1870 | 2774 | 11,166 | 5211 | 2428 | 19,026 | 6.0 | 4.0 | 3.7 | 7.8 | |
| Bra018384 | 2575 | 5095 | 6789 | 5906 | 5362 | 14,692 | 2.6 | 1.3 | 2.5 | 2.7 | |||
| Bra031725 | 1996 | 4409 | 5909 | 6292 | 5275 | 13,450 | 3.0 | 1.3 | 2.1 | 2.5 | |||
| AT4G27890 | HSP20-like chaperones superfamily protein | Bra040837 | 77 | 29 | 232 | 128 | 199 | 637 | 3.0 | 8.1 | 5.0 | 3.2 | |
| AT5G51440 | HSP20-like chaperones superfamily protein | Bra029174 | 406 | 376 | 5772 | 1657 | 292 | 7627 | 14.2 | 15.3 | 4.6 | 26.1 | |
| AT5G59720 | Heat shock protein 18.2 (HSP18.2) | Bra002539 * | 73 | 259 | 747 | 206 | 58 | 5114 | 10.2 | 2.9 | 24.8 | 88.5 | |
| Bra006697 * | 1129 | 779 | 2431 | 765 | 426 | 5739 | 2.2 | 3.1 | 7.5 | 13.5 | |||
| Bra020295 * | 1659 | 383 | 1167 | 2436 | 197 | 5383 | 0.7 | 3.0 | 2.2 | 27.3 | |||
| AT5G12020 | 17.6 kDa class II heat shock protein (HSP17.6II) | Bra006137 * | 52 | 271 | 2513 | 909 | 310 | 2036 | 48.6 | 9.3 | 2.2 | 6.6 | |
| Bra008920 | 3065 | 1599 | 5017 | 1267 | 1426 | 3693 | 1.6 | 3.1 | 2.9 | 2.6 | |||
* Notable genes possibly related to the HT response in B. rapa.
2.1.5. Genes Upregulated by Both LT and Warming Treatment
Since both LT (transfer from 22 to 12 °C) and warming (transfer from 12 to 28 °C) are considered to be temperature-stress treatments, we analyzed genes responsive to LT and warming conditions (Table 1; Tables S7 and S15). We expected that genes upregulated by LT would also be upregulated by warming, but only a small number of genes in these categories overlapped: 40 and 59 genes in Chiifu and Kenshin, respectively (Table 1; Table S15). Interestingly, only one gene, BrHSFA2 (Bra000557, an ortholog of AT2G26150), was upregulated under both conditions as well as in both inbred lines. Therefore, BrHSFA2 represents a candidate temperature-specific regulator in Chinese cabbage.
In contrast to the single upregulated gene, many genes were downregulated by both LT and warming (Table S16). Whereas no gene was upregulated under both LT and warming conditions in Chiifu, 28 genes were downregulated under both conditions in Kenshin, implying that Kenshin is more sensitive to changes in temperature. The expression levels of these 28 genes were highest under normal growth conditions and lowest at 28 °C. Well-known genes in this category include genes encoding HY5-homolog (HYH; AT3G17609; Bra022225, Bra021258), gibberellin 2-oxidase 1 (GA2OX1; AT1G78440; Bra008362), CONSTANS-like 1 (COL1; AT5G15850; Bra023541), and CONSTANS-like 2 (COL2; AT3G02380; Bra021464, Bra001043).
2.2. GO Analysis of Warming- and Minor Warming-Responsive Genes
To identify genes associated with acclimation or adaptation to HT, we functionally classified upregulated genes in Kenshin under warming conditions, in Kenshin under both warming and minor-warming conditions, and in both lines under warming and minor-warming conditions (Tables S12–S14) via GO analysis (Table 2). Unexpectedly, no gene was identified in the “heat acclimation” or “response to heat” category among genes only expressed in Kenshin in response to warming conditions, but these categories were enriched among upregulated genes in both lines. Only one “heat acclimation”-related gene (BrCYP71B2) was identified in Kenshin under both minor-warming and warming conditions. However, several genes in putative HT adaptation-related gene categories were identified, such as 6 “heat acclimation”, 13 “response to heat”, and “chromosome organization” genes, among genes upregulated by warming and minor warming in both lines. Genes in the “heat acclimation” category included BrHSP18.2, BrROF2 (FKBP-type peptidyl-prolyl cis-trans isomerase family protein), BrHSP20-L, BrHSFA2, BrMge1 (Mitochondrial GrpE2), and an unknown gene. Genes in the “response to heat” category included most sHSPs, and “chromosome organization” genes included BrHON4, BrSMP1 (Swellmap 1), BrSWIB, BrENTG/VHS family protein, and BrSWC6 (SWR1 complex subunit 6) (Table S14). Categories that were specifically enriched in Kenshin under warming conditions included “lipid biosynthetic process”, “response to hormone stimulus” (most were auxin responsive), “intracellular membrane bound organelle” (most of unknown function), and “signal transduction” (most were defense related) (Table S12).
2.3. Identification of Genes Associated with HT Adaptation in Kenshin
To identify HT adaptation-related genes, we adopted several selection criteria based on the results shown in Table 2. The selection criteria were: (1) expression levels in Kenshin at 22 °C over 2-fold higher than those in Chiifu (we hypothesized that genes essential for long-term adaptation to HT would exhibit high basal levels of expression); and (2) expression levels under both warming and minor-warming conditions at least 2-fold higher in Kenshin than in Chiifu. Sixty-four genes were identified (Table 3), including 16 genes that were upregulated in both lines under minor-warming and warming conditions (asterisks in Table 3): 10 HSR genes, 2 TF (transcription factor) genes, 1 SF (splicing factor) gene, and 3 other genes. The two transcription factor genes were BrPIF6 (Bra007660; phytochrome-interacting factor 3-like 2; PIL2/PIF6) and Bra006853 (MYB-like transcription factor family protein). Genes in other categories included Bra037453 (disease resistance protein (CC-NBS-LRR class) family) for “response to stress”, BrGLN1.3 (Bra021276; glutamine synthetase 1.3) for “ligase activity”, and BrROT3 (Bra011678; cytochrome P450 superfamily protein (ROT3)) for “lipid biosynthesis process”. These 16 genes might play important roles in HT adaptation in B. rapa. We subjected three of these genes to further analysis: BrHSFA2, BrHSP18.2s, and BrSMP1 (Bra023741).
2.4. Comparison of HT-Related Gene Expression between B. rapa and Arabidopsis
Genes involved in thermotolerance (basal thermotolerance, acquired thermotolerance, and warming tolerance) and the associated marker genes are well known in the model plant Arabidopsis. To determine whether the same set of genes functions in B. rapa, we compared the expression patterns of these genes and other HSP genes with our microarray data (Table 4). Warming genes (PHYB, HSP70, and PIF4) identified in Arabidopsis were highly expressed in all B. rapa samples, with no notable increase in expression upon warming treatment, implying that genes responsible for long-term HT adaptation in B. rapa are different from Arabidopsis warming genes. In other cases, we assumed differences among samples, such as two contrasting lines in B. rapa vs. an ecotype of Arabidopsis. BrPIF4 (Bra000283) expression appeared to be somewhat related to HT adaptation in B. rapa. Two Chinese cabbage genes homologous to acquired thermotolerance-related genes in Arabidopsis, BrROF2 and BrHSFA2, appear to be critical for warming adaptation in B. rapa. The expression levels of several HSP genes were also consistent with warming treatment, pointing to their possible involvement in adaptation to HT.
2.5. Confirmation of Microarray Data via qRT-PCR
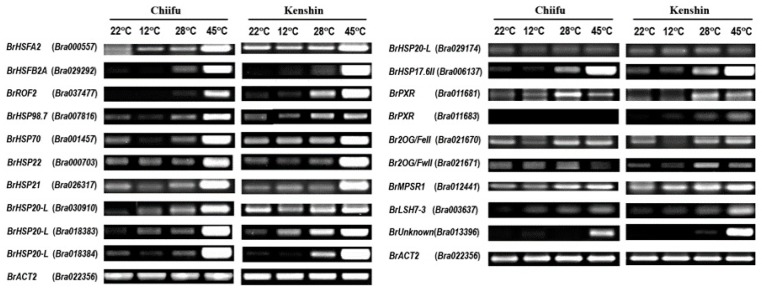
To confirm the expression levels of the genes detected by microarray analysis, we performed RT-PCR analysis of several selected genes (Figure 5). Although RT-PCR appears to be less sensitive than microarray analysis, RT-PCR results are often used to support microarray data. The expression levels of most of these genes increased upon warming treatment (28 °C), with maximum levels detected at 45 °C. These genes included three heat-acclimation-related genes (BrHSFA2, BrHSFB2A, and BrROF2), various HSP genes (especially sHSPs), peroxidase family genes, and others. Several genes showed high basal expression levels that further increased upon warming conditions: BrHSP98.7, BrHSP70, BrHSP21, three BrHSP20Ls (Bra30910, Bra01883, Bra01884), and BrMPSR1. As expected, all of these genes showed higher basal expression levels in Kenshin than in Chiifu.
Figure 5.
RT-PCR analysis of selected genes identified by microarray analysis. The expression levels of these genes obtained by microarray analysis are summarized in Table S16.
2.6. Expression of BrHSFA2 and BrHSP18.2
Based on our data (Table 3 and Table 4) and previous reports [22,23,24], we selected BrHSFA2 and BrHSP18.2A-C for further analysis of their possible involvement in response to warming and HT conditions. To examine whether BrHSFA2 undergoes alternative splicing upon HT exposure, as does Arabidopsis HSFA2, and whether the intron sequences of this gene are the same in Kenshin and Chiifu, we cloned and sequenced at least 10 clones of the BrHSFA2 intron region including part of Exon 1 and Exon 2 from both lines. These 548 bp fragments, including the 337 bp intron sequence, were 100% identical between Chiifu and Kenshin (NCBI accession MH310901, MH310902). We then compared BrHSFA2 with the homologous sequence from Arabidopsis to investigate whether BrHSFA2 also undergoes alternative splicing (Figure S1). As shown in Figure S1B, BrHSFA2 might contain a mini-exon with a TAG stop codon, which produces a truncated version of the BrHSFA2 polypeptide via alternative splicing. The truncated version of BrHSFA2 has different C-terminal amino acids from Arabidopsis HSFA2 (Figure S1C).
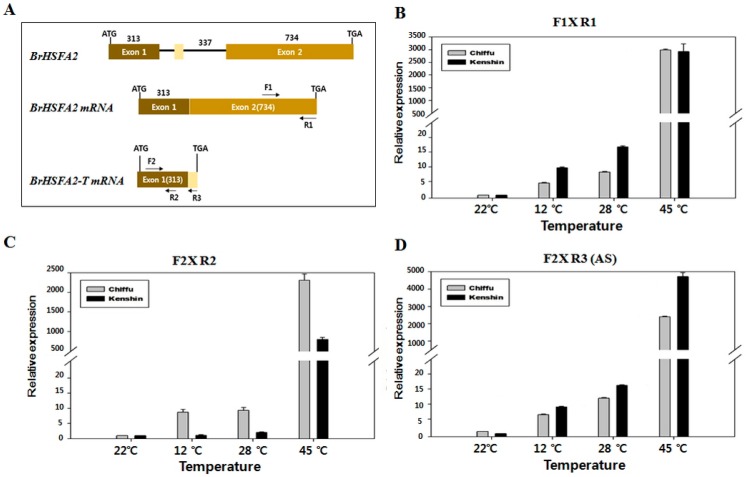
To confirm that alternative splicing occurs in BrHSFA2, we carried out qRT-PCR with a reverse primer consisting of possible mini-exon-derived mRNA (Table S1; Figure 6). The levels of an alternatively spliced form of the transcript were higher in Kenshin than in Chiifu upon warming conditions (Figure 6D), while the total transcript levels (full-length + alternatively spliced form) were higher in Chiifu (Figure 6C). The level of full-length mRNA was higher in Kenshin than in Chiifu (Figure 6B). These results indicate that alternative splicing occurs in the intron of BrHSFA2, that this process facilitates the expression of full-length BrHSFA2 as in Arabidopsis, and that the levels of the alternatively spliced form of this gene are higher in Kenshin (adapted to HT) than in Chiifu. To examine any association of the alternative splicing of BrHSFA2 with BrSMP1, encoding a spliceosome component and a candidate gene involved in HT adaptation in Kenshin (Table 3), we examined the expression of BrSMP1 under the same conditions (Figure S3). The expression pattern of BrSMP1 upon warming and HS was proportional to BrHSFA2 expression, suggesting the possible involvement of BrSMP1 in alternative splicing of BrHSFA2 upon HT treatment.
Figure 6.
BrHSFA2 expression in Chiifu and Kenshin during warming and heat shock treatments. qRT-PCR was performed with primer sets described in Table S1 and data analysis was carried out using qPCR value of three replicates. (A) Genomic organization of BrHSFA2 and possible mRNAs with primer positions indicated; (B) Full-length BrHSFA2 mRNA levels; (C) BrHSFA2 mRNA containing both full-length and truncated (alternatively spliced) forms; (D) Truncated form of BrHSFA2 mRNA.
HSFA2 is responsible for maintaining HS memory up to two days in Arabidopsis by maintaining histone methylation, thereby enabling the quick induction of HSR genes upon recurring HS [23,24]. HSFA2 has the most pronounced effect on Arabidopsis HSP18.2 (Hsp18.1-CI/AT5G59720) [23]. B. rapa possesses three homologs corresponding to AtHSP18.2, BrHSP18.2A (Bra002539), BrHSP18.2B (Bra020295), and BrHSP18.2C (Bra006697), as listed in order from the highest to lowest identity with AtHSP18.2. These three genes encode highly identical polypeptides (93–96% identity) (Figure S2), but we successfully generated primer sets to distinguish each gene (Table S1). A gradual increase in temperature (by 5 °C every 2 h [HS]) strongly upregulated all three genes at 37 °C, with the greatest increase observed for BrHSP18.2A and BrHSP18.2B in Kenshin (Table 5). However, under warming conditions, compared with 27 °C during HS treatment, tremendous increases were observed in the expression of BrHSP18.2A and BrHSP18.2B in Kenshin, but there was a several-fold higher increase in BrHSP18.2C expression in Chiifu than in Kenshin. These results appear to reflect the different responses of B. rapa to HT acclimation compared with Arabidopsis, as well as differences between Kenshin and Chiifu.
Table 5.
Expression of BrHSP18.2 family genes in Chiifu and Kenshin under various temperature conditions. Expression level (fold change) was calculated based on qRT-PCR values of three replicates using BrACT2 as a standard. Heat shock treatment was performed by increasing the temperature 5 °C every 2 h.
| Gene | Line | Expression (Fold Change) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Heat Shock | Warming | |||||||||
| 22 °C | 27 °C | 32 °C | 37 °C | 42 °C | 22 °C | 12 °C | 28 °C | 45 °C | ||
| BrHSP18.2A | Chiifu | 1.0 | 2.2 | 10.3 | 3220.4 | 4132.9 | 1.0 | 0.5 | 8.4 | 7154.1 |
| Kenshin | 1.0 | 0.8 | 5.5 | 4921.5 | 49,617.7 | 1.0 | 0.5 | 106.7 | 34,142.0 | |
| BrHSP18.2B | Chiifu | 1.0 | 0.5 | 8.7 | 74.4 | 357.9 | 1.0 | 1.2 | 1.3 | 241.4 |
| Kenshin | 1.0 | 3.9 | 5.7 | 160.5 | 1738.6 | 1.0 | 1.8 | 42.3 | 31,296.8 | |
| BrHSP18.2C | Chiifu | 1.0 | 1.4 | 13.8 | 140.0 | 646.8 | 1.0 | 1.8 | 7.8 | 7540.8 |
| Kenshin | 1.0 | 2.3 | 4.0 | 84.1 | 2960.7 | 1.0 | 1.5 | 3.1 | 733.5 | |
2.7. BrHSP18.2 Promoter Analysis
The differential expression levels of the three BrHSP18.2s upon warming treatment prompted us to analyze cis-elements in their promoters. We designed a common forward primer based on a comparison of known B. oleracea genes, B. napus genes, three B. rapa genes (BrHSP18.2A: Bra002539, BrHSP18.2B: Bra020295, BrHSP18.2C: Bra006697), and AT5G59720, as well as specific reverse primers for each gene (Table S1). We obtained promoter sequences of different sizes (from the ATG start codon): 1257 bp for BrHSP18.2A, 969 bp for BrHSP18.2B, and 1199 and 1236 bp for BrHSP18.2C_Chiifu and Kenshin, respectively (NCBI accession MH310903-8). The promoter sequences of BrHSP18.2A and B were identical between Chiifu and Kenshin, but the promoter sequences of BrHSP18.2C were not similar between the two inbred lines. However, the promoters of the three genes were different from each other but with partially conserved regions.
Despite sequence difference among the BrHSP18.2A, B, and C promoters, four HSE-binding modules [25] were present between −53 and −194 upstream of the ATG start codon: two head-to-head (nGAAnnTTCn) and two tail-to-tail (nTTCnnGAAn) modules (Figure S4). Two modules at −194 to −180 were overlapping. In Arabidopsis, eight HSEs (a(g,t,c)GAAn, a(g,t,c)GnAn, or a(g,t,c)Gann) have been detected between −97 and −53 bp in HSP18.2, and six HSE deletions were detected, leading to a loss of promoter activity [26]. In BrHSP18.2s, seven HSEs were present. The finding that BrHSP18.2A, B, and C possess sufficient numbers of HSEs for HSF binding, as well as possessing identical HSEs, points to the importance of having sufficient numbers of HSFs (or other elements) to control HSP expression levels.
3. Discussion
Plants exposed to HT exhibit reduced growth and development, as well as changes in signaling cascades or gene expression, representing adaptive responses to HT [27]. Global climate change has prompted breeders to develop thermotolerant crop varieties, including vegetable crops. Identifying genes that regulate plant responses to warming (or HT adaptation) and elucidating the mechanisms underlying their functions will be crucial for coping with the effects of global warming on agriculture. To identify and characterize warming-associated genes from Chinese cabbage, we subjected Chiifu and Kenshin, two contrasting inbred lines with respect to geographic origin and temperature responsiveness, to transcriptome analysis with a newly developed 3′-tiling microarray (135 K) covering the whole Brassica rapa genome using newly designed temperature treatments. Although the warming condition is different from the heat stress, some of HSR genes like sHSPs showed similar upregulated patterns as described in previous work [17]. However, this analysis yielded several novel findings, including the discovery of putative HT-adaptive genes, alternative splicing of BrHSFA2, and the expression patterns of its target genes.
3.1. Transcriptome Analysis
Of the three treatments, both warming and minor-warming conditions upregulated genes enriched in five biological process categories in both inbred lines, including “response to heat”, “heat acclimation”, and “response to temperature stimulus” (Figure 2 and Figure 3), indicating that the general responses of both lines are similar. However, genes involved in “response to auxin stimulus” were upregulated by both warming and minor warming in Kenshin, but not in Chiifu. This category of genes might be important for HT adaptation in Chinese cabbage. There is increasing evidence for an association between HT and auxin responses—HT reduces auxin biosynthesis [28], and HSPs such as sHSP22 [29] and cytosolic HSP90 [30] regulate auxin responses.
Our transcriptome analysis indicated that BrHSFA2 was upregulated or induced in both lines under all three treatment conditions (LT, warming, and minor warming). Arabidopsis HSFA2 is induced by HT and plays a role in the maintenance of HS memory [26,31,32]. Arabidopsis HSFA2 expression is also associated with H2O2/ROS stress [33] and salt/osmotic stress [34], suggesting that it plays an important role in responses to various stress conditions. The induction of BrHSFA2 expression by LT treatment suggests that BrHSFA2 targets LT-responsive genes besides HSR genes upon exposure to HT.
3.2. Functional Classification of Putative Warming Genes
As shown in Table 2 and Tables S12–S14, most genes belonging to the GO categories “heat acclimation”, “response to heat”, and “chromosome organization” were upregulated in both inbred lines, but their levels were higher in Kenshin than in Chiifu. These results suggest that the minor differences in expression of these genes in B. rapa could result from long-term adaptation to HS or that the genes identified in Arabidopsis could be associated with short-term HS adaptation. BrCYP71B2 (involved in “heat acclimation”) was upregulated in Kenshin under both minor-warming and warming conditions; its homolog is also upregulated upon HS in Arabidopsis [35], indicating its possible involvement in the HSR in B. rapa. Some putative HT adaptation-related genes have well-known functions in Arabidopsis. For example, Mge1 is induced by heat (under the control of HsfA1) and confers thermotolerance under priming conditions [36]. Arabidopsis SMP1 and SWC6 are associated with splicing [37] and chromatin remodeling [38], respectively. These findings suggest that these genes might play a role in HT adaptation in Kenshin. Kenshin-specific genes were classified into four categories (“lipid biosynthetic process”, “response to hormone stimulus”, “intracellular membrane bound organelle”, and “signal transduction”) (Table 2; Table S12), but none were found to be involved in HT adaptation, except for auxin-responsive genes. Further studies are needed to investigate the roles of these genes in B. rapa.
3.3. Putative HT Adaptation-Related Genes in Kenshin
To identify and characterize putative HT adaptation-related genes in Kenshin, we applied new cutoff criteria (Table 3) compared with known genes from Arabidopsis (Table 4) and confirmed their expression by RT-PCR (Figure 5). Unexpectedly, B. rapa warming genes appeared to differ from those of Arabidopsis, suggesting that different warming adaptation mechanisms or different sets of genes might function in B. rapa upon warming conditions. Arabidopsis warming genes such as PHYB, HSP70, and PIF4 were highly expressed in all B. rapa samples, with no notable increase upon warming treatment. Only the expression pattern of BrPIF4 (Bra000283) appeared to be somewhat related to HT adaptation, and two homologs of acquired thermotolerance-related genes in Arabidopsis, BrROF2 and BrHSFA2, appeared to be responsive to warming in B. rapa. PIF4, a basic helix-loop-helix (bHLH) transcription factor, is a central regulator of ambient temperature signaling in Arabidopsis [9]. PIF4-mediated thermomorphogenesis is associated with the circadian clock [39,40], auxin [9,41,42], other phytohormones [43,44], and epigenetic modification [6]. Quint et al. (2016) [11] indicated that PIF controls thermomorphogenesis via three molecular circuitries: (1) transcriptional regulation of circadian clock genes; (2) post-translation regulation by phosphorylation and degradation; and (3) phytohormonal control through interactions at various levels. These findings and our expression data suggest that BrPIF4 plays a role in the adaptation of Kenshin to HT.
Most warming-responsive genes in Kenshin are orthologs of Arabidopsis genes involved in acquired thermotolerance: HSR genes, sHSPs, peroxidase family genes, and disease-resistance genes (Table 3; Figure 5). The expression of other HSP genes and BrMPSR1 also increased in response to warming conditions, suggesting their possible involvement in HT adaptation (Figure 5). The roles of a few sHSPs in heat tolerance have been examined, including genes encoding HSP 21 (HSP21; Bra026317, AT4G27670) [45] and 17.6 kDa class II HSP (HSP17.6II; AT5G12020; Bra006137, Bra008920) [29]. However, many sHSPs, such as HSP21, HSP22.0, HSP18.2, and ASCORBATE PEROXIDASE 2 (APX2), are HS memory-related genes [22] and are targeted by HSFA2 to help maintain HS memory [31,32]. Class III peroxidases (PRXs) are plant-specific enzymes encoded by multigene families that are involved in lignification, cell elongation, stress responses, and seed germination [46]. Ascorbate peroxidase (APX), a key antioxidant enzyme, participates in various abiotic stress responses and in maintaining cellular homeostasis [47]. In Arabidopsis, MPSR1 (Misfolded Protein Sensing RING E3 Ligase 1) is involved in the rapid degradation of misfolded proteins due to protein-damaging stress, thereby controlling proteotoxic stress in the cytoplasm [48]. In B. rapa, two MPSR1 genes (BrMPSR1-1 (Bra012441) and BrMPSR1-2(Bra016290)) appear to be regulated at the transcriptional level or regulated in evolutionarily divergent ways. These genes might also participate in HT tolerance in Kenshin. Together, these findings support the notion that these genes play a role in long-term adaptation to HT in Kenshin.
3.4. BrHSFA2 and Its Target BrHSP18.2s
The expression of Arabidopsis HSFA2 is dependent on HS (the expression of which is amplified by the production of its alternatively spliced form), increases the expression of target HSR genes such as HSP18.2, and confers acquired thermotolerance or HS memory. In the current study, BrHSFA2 expression and splicing, and the expression of its target gene, BrHSP18.2s (Figure 6; Table 5), followed a similar pattern to that of Arabidopsis under warming conditions, implying that the warming response of B. rapa is similar to acquired thermotolerance in Arabidopsis.
Acquired thermotolerance by exposure to moderate HS confers tolerance to normally lethal HT [7]; this thermotolerance is maintained as HS memory for several days [32,49,50]. Three HS memory maintenance-related genes have been identified in Arabidopsis, which maintain this memory for several days after the plant returns to nonstress temperature conditions: two days for HSFA2 [23,24,28], three days for heat stress-associated 32 kD protein gene (Hsa32) [49], and three days for miR156 [22]. The maintenance of HS memory results from the induced hypermethylation of target genes (HSR genes), although not all target genes are hypermethylated [51].
Arabidopsis HSFA2 is a key regulator of responses to various types of stress including heat, high light, and ROS stress and is required for extending acquired thermotolerance by maintaining the expression of HSP genes [26,28,31]. HSFA2 is a regulatory component responsive to the accumulation of misfolded proteins in the cytosol [52]. HSFA2 also induces abscisic-acid-mediated heat tolerance by upregulating HSPs in both a monocot (fescue) and a dicot (Arabidopsis) [53]. HSFA2 is responsible for maintaining HS memory up to two days by maintaining histone methylation, thereby inducing HSR gene expression upon recurring HS [23,24]. Many small HSP genes, such as HSP21, HSP22.0, HSP18.2, and ASCORBATE PEROXIDASE 2 (APX2), are HS memory-related genes in Arabidopsis [22,24], whereas Hsp70 (AT3G12580) and Hsp101 (AT1G74310) are non-HS memory-related genes [24]. Arabidopsis HSFA2 produces an alternatively spliced form (truncated form) upon HS, and this truncated form in turn increases HSFA2 transcription levels [54]. This scenario appears to operate in B. rapa as well, where BrHSFA2 undergoes alternative splicing, is upregulated, and induces/maintains target gene expression/memory. Alternative splicing by a spliceosome complex is an important mechanism in the sensing of (and adaptation to) small variations in ambient temperature [14], as well as acquired thermotolerance conditions [48], in Arabidopsis. These findings imply that temperature changes, including HS and priming, lead to alternative splicing. Alternative splicing might contribute to long-term adaptation to HT in Chinese cabbage, in which thermotolerance does not appear to be due to morphological and architectural changes caused by high ambient temperatures, as found in other plants (thermomorphogenesis) [11].
3.5. BrHSP18.2s Promoters and Their Possible Control
The induction of Arabidopsis HSP18.2 (Hsp18.1-CI/AT5G59720) expression by HSFA2, a major thermotolerance HSF [24,26,31], is related to the role of HSFA2 in sustaining H3K4 methylation [24]. The promoter activity of ArabidopsisHSP18.2 is highest at 35 °C [55]. This promoter contains eight HSE modules between −97 and −53 bp; the deletion of two modules maintains promoter activity, but a deletion of six modules causes a dramatic reduction in promoter activity [26]. All BrHSP18.2 promoters contain seven HSE modules, implying that there is no difference in BrHSFA2 binding among the three BrHSP18.2 promoters. In other organisms, at least two nGAAn units arranged head-to-head (nGAAnnTTCn) or tail-to-tail (nTTCnnGAAn) are required in the promoters of HSPs for efficient HSF binding [25]. Four HSE units were found in all BrHSP18.2s promoters, one of which is overlapping (Figure S4), indicating that BrHSP18.2A to C contain sufficient numbers of HSEs for TF binding. The binding of HSFA2 with several target genes including HSP18.2 has been assessed [26], showing that two modules of a TATA-proximal HSE (nGAAnnTTCn) are essential for transcriptional activation by HSFA2. These modules are also conserved in all BrHSP18.2s promoters, suggesting that the expression differences among BrHSP18.2s upon HT exposure or warming conditions might be due to the presence of different numbers of HSFs such as BrHSFA2 (which is controlled by alternative splicing) and/or other factor(s).
4. Materials and Methods
4.1. Plant Materials
Seeds of two Chinese cabbage (Brassica rapa ssp. pekinensis) inbred lines, Chiifu and Kenshin, were kindly provided by Woori Seed Co., Sejong City, Korea. The seeds were sown in a 32-hole tray (6 × 6 × 6 cm × 32 holes) and grown for approximately 3 weeks in a growth chamber at 22 °C under a 16 h light/8 h dark photoperiod with a photon flux density of 140 μmol m−2 s−1. For warming treatment, the plants were subjected to 12 °C for 2 days under the same photoperiod and transferred to 28 °C for 3 h. For extreme HS treatment, warming-exposed plants were further incubated at 45 °C in a growth chamber for 3 h. The samples were collected at the end of each treatment (Figure 1). Humidity of growth chambers was set to 70 ± 10%. Shoots from five individual plants were sampled and quickly frozen in liquid nitrogen. To prepare the other experimental samples, plants grown for 3 weeks were subjected to various temperature treatments.
4.2. Br135K Microarray Analysis
The Br135K microarray (Brapa_V3_microarray, 3′-Tiling microarray) is a high-density DNA array prepared with Maskless Array Synthesizer (MAS) technology by NimbleGen (http://www.nimblegen.com/) [55]. Probes were designed from 41,173 genes from Brassica rapa accession Chiifu-401-42 (http://brassicadb.org/brad/). All three probes were 60 mers with 30 bp overlaps in 120 bp regions (60 bp of coding sequence plus 60 bp of the 3’UTR for each gene), representing 123,647 features. Fifty features from five markers (GUS, GFP, Bar, Kan, Hyg) were also included. Total and polysomal RNA were extracted using an RNeasy Mini kit (Qiagen, GmbH, Hilden, Germany) and the RNA protect Reagent (Qiagen), and contaminating DNA was removed by on-column DNase digestion with RNase-free DNase (Promega, Madison, WI, USA). Labeling, data processing, and background correction were performed as described previously [56]. To assess the reproducibility of the microarray analysis, the experiment was repeated using independently prepared total RNA samples from two biological replicates. To obtain insights regarding the putative biological functions and biochemical pathways of the DEGs, enrichment analysis was carried out by searching the GO [57], agriGO [58], and Kyoto Encyclopedia of Genes and Genomes [59] databases.
4.3. RNA Extraction, RT-PCR, and qRT-PCR
Total RNA was extracted from the plant samples using an RNeasy Mini kit (Qiagen). The RNA was treated with RNase-free DNase (Promega) to remove genomic DNA contamination. RT-PCR was performed using an Avian Myeloblastosis Virus (AMV) One-step RT-PCR kit (Takara, Kusatsu, Shiga, Japan). The gene-specific primers used to analyze the selected genes are listed in Table S1. For qRT-PCR, the RNA was subjected to first-strand cDNA synthesis using an Ace-α kit with Oligo-dT primers (Toyobo, Osaka, Japan). The primer sequences were designed according to sequences from the Brassica database (BRAD, http://brassicadb.org/brad/). PCR was performed using SYBR® Green Realtime PCR Master Mix-Plus (Toyobo, Japan) under the following cycling conditions: 30 s at 95 °C followed by 30 cycles of 95 °C for 5 s, 58 °C for 10 s, and 72 °C for 15 s.
4.4. Gene Cloning and Sequence Analysis
To analyze the intron sequence of BrHSFA2 and promoter sequences of B. rapa small HSP18.2 genes (BrHSP18.2s), genomic DNA was cloned and analyzed. Genomic DNA was isolated from Chiifu and Kenshin leaves using a DNeasy Plant Mini kit (Qiagen GmbH, Hilden, Germany). Primers were designed based on sequences listed in the BRAD website (Table S1). Genomic PCR was performed under the following conditions: denaturation (5 min at 94 °C), 30 cycles of amplification (30 s at 94 °C, 30 s at 52 °C, and 3 min at 72 °C), and a final extension (7 min at 72 °C). The PCR products were purified using a MEGA-Spin Gel Extraction kit (Intron Biotech. Inc., Sungnam, Korea) and cloned into the TA-vector using a T&A Cloning kit (RBC Bioscience Corp., New Taipei City, Taiwan). Escherichia coli (DH5α) cells were transformed with plasmid DNA carrying the desired insert. Plasmid DNA was purified using DNA-Spin (Intron Biotech. Inc., Sungnam, Korea) prior to sequencing (Macrogen, Seoul, Korea). To eliminate PCR and sequencing errors, at least 10 clones per gene were sequenced and analyzed. Any possible PCR and/or sequencing errors were eliminated by aligning independent sequences (http://www.genome.jp/tools-bin/clustalw).
5. Conclusions
This is the first report of the effects of long-term adaptation to warmer growth conditions or HT-adaptation of the gene expression profile in crops. We were able to derive the following conclusions from this study. Many DEGs were overlapping between minor-warming and warming conditions and in both lines examined, Chiifu and Kenshin. Most HT adaptation-associated genes in Chinese cabbage are homologous to acquired thermotolerance-related genes in Arabidopsis. Sixteen putative HT adaptation-related genes were identified: 10 HSR genes (including BrHSFA2 and sHSPs), 2 TF genes (BrPIF6 and a BrMyB), 1 SF gene (Pre-mRNA splicing Prp18-interacting factor, BrSMP1), and 3 other genes. Three additional genes, BrPIF4, BrROF2, and BrMPSR1, were also identified as candidate genes involved in HT adaptation. The degree of expression of HSR genes such as BrHSP18.2s appears to be related to the levels of HSF protein such as BrHSFA2 rather than their own promoter activity. Adaptation to HT in Chinese cabbage appears to be due to changes in the auxin response, increases in the alternative splicing of BrHSFA2 to amplify its expression, HS memory of HSR genes, and their increases in expression upon recurring HT. The genes identified in this study could be utilized in molecular breeding and marker development after further analysis.
Acknowledgments
This work was supported by a grant from the Research Fund of Chungnam National University (CNU), Daejeon, Korea, to Yoonkang Hur (2017-1828-01).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/6/1727/s1.
Author Contributions
Conceptualization: H.Y., C.-T.H., Y.H. Data curation: J.Y.A., X.D., C.-T.H. Formal analysis: J.Y.A., M.S. Funding acquisition: Y.H. Investigation: J.Y.A., X.D. Methodology: H.S., M.S., K.Y. Software: X.D., J.Y.A. Supervision: Y.H. Validation: J.Y.A., X.D., H.Y., Y.H. Writing-original draft: X.D., J.Y.A. Writing-review & editing: H.Y., C.-T.H., Y.H.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bita C.E., Gerats T. Plant tolerance to high temperature in a changing environment, scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013;4:273. doi: 10.3389/fpls.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Driedonks N., Rieu I., Vrienzen W.H. Breeding for plant heat tolerance at vegetative and reproductive stages. Plant Reprod. 2016;29:67–79. doi: 10.1007/s00497-016-0275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fragkostefanakis S., Röth S., Schleiff E., Scharf K.D. Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ. 2015;38:1881–1895. doi: 10.1111/pce.12396. [DOI] [PubMed] [Google Scholar]
- 4.Kole C., Muthamilarasan M., Henry R., Edwards D., Sharma R., Abberton M., Batley J., Bentley A., Blakeney M., Bryant J., et al. Application of genomics-assisted breeding for generation of climate resilient crops, progress and prospects. Front. Plant Sci. 2015;6:563. doi: 10.3389/fpls.2015.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkindale J., Vierling E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008;146:748–761. doi: 10.1104/pp.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar S.V., Wigge P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Mittler R., Finka A., Goloubinoff P. How do plants fee the heat? Trends Biochem. Sci. 2012;37:118–125. doi: 10.1016/j.tibs.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Yeh C.H., Kaplinsky N.J., Hu C., Charng Y.Y. Some like it hot, some like it warm, phenotyping to explore thermotolerance diversity. Plant Sci. 2012;195:10–23. doi: 10.1016/j.plantsci.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koini M.A., Alvey L., Allen T., Tilley C.A., Harberd N.P., Whitelam G.C., Franklin K.A. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 10.Kumar S.V., Lucyshyn D., Jaeger K.E., Alos E., Alvey E., Harberd N.P., Wigge P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quint M., Delker C., Franklin K.A., Wigge P.A., Halliday K.J., van Zanten M. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants. 2016;2:15190. doi: 10.1038/nplants.2015.190. [DOI] [PubMed] [Google Scholar]
- 12.Proveniers M.C., van Zanten M. High temperature acclimation through PIF4 signaling. Trends Plant Sci. 2013;18:59–64. doi: 10.1016/j.tplants.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Jung J.H., Domijan M., Klose C., Biswas S., Ezer D., Gao M., Khattak A.K., Box M.S., Charoensawan V., Cortijo S., et al. Phytochromes function as thermosensors in Arabidopsis. Science. 2016;354:886–889. doi: 10.1126/science.aaf6005. [DOI] [PubMed] [Google Scholar]
- 14.Verhage L., Severing E.I., Bucher J., Lammers M., Busscher-Lange J., Bonnema G., Rodenburg N., Proveniers M.C., Angenent G.C., Immink R.G. Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS ONE. 2017;12:e0172950. doi: 10.1371/journal.pone.0172950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hossain M.M., Inden H., Asahira T. Interspecific hybrids between Brassica campestris L. and B. oleracea L. through embryo and ovary culture. Mem. Coll. Agric. Kyoto Univ. 1989;135:21–30. [Google Scholar]
- 16.Yamagishi H., Hossain M.M., Yonezawa K. Morphology, fertility and cross-compatibility of somatic hybrids between Brassica oleracea L. and B. campestris L. Sci. Horticult. 1994;58:283–288. doi: 10.1016/0304-4238(94)90098-1. [DOI] [Google Scholar]
- 17.Dong X., Yi H., Lee J., Nou I.S., Han C.T., Hur Y. Global gene-expression analysis to identify differentially expressed genes critical for the heat stress response in Brassica rapa. PLoS ONE. 2015;10:e0130451. doi: 10.1371/journal.pone.0130451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinnusamy V., Zhu J., Zhu J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Scharf K.D., Berberich T., Ebersberger I., Nover L. The plant heat stress transcription factor (Hsf) family, structure, function and evolution. Biochim. Biophys. Acta. 2012;1819:104–119. doi: 10.1016/j.bbagrm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhu J.K. Abiotic stress signaling and response in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T., Li Y., Daun W., Huang F., Hou X. Cold acclimation alters DNA methylation patterns and confers tolerance to heat and increase growth rate in Brassica rapa. J. Exp. Bot. 2017;68:1213–1224. doi: 10.1093/jxb/erw496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stief A., Altmann S., Hoffmann K., Pant B.D., Scheible W.R., Bäurle I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell. 2014;26:1792–1807. doi: 10.1105/tpc.114.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lämke J., Brzezinka K., Bäurle I. HSFA2 orchestrates transcriptional dynamics after heat stress in Arabidopsis thaliana. Transcription. 2016;7:111–114. doi: 10.1080/21541264.2016.1187550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lämke J., Brzezinka K., Altmann S., Bäurle I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 2016;35:162–175. doi: 10.15252/embj.201592593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perisic O., Xiao H., Lis J.T. Stable binding of Drosophia heat shock factor to head-to-head and tail-to-tail repeats of conserved 5 bp recongnition unit. Cell. 1989;59:797–806. doi: 10.1016/0092-8674(89)90603-X. [DOI] [PubMed] [Google Scholar]
- 26.Nishizawa-Yokoi A., Yoshida E., Yabuta Y., Shigeoka S. Analysis of the regulation of target genes by an Arabidopsis heat shock transcription factor, HsfA2. Biosci. Biotechnol. Biochem. 2009;73:890–895. doi: 10.1271/bbb.80809. [DOI] [PubMed] [Google Scholar]
- 27.Hasanuzzaman M., Nahar K., Alam M.M., Roychowdhury R., Fujita M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013;14:9643–9684. doi: 10.3390/ijms14059643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshino T., Miura S., Kikuchi S., Hamada K., Yano K., Watanabe M., Higashitani A. Auxin depletion in barley plants under high-temperature conditions represses DNA proliferation in organelles and nuclei via transcriptional alterations. Plant Cell Environ. 2011;34:284–290. doi: 10.1111/j.1365-3040.2010.02242.x. [DOI] [PubMed] [Google Scholar]
- 29.Li G., Li J., Hao R., Guo Y. Activation of catalase activity by a peroxisome-localized small heat shock protein Hsp17.6CII. J. Genet. Genom. 2017;44:395–404. doi: 10.1016/j.jgg.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe E., Mano S., Hara-Nishimura I., Nishimura M., Yamada K. HSP90 stabilizes auxin receptor TIR1 and ensures plasticity of auxin responses. Plant Signal. Behav. 2017;12:e1311439. doi: 10.1080/15592324.2017.1311439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishizawa A., Yabuta Y., Yoshida E., Maruta T., Yoshimura K., Shigeoka S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006;48:535–547. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- 32.Charng Y.Y., Liu H.C., Liu N.Y., Chi W.T., Wang C.N., Chang S.H., Wang T.T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller G., Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot. 2006;98:279–288. doi: 10.1093/aob/mcl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogawa D., Yamaguchi K., Nishiuchi T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 2007;58:3373–3383. doi: 10.1093/jxb/erm184. [DOI] [PubMed] [Google Scholar]
- 35.Lim C.J., Yang K.A., Hong J.K., Choi J.S., Yun D.J., Hong J.C., Chung W.S., Lee S.Y., Cho M.J., Lim C.O. Gene expression profiles during heat acclimation in Arabidopsis thaliana suspension-culture cell. J. Plant Res. 2006;119:373–383. doi: 10.1007/s10265-006-0285-z. [DOI] [PubMed] [Google Scholar]
- 36.Hu C., Lin S.Y., Chi W.T., Charng Y.Y. Recent gene duplication and subfunctionalization produced a mitochondrial GrpE, the nucleotide exchange factor of the Hsp70 complex, specialized in thermotolerance to chronic heat stress in Arabidopsis. Plant Physiol. 2012;158:747–758. doi: 10.1104/pp.111.187674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clay N.K., Nelson T. The recessive epigenetic swellmap mutation affects the expression of two stop II splicing factors required for the transcription of the cell proliferation gene STRUWWELPETER and for the the timing of cell cycle arrest in the Arabidopsis leaf. Plant Cell. 2005;17:1994–2008. doi: 10.1105/tpc.105.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lázaro A., Gómez-Zambrano A., López-González L., Piñeiro M., Jarillo J.A. Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 2008;59:653–666. doi: 10.1093/jxb/erm332. [DOI] [PubMed] [Google Scholar]
- 39.Nomoto Y., Kubozono S., Miyachi M., Yamashino T., Nakamichi N., Mizuno T. A circadian clock- and PIF4-mediated double coincidence mechanism is implicated in the thermosensitive photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1965–1973. doi: 10.1093/pcp/pcs141. [DOI] [PubMed] [Google Scholar]
- 40.Nomoto Y., Kubozono S., Miyachi M., Yamashino T., Nakamichi N., Mizuno T. Circadian clock and PIF4-mediated external coincidence mechanism coordinately integrates both of the cues from seasonal changes in photoperiod and temperature to regulate plant growth in Arabidopsis thaliana. Plant Signal. Behav. 2013;8:e22863. doi: 10.4161/psb.22863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franklin K.A., Lee S.H., Patel D., Kumar S.V., Spartz A.K., Gu C., Ye S., Yu P., Breen G., Cohen J.D., et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun J., Qi L., Li Y., Chu J., Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stavang J.A., Gallego-Bartolomé J., Gómez M.D., Yoshida S., Asami T., Olsen J.E., García-Martínez J.L., Alabadí D., Blázquez M.A. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 2009;60:589–601. doi: 10.1111/j.1365-313X.2009.03983.x. [DOI] [PubMed] [Google Scholar]
- 44.Oh E., Zhu J.Y., Wang Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernfur K., Rutsdottir G., Emanuelsson C. The chloroplast-localized small heat shock protein Hsp21 associates with the thylakoid membranes in heat-stressed plants. Protein Sci. 2017;26:1773–1784. doi: 10.1002/pro.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shigeto J., Tsutsumi Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2015;209:1395–1402. doi: 10.1111/nph.13738. [DOI] [PubMed] [Google Scholar]
- 47.Pandey S., Fartyal D., Agarwal A., Shukla T., James D., Kaul T., Negi Y.K., Arora S., Reddy M.K. Abiotic stress tolerance in plants: Myriad roles of ascorbate peroxidase. Front. Plant Sci. 2017;8:581. doi: 10.3389/fpls.2017.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J.H., Cho S.K., Oh T.R., Ryu M.Y., Yang S.W., Kim W.T. MPSR1 is a cytoplasmic PQC E3 ligase for eliminating emergent misfolded proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2017;114:E10009. doi: 10.1073/pnas.1713574114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charng Y.Y., Liu H.C., Liu N.Y., Hsu F.C., Ko S.S. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 2006;140:1297–1305. doi: 10.1104/pp.105.074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meiri D., Breiman A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009;59:387–399. doi: 10.1111/j.1365-313X.2009.03878.x. [DOI] [PubMed] [Google Scholar]
- 51.Lämke J., Bäurle I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017;18:124. doi: 10.1186/s13059-017-1263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugio A., Dreos R., Apricio F., Maule A.J. The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell. 2009;21:642–654. doi: 10.1105/tpc.108.062596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X., Zhuang L., Shi Y., Huang B. Up-regulation of HSFA2c and HSPs by ABA contributing to improved heat tolerance in tall fescue and Arabidopsis. Int. J. Mol. Sci. 2017;18:1981. doi: 10.3390/ijms18091981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J., Sun N., Liu M., Liu J., Du B., Wang X., Qi X. An autoregulatory loop controlling Arabidopsis HsfA2 expression, role of heat shock-induced alternative splicing. Plant Physiol. 2013;162:512–521. doi: 10.1104/pp.112.205864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahashi T., Naito S., Komeda Y. The Arabidopsis HSP18.2promoter/GUS gene fusion in transgenic Arabidopsis plants, a powerful tool for the isolation of regulatory mutants of the heat-shock response. Plant J. 1992;2:751–761. doi: 10.1111/j.1365-313X.1992.tb00144.x. [DOI] [Google Scholar]
- 56.Jung H.J., Dong X., Park J.I., Thamilarasan S.K., Lee S.S., Kim Y.K., Lim Y.P., Nou I.S., Hur Y. Genome-wide transcriptome analysis of two contrasting Brassica rapa doubled haploid lines under cold-stresses using Br135K oligomeric chip. PLoS ONE. 2014;9:e106069. doi: 10.1371/journal.pone.0106069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology, tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du Z., Zhou X., Ling Y., Zhang Z., Su Z. agriGO, A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.