Abstract
Frontotemporal dementia (FTD) is the second most common senile neurodegenerative disease. FTD is a heterogeneous disease that can be classified into several subtypes. A mutation in CHMP2B locus (CHMP2Bintron5), which encodes a component of endosomal sorting complex required for transport-III (ESCRT-III), is associated with a rare hereditary subtype of FTD linked to chromosome 3 (FTD-3). ESCRT is involved in critical cellular processes such as multivesicular body (MVB) formation during endosomal–lysosomal pathway and autophagy. ESCRT mutants causes diverse physiological defects primarily due to accumulation of endosomes and defective MVBs resulting in misregulation of signaling pathways. Charged multivesicular body protein 2B (CHMP2B) is important for neuronal physiology which especially rely on precise regulation of protein homeostasis due to their post-mitotic status. Drosophila has proven to be an excellent model for charaterization of mechanistic underpinning of neurodegenerative disorders including FTD. In this review, current understanding of various FTD-related mutations is discussed with a focus on Drosophila models of CHMP2Bintron5-associated FTD.
Keywords: frontotemporal dementia, endosomal-lysosomal pathway, ESCRT, CHMP2B, CHMP2Bintron5, animal models, Drosophila
1. Introduction
Frontotemporal dementia (FTD) is the second most common form of dementia, and the second most common neurodegenerative disease in individuals under the age of 65 [1]. It is characterized by progressive impairment of language and executive functioning, as well as behavioral modifications, due to degeneration of the frontal and temporal lobes of the brain [2,3]. FTD is a heterogeneous disease because it can result from a multitude of genetic mutations [4]. FTD also shares similarities with various other neurodegenerative disorders, such as progressive supranuclear palsy (PSP) syndromes, corticobasal degeneration (CBD), and argyrophilic disease (AGD) and amyotrophic lateral sclerosis (ALS), suggesting common neuropathological mechanisms to neurodegeneration [2,3]. FTD is divided into different subtypes which encompass a range of molecular and behavioral deficits. One subtype is the behavioral variant of FTD (bvFTD), which is typically classified by gradual changes in behavior, cognition, or both. Another subtype is primary progressive aphasia (PPA), which is a language disorder that results from temporal lobe atrophy. This disorder is further divided into additional subgroups which are categorized based on a spectrum of the areas of language affected: logopenic variant PPA (lvPPA), nonfluent variant PPA (nfvPPA), and semantic dementia (SD) [2,3,4].
An additional subtype of FTD, called frontotemporal dementia linked to chromosome 3 (FTD-3), is dominantly inherited and is characterized by selective cortical neuron degeneration. FTD-3 patients display cognitive impairment, social behavior deficits, and dystonia, which typically appear between the ages of 46 and 67 [1]. For this reason, FTD-3 is classified under the FTD subtype of frontotemporal lobar dementia [5]. One of the genetic defects associated with FTD-3 is a mutation in the CHMP2B gene; a single nucleotide mutation (G to C) at the splice site of CHMP2B exon 6. This variation establishes two abnormal transcripts, CHMP2Bintron5 and CHMP2BΔ10, both of which encode for proteins with a defective carboxyl terminus [5,6,7].
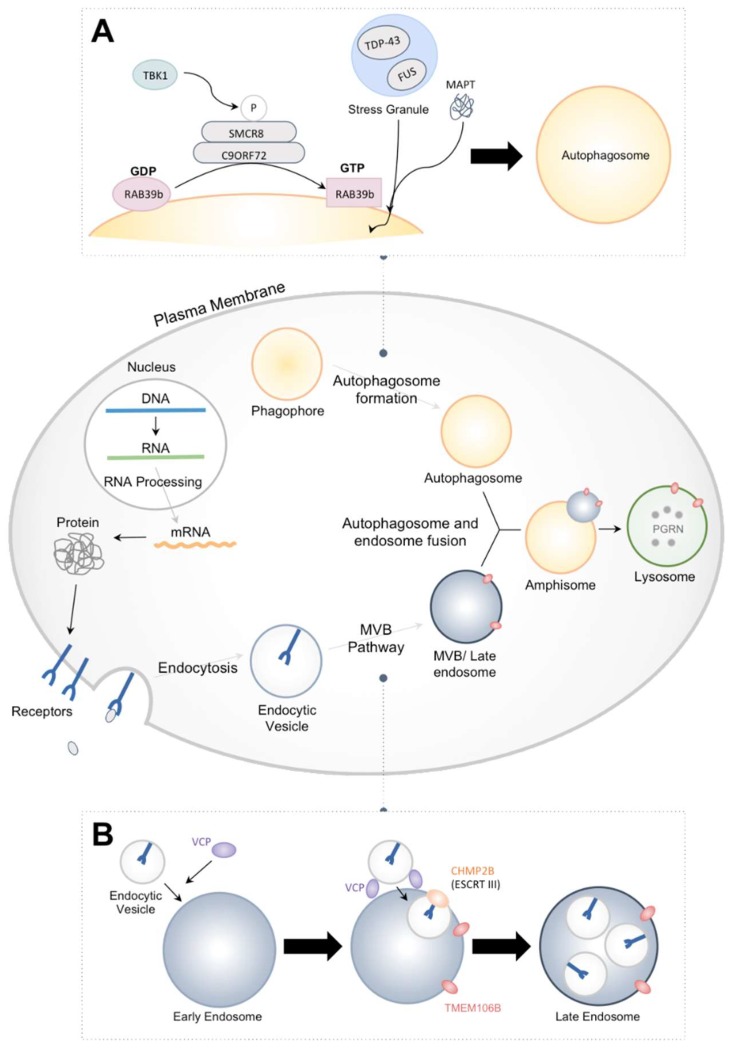
This review will focus on the most recent findings concerning FTD-associated mutations, particularly on the CHMP2Bintron5 mutation and its effect on misregulation of receptor-mediated signaling due to perturbation of the endosome–lysosome pathway and autophagosome–lysosome pathway (Figure 1). Drosophila and mouse models have made great strides in discovering the underlying mechanisms of the CHMP2Bintron5-mediated phenotypes.
Figure 1.
Role of proteins associated with frontotemporal dementia (FTD) in endosomal–lysosomal and autophagy pathway. FTD associated proteins, highlighted in yellow, are involved in both autophagy and endosomal–lysosomal pathways. (A) Phagophores in the cytoplasm undergo autophagosome formation with the assistance of C9ORF72, TANK binding kinase 1 (TBK1), microtubule-associated protein tau (MAPT), transactive DNA-binding protein (TDP-43), and fused in sarcoma (FUS) proteins, as well as various other protein complexes not included in the figure above. (B) Endocytic vesicles bearing receptor cargo transition through maturation stages to form MVBs and eventually fuse with the lysosomes. ESCRT complexes and additional proteins, including valosin containing protein (VCP) and transmembrane protein 106B (TMEM106B), contribute in the development of the endosome and MVB pathway in the endosomal–lysosomal pathway.
2. Mutations Associated with FTD
Aside from the mutant CHMP2Bintron5, numerous mutations in many other loci are associated with FTD (Table 1; for review of molecular genetics of FTD, see Rainero et al. [8]). In this section we have provided a brief overview of the mutations associated with FTD. We have also briefly included the major conclusions obtained from the Drosophila model of the mutations. The most common mutations occur in the genes C9ORF72 (chromosome 9 open reading frame 72), PGRN (progranulin), and MAPT (Microtubule associated protein tau). Alterations in the C9ORF72 locus is the most prevalent cause of FTD (25% of all FTD cases) [1]. The GGGGCC hexanucleotide repeat expansion located in intron 1 of this gene is the source of pathogenicity [9,10,11]. While 2 to 24 repeats are typical in normal individuals, hundreds to thousands of these repeats are present in the affected individuals. The expanded repeats cause abnormal translation of C9ORF72 transcript resulting in toxic dipeptide repeats (DPRs) through a mechanism known as repeat associated non-ATG translation (RAN) [9,12]. Comprehensive analysis by Mizielinska et al., using a Drosophila model, has shown that a toxic DPR secondary structure is not necessary to cause neurodegeneration in adult flies; instead, the aberrant translation of RNA is sufficient [13]. DPRs are particularly important in enhancing or suppressing nuclear import or export of proteins. Additionally, C9ORF72, along with SMCR8 and WDR41, are associated with autophagosome formation, and loss of C9ORF72 is associated with defects in autophagosome formation in cell culture and mouse models (Figure 1A) [14,15].
Table 1.
Classification of FTD-associated loci.
| Gene Name | C9ORF72 | MAPT | CHMP2B | FUS | VCP | TARDBP (TDP-43) | PGRN | TBK1 | TMEM106B |
|---|---|---|---|---|---|---|---|---|---|
| Full Name | Chromosome 9 open reading frame 72 | Microtubule—associated protein tau | Chromatin modifying protein 2B | Fused in sarcoma | Valosin containing protein | Transactive DNA-binding protein | Progranulin | TANK-binding kinase 1 | Transmembrane protein 106B |
| Location | 9p21.2 | 17q21.32 | 3p11.2 | 16q11.22 | 9p13.3 | 1p36.22 | 17q21.32 | 12q14.2 | 7q21.3 |
| Incidence Rate | 25% | Familial: 10–20% Sporadic: 0–3% |
Rare | Rare | 1.6% | Rare (<20 cases) | Familial: 5–20% Sporadic: 1–5% |
1.1% | Unknown |
| Normal Function |
|
|
|
|
|
|
|
|
|
| Deficits Caused |
|
|
|
|
|
|
|
|
|
| Drosophila Model | Yes 13 | Yes 22 | Yes 41, 81, 84–86 | Yes 32 | Yes 34 | Yes 32 | No | No | No |
| Drosophila Homolog | None | tau/ CG45110 | CG4618 | caz | ter94 | TBPH | N/A | N/A | N/A |
Another locus associated with FTD is GRN which encodes for PGRN, a growth factor involved in multiple pathways including related to inflammation [16,17]. FTD associated with nonsense, splice-site, or frameshift variants of GRN is inherited in an autosomal dominant mode [5,18,19]. PGRN has a fundamental role in lysosome functioning and in tissue development and inflammation [1,18]. The reduction of PGRN levels from these mutations have been known to result in haploinsufficiency, a major cause of FTD [5,18]. Interestingly, there are no obvious homologs of GRN in Drosophila [20].
The MAPT gene encodes for the protein Tau which is responsible for stabilizing and assembling microtubules and also regulating vesicle transport regulated by kinesin [1,18,21]. There are currently 54 identified pathogenic MAPT mutations associated with the onset of neurodegenerative diseases, including FTD. Presence of a mutation in MAPT generally prevents binding between Tau and Tubulin, subsequently causing hyperphosphorylated Tau accumulation—a common neurodegenerative disease precursor [1]. Drosophila models of MAPT have successfully identified various Tau-induced neurodegeneration modifiers, including eight Tau toxicity suppressors and 16 enhancers [22].
TMEM106B serves as a genetic modifier most notably in patients carrying a mutation in C9ORF72 and PGRN. While the S185 isoform of TMEM106B provides some protection against neurodegeneration, the isoform T185 raises risk [23]. Overexpression of TMEM106B results in the disruption of lysosome functioning as well as impairment of protein trafficking regulation [18]. Research presented by Jun et al. and Rostgard et al. propose a potential link between TMEM106B and autophagy, and more specifically the ESCRT pathway and CHMP2B [23,24]. Overall, the Drosophila models have been particularly useful in identifying the pathogenicity of the mutated proteins as well as in drug targeting investigation [19].
Mutations in CHMP2B, (chromatin modifying protein 2B), FUS (fused in sarcoma), TARDBP (Transactive DNA-binding protein), VCP (Valosin containing protein), TBK-1 (TANK-binding kinase 1) have also been associated with FTD but are much rarer in incidence (Table 1). Together, mutations in these genes account for fewer than 3% of FTD cases. Interestingly, mutations in FUS, TARDBP, VCP, and TBK-1 are also associated with ALS [8,19]. The TARDP locus codes for TDP-43 which is involved in multiple RNA-related gene expression events including transcription, splicing, transport (including transport of RNA granules to dendrites [25]), and translation [26]. Identification of TDP-43 variants as a major component in inclusions associated with neurodegenerative diseases including ALS and FTD led to increased interest to characterize its function and role in neuroanal pathology [27,28,29]. The Drosophila models of TDP-43–associated FTD/ALS were able to recreate most aspects of human proteinopathies. Further, these models also provided additional levels of characterization, for instance, neurodegeneration was not always dependent on formation of inclusions [30,31].
The FUS locus encodes for an RNA binding protein that is a component of stress granules, with the primary function of DNA repair and regulation of RNA splicing [26]. FUS share remarkable structural and functional similarity with TDP-43. Mutations in FUS result in cytoplasmic aggregate formation as well as stabilization of stress granules, restricting the disassembly of the granules [18,26]. Drosophila model of mutant FUS-associated toxicity recreate ALS/FTD pathology e.g., aggregation of ubiquitinated proteins, neurodegeneration, behavioral deficits in locomotion, and early mortality [32].
VCP encodes for an ATPase that has a wide range of cellular functions including degradation of endoplasmic-reticulum-associated proteins, delivery and unfolding of ubiquitinated proteins, and regulation of endosome morphology [2,18]. VCP mutations have also been found to inhibit autophagy-mediated turnover of stress granules [18]. Drosophila homolog of VCP plays a role in dendritic pruning by misregulating RNA-binding proteins including TDP-43 [33]. Interestingly, ectopic expression of disease-associated mutant VCP in Drosophila cause neurodegeneration due to mislocalization of TDP-43 to cytoplasm [34]. Further, VCP was also shown to genetically interact with FUS in Drosophila. Taken together, Drosophila models employing ectopic overexpression of disease-associated mutations and analysis of loss of function of Drosophila homologs of disease-associated loci i.e., cabeza (FUS), tbph (TDP-43), and ter94 (VCP) have shown genetic interaction between FUS, TDP-43, and VCP. Such studies further highlight the utility of Drosophila models to identify and characterize gene networks that contribute in complex neurological diseases [32,35,36].
Recently, haploinsufficiency of TBK1 locus was associated with FTD and ALS [37]. Reduction in TBK1 levels due to mutations show deficits in autophagy pathway [38,39]. TBK1 influences autophagy pathway by regulating the activity/binding of p62 (nucleoporin 62) and OPTN (optineurin)—autophagy adaptor proteins. Consequently, P62 and OPTN also engage in the pathology of the ALS/FTD spectrum [39,40]. Interestingly, there is an interaction between TBK1 and OPTN with the Ras-related protein Rab8 which also interacts with the C9ORF72 complex [14]. Rab8-mediated signaling is also misregulated in a Drosophila model of FTD associated with mutant CHMP2B (next section of this review) [41]. Overall, Drosophila models of FTD-associated mutations have significantly contributed in identification, characterization, or both, of mechanistic details about the incidence and progression of the disorder.
3. FTD Associated with Mutant CHMP2B
3.1. Structure and Function of CHMP2B
CHMP2B is a subunit of the multi-protein ESCRT-III complex (Figure 1). The ESCRTs are composed of multiple highly conserved heteromeric protein sub-complexes, namely ESCRT-0 (hepatocyte growth factor-regulated substrate (HRS), signal-transducing adaptor molecule (STAM1-2)), ESCRT-I (tumor susceptibility gene (TSG101), vacuolar protein sorting 28 (Vps28), Vps37, MVB12, ubiquitin-associated protein UBAP), ESCRT-II ((ELL-associated protein 20 (EAP20), EAP30, EAP45), ESCRT-III (CHMP1A-B, CHMP2A-B, CHMP3, CHMP4, CHMP5, CHMP6, CHMP7), and Vps4 [42]. Although human isoforms are of ESCRTs are listed above in parenthesis, all ESCRT components are often interchangeably referred by nomenclature from yeast. Seminal work in the Saccharomyces model system have led to discovery and mostly contributed to the characterization of the ESCRT pathway [43]. The ESCRT machinery is involved in membrane remodeling and scission during various critical cellular processes, such as multivesicular body (MVB) formation in endosomal–lysosomal pathway [44], retrovirus budding [45], and cytokinesis [46]. Recently, ESCRT was also shown to mediate other important cellular activities for example repair and homeostasis of plasma and nuclear membranes, protein secretion, and dendritic pruning (For review, see Christ et al., 2017) [47]. This section focuses on ESCRT-mediated endosomal sorting and macroautophagy, which are two processes so far shown to be primarily affected by CHMP2B mutation. In addition, the role of ESCRT in the nervous system is discussed.
The MVB pathway is initiated by the ESCRT-0 complex. By binding both phosphatidylinositol 3-phosphate (PtdIns3P) and ubiquitin, ESCRT-0 binds to the endosomal membrane and clusters the ubiquinated cargo protein. ESCRT-0 also recruits ESCRT-I, which in turn recruits ESCRT-II. The two complexes induce membrane budding and confine the cargo. ESCRT-II then initiates ESCRT-III assembly by binding and activating CHMP6 (Vps20). The active CHMP6 instigates CHMP4 (Snf7) homo-oligomerization, which is capped by CHMP3 (Vps24). CHMP3 finally recruits CHMP2 (A and B) and thus completes the assembly of ESCRT-III [4,42,48,49,50,51]. In addition, interactions between Vps28 of ESCRT-I and CHMP6 of ESCRT-III were observed, suggesting existence of direct connections between ESCRT-I and III [52,53]. Other accessory proteins, such as ALG-2-interacting-protein-X (ALIX), also contribute to facilitate the recruitment of ESCRT-III [48]. CHMP2, together with other ESCRT-III subunits, subsequently recruits AAA ATPase Vps4 by binding the MIT domain on the N-terminus. The recruited Vps4 assembles to a dodecomer and binds its cofactor Vta1 to form the Vps4-Vta1 complex [42,51]. Vps4 provides the ATPase input in the membrane scission, and it also rapidly depolymerizes ESCRT-III filaments, a process essential to the recycling of ESCRT-III subunits [54,55,56].
CHMP2B is a highly conserved protein of 213 amino acids that contains coiled-coil, Snf-7, and acidic C-terminus domains [7]. The coiled-coil domain functions as a molecular spacer and mediates membrane tethering of vesicles [57]. The acidic C-terminus domain contains a microtubule interacting and transport (MIT)-interacting motif (MIM), which can form an amphipathic helix that binds the Vps4 MIT domain [58,59]. The recruited Vps4 catalyzes the disassembly of ESCRT-III [60]. In the cytosol, CHMP2B and other ESCRT-III subunits exist in an autoinhibition monomer. When autoinhibition is released, the subunits can polymerize into circular or helical filaments on the cellular membrane [51,61,62,63,64,65]. In particular, CHMP2B plays a structural role in membrane scaffolding directly. Overexpression of CHMP2B can form full-length polymers of the protein and create membrane protrusions [60].
Cargos sorted by the ESCRT machinery are mostly ubiquinated transmembrane proteins i.e., cell-surface receptors. The ESCRT machinery therefore plays a role in the degradation and recycling of receptors, and thus regulates the strength and timing of the signaling pathways. Receptor tyrosine kinases (RTKs) such as epidermal growth factor receptor (EGFR) and G-protein coupled receptors (GPCRs) are the two best studied cases [66,67,68,69]; other receptors regulated by ESCRTs include integrin, cytokine receptors, Toll-like receptors, and Notch receptors (For review, see Szymanska et al., 2018; receptors identified using animal model of CHMP2B mutations are discussed in next section) [70,71,72,73,74]. Several studies in Drosophila using mutant ESCRT subunits have shown a role of ESCRTs in tumorigenesis because of their ability to sequester Notch and JAK-STAT pathways [75,76,77,78]. These studies have raised an intriguing possibility of utilizing ESCRTs for tumor-suppression strategies.
In addition to endocytosis, ESCRTs also plays a role in macroautophagy (often referred to as autophagy). Autophagy is a conserved process of degradation of cytosolic proteins and organelles induced by various stresses, such as starvation. The initial step of autophagy is the formation of the cargo-containing double-membrane autophagosomes. The autophagosomes then fuse with lysosomes to form autolysosomes, where the cargos are degraded (Figure 1B). In this process, ESCRT machinery and endosomes are involved through the formation of amphisome, the result of autophagosomes and endosomes fusion (For review, see Lefebvre et al., 2018) [79,80]. In addition to the amphisome formation, ESCRTs also plays a role in autophagosome maturation by interacting with SNAREs. In particular, CHMP2B was shown to interact with Syntaxin 13 [81]. Recent studies also showed other interactions between specific ESCRT proteins and autophagy related proteins, suggesting a close connection between the two pathways (For review, see Lefebvre et al., 2018) [80].
ESCRT functions are crucial in the nervous system. During embryogenesis of the central nervous system, ESCRT controls the survival of neural progenitors. In the later stages of development, ESCRT regulates the outgrowth and pruning. In the mature nervous system, ESCRTs plays a role in regulating synaptic transmissions. (For review, see Sadoul et al., 2018) [82]. In particular, CHMP2B is involved in regulating synaptic plasticity in dendrite spines [6,83].
3.2. Defects Caused by CHMP2Bintron5
At the molecular level, the G-C mutation in the CHMP2B transcript causes the inclusion of the 201-bp intron 5, which results in addition of a valine residue followed by a stop codon. Hence, CHMP2Bintron5 features a C-terminus truncation, with the last 36 amino acids replaced by a valine [7]. The truncated part of C-terminal region contains the MIM domain, which is responsible for Vps4 recruitment, so CHMP2Bintron5 cannot interact with Vps4 [60]. As a result, the membrane scission function of ESCRT is impaired, and therefore the MVB pathway and the autophagy pathway are disrupted. Also, since the C-terminal domain is essential to the autoinhibition of CHMP2B, the mutant protein is constitutively active, resulting in polymerization that deform the membrane [60].
Disruption of the endosomal–lysosomal pathway by CHMP2Bintron5 primarily causes misregulation of receptor turnover. The impaired degradation of receptors results in upregulation of receptor-mediated signaling pathways. Because of the relative ease of ectopic expression of genes with control over spatio-temporal attributes and the capacity to conduct genetic screens, studies using the Drosophila model of CHMP2Bintron5 have led to the identification of multiple misregulated receptor-mediated signaling. Ectopic expression of CHMP2Bintron5 primarily during photoreceptor cell diffrentiation in the Drosophila eye imaginal disc resulted in excessive melanization in the eyes due to upregulation of Toll receptor-mediated signaling, a conserved pathway of innate immune response [84]. Additionally, ectopic expression of CHMP2Bintron5 during eye tissue specification during larval stages caused tumorigenic deformities in the adults eyes due to upregulation of Notch pathway—a fundamental cellular interaction pathway involved in cell fate determination and differentiation during development [85]. Furthermore, Transforming Growth Factor beta (TGF-β) and c-Jun N-terminal kinases (JNK), Rab8 signaling are upregulated when CHMP2Bintron5 is expressed in Drosophila neuromuscular junctions (NMJ). The resulting synaptic overgrowth at the NMJ synapse is a distinct feature of neurodegeneration [41]. Recently, the pro-apoptotic protein POSH/SH3RF1 was shown to mediate the JNK and NF-κB dependent apoptosis in CHMP2Bintron5–mediated toxicity in Drosophila NMJ [86].
ESCRT machinery also plays a role in autophagosome formation. Therefore, CHMP2Bintron5 also induces defects in autophagy. Recent genetic interaction evidence suggests that the mutant protein inhibits phagophore maturation through Syntaxin 13 [81]. Ectopic expression of CHMP2Bintron5 was shown to cause accumulation of autophagosomes, which contained aggregates of ubiquinated proteins [87,88].
Although the Drosophila model provides major advantages in identification and characterization of genetic modifiers and pathological mechanisms of mutations associated with FTD, alternative models e.g., knock out and trangenic mice, primary neuronal culture, and neuronal culture derived from induced pluripotent stem cell (iPSC) and Human embryonic stem cell (hESC) lines have significantly contributed in understanding to further progress discoveries in these areas [19]. When expressed in mouse neurons, CHMP2Bintron5 was shown to cause behavioral and histological features of FTD and ALS. In particular, CHMP2Bintron5 mouse exhibited behavioral deficits as well as progressive loss of motor activities [89]. On the cellular level, CHMP2Bintron5 effects dendrites by inducing decrease in spine density and reduction in mushroom spine morphology [6]. In mouse models, in addition to ubiquinated protein aggregation, CHMP2Bintron5 also induces gliosis and axonal swelling [90]. Recently, using cortical mouse neurons Transmembrane protein 106B (TMEM106B) was identified as a modifier of CHMP2Bintron5 toxicity through the autophagy pathway [23]. This research identified an association between TMEM106B and CHMP2B, particularly the localization of TMEM106B variants in Rab5 and Rab7 positive endosomes, suggesting possible engagement of ESCRT pathways [23].
CHMP2Bintron5 transfected cell lines showed reduced fusion of endosomes with lysosomes and delayed degradation of Epidermal Growth Factor (EGF) and EGFR [91]. Human embryonic stem cell (hESC)-derived postmitotic neurons were used to show the functions of ESCRT-III subunit Snf7-1 and Snf7-2, which play key roles in the neurotoxicity of CHMP2Bintron5 [92]. A human-induced pluripotent stem cell (iPSC) line with the CHMP2Bintron5 mutation was also generated recently using the CRISPR-Cas9 system [93]. The forebrain-type cortical type neuron differentiated from the iPSC line exhibited defects in endosome and mitochondria, increased oxidative stress, as well as perturbed iron homeostasis [94].
Another pathology caused by CHMP2Bintron5 in mouse models is a lysosome storage disorder of progressive neuronal autofluorescent aggregate formation [95]. In addition to defects in protein trafficking and autophagy, there is also evidence that CHMP2Bintron5 causes defects in the miRNA pathway. In particular, it was shown that CHMP2Bintron5 reduces the level of miR-124, which leads to decrease in AMPA receptor (AMPAR) abundance [96].
4. Future Perspectives
Since the first systematic characterization of CHMP2Bintron5 in 2005, there have been great strides in the understanding of the function of CHMP2B and its role in pathological mechanisms associated with FTD [39]. However, there is still much that can be discovered on this topic. One area that has seen increased interest is in identification of receptor-mediated signaling pathways and their modifiers affected by CHMP2Bintron5. Using the Drosophila model, genetic screens has identified signaling pathways and their modifiers such as Toll, Notch, Syntaxin 13, Rab8, JNK, TGF-β pathways [21,41,81,84,86]. Although preliminary information is known about the function of these modifiers in their respective pathways, further investigation would provide an even better understanding of their role in these pathways and in the development of FTD in general.
Additionally, Drosophila has been used for a number of FTD-related mutations, there has still yet to be a model of the mutations PGRN, TBK1, and THEM106B. Characterization of these mutations using the Drosophila model would provide insight into how these mutations affect the typical functioning of the pathways they are involved in, and thus their role in FTD pathology.
With increasing advances in biotechnology, future work in in identification and characterization of molecular-genetic markers of FTD is warranted. Next-Generation Sequencing (NGS) has made significant contributions to phenotypic dissections and molecular genetics of neurological diseases [97]. Particularly, RNA sequencing (RNA-seq) analysis was utilized in the study of FTD3 to confirm defects in endosome and mitochondria and to reveal the imbalance of iron homeostasis [94]. This method is particularly beneficial as it is able to detect transcripts regardless of whether or not there is an existing characterized genomic sequence. Also, it results in very minimal background allowing for a large dynamic expression range for transcript detection [98]. Human iPSC-derived neuronal culture systems also offers exciting possibilities for longitudinal analysis for molecular pathology and drug development of FTD and other neurodegenerative disorders. Animal models of FTD will continue to contribute in improving overall understanding of the disease and development of effective therapeutic treatment of FTD. Furthermore, studies with these models will provide insights into mechanisms responsible for the same mutations manifesting into different aspects of the FTD-ALS spectrum.
5. Conclusions
In this review we have identified the recent findings of FTD related mutations with emphasis on CHMP2Bintron5, in order to understand their pathological neurodegeneration mechanisms. Although rare in FTD incidence, CHMP2Bintron5 mutation causes various molecular and cellular deficits in the endolysosomal and autophagosome–lysosome pathways. Drosophila have provided an inexpensive, genetically tractable, and high throughput animal model into understanding the defects caused by CHMP2Bintron5 mutation as well as other FTD associated mutations.
Acknowledgments
We would like to thank Peter Joshua Kavaler for reviewing and providing comments on the manuscript. S. Tariq Ahmad is supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103423 and science division grant from Colby College.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Olszewska D.A., Lonergan R., Fallon E.M., Lynch T. Genetics of frontotemporal dementia. Curr. Neurol. Neuroscie. Rep. 2016;16:107. doi: 10.1007/s11910-016-0707-9. [DOI] [PubMed] [Google Scholar]
- 2.Pan X.-D., Chen X.-C. Clinic, neuropathology and molecular genetics of frontotemporal dementia: A mini-review. Transl. Neurodegener. 2013;2:8. doi: 10.1186/2047-9158-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young J.J., Lavakumar M., Tampi D., Balachandran S., Tampi R.R. Frontotemporal dementia: Latest evidence and clinical implications. Ther. Adv. Psychopharmacol. 2018;8:33–48. doi: 10.1177/2045125317739818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krasniak C.S., Ahmad S.T. The role of CHMP2B(Intron5) in autophagy and frontotemporal dementia. Brain Res. 2016;1649:151–157. doi: 10.1016/j.brainres.2016.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindquist S.G., Brændgaard H., Svenstrup K., Isaacs A.M., Nielsen J.E. on behalf of the FReJA Consortium. Frontotemporal dementia linked to chromosome 3 (FTD-3)—Current concepts and the detection of a previously unknown branch of the danish FTD-3 family. Eur. J. Neurol. 2008;15:667–670. doi: 10.1111/j.1468-1331.2008.02144.x. [DOI] [PubMed] [Google Scholar]
- 6.Belly A., Bodon G., Blot B., Bouron A., Sadoul R., Goldberg Y. CHMP2B mutants linked to frontotemporal dementia impair maturation of dendritic spines. J. Cell Sci. 2010;123:2943–2954. doi: 10.1242/jcs.068817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skibinski G., Parkinson N.J., Brown J.M., Chakrabarti L., Lloyd S.L., Hummerich H., Nielsen J.E., Hodges J.R., Spillantini M.G., Thusgaard T., et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 8.Rainero I., Rubino E., Michelerio A., D’Agata F., Gentile S., Pinessi L. Recent advances in the molecular genetics of frontotemporal lobar degeneration. Funct. Neurol. 2017;32:7–16. doi: 10.11138/FNeur/2017.32.1.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann D., Parlato R. C9ORF72-associated neurodegeneration in ALS-FTD: Breaking new ground in ribosomal RNA and nucleolar dysfunction. Cell Tissue Res. 2018 doi: 10.1007/s00441-018-2806-1. [DOI] [PubMed] [Google Scholar]
- 10.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almeida S., Gao F.B. Lost & found: C9ORF72 and the autophagy pathway in ALS/FTD. EMBO J. 2016;35:1251–1253. doi: 10.15252/embj.201694578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizielinska S., Grönke S., Niccoli T., Ridler C.E., Clayton E.L., Devoy A., Moens T., Norona F.E., Woollacott I.O.C., Pietrzyk J., et al. C9ORF72 repeat expansions cause neurodegeneration in drosophila through arginine-rich proteins. Science (N. Y.) 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sellier C., Campanari M.L., Julie Corbier C., Gaucherot A., Kolb-Cheynel I., Oulad-Abdelghani M., Ruffenach F., Page A., Ciura S., Kabashi E., et al. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 2016;35:1276–1297. doi: 10.15252/embj.201593350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farg M.A., Sundaramoorthy V., Sultana J.M., Yang S., Atkinson R.A., Levina V., Halloran M.A., Gleeson P.A., Blair I.P., Soo K.Y., et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 2014;23:3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S., et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 17.Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J.J., et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan P.M., Zhou X., Hu F. Autophagy-lysosome dysfunction in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. In: Sharma P.D., editor. Lysosomes—Associated Diseases and Methods to Study Their Function. InTech; Rijeka, Croatia: 2017. Chapter 4. [Google Scholar]
- 19.Boxer A.L., Gold M., Huey E., Gao F.-B., Burton E.A., Chow T., Kao A., Leavitt B., Lamb B., Grether M., et al. Frontotemporal degeneration, the next therapeutic frontier: Molecules and animal models for FTD drug development (part 1 of 2 articles) Alzheimer Dement. J. Alzheimer Assoc. 2013;9:176–188. doi: 10.1016/j.jalz.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cenik B., Sephton C.F., Kutluk Cenik B., Herz J., Yu G. Progranulin: A proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J. Biol. Chem. 2012;287:32298–32306. doi: 10.1074/jbc.R112.399170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rademakers R., Cruts M., Van Broeckhoven C. The role of tau (MAPT) in frontotemporal dementia and related tauopathies. Hum. Mutat. 2004;24:277–295. doi: 10.1002/humu.20086. [DOI] [PubMed] [Google Scholar]
- 22.Shulman J.M., Feany M.B. Genetic modifiers of tauopathy in drosophila. Genetics. 2003;165:1233–1242. doi: 10.1093/genetics/165.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jun M.-H., Han J.-H., Lee Y.-K., Jang D.-J., Kaang B.-K., Lee J.-A. TMEM106B, a frontotemporal lobar dementia (FTLD) modifier, associates with FTD-3-linked CHMP2B, a complex of ESCRT-III. Mol. Brain. 2015;8:85. doi: 10.1186/s13041-015-0177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rostgaard N., Roos P., Budtz-Jørgensen E., Johannsen P., Waldemar G., Nørremølle A., Lindquist S.G., Gydesen S., Brown J.M., Collinge J. TMEM106B and apoe polymorphisms in CHMP2B-mediated frontotemporal dementia (FTD-3) Neurobiol. Aging. 2017;59:221.e1–221.e7. doi: 10.1016/j.neurobiolaging.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 25.Wang I.F., Wu L.S., Chang H.Y., Shen C.K. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J. Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- 26.Lagier-Tourenne C., Polymenidou M., Cleveland D.W. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geser F., Martinez-Lage M., Kwong L.K., Lee V.M., Trojanowski J.Q. Amyotrophic lateral sclerosis, frontotemporal dementia and beyond: The TDP-43 diseases. J. Neurol. 2009;256:1205–1214. doi: 10.1007/s00415-009-5069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 29.Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 30.Miguel L., Frebourg T., Campion D., Lecourtois M. Both cytoplasmic and nuclear accumulations of the protein are neurotoxic in drosophila models of TDP-43 proteinopathies. Neurobiol. Dis. 2011;41:398–406. doi: 10.1016/j.nbd.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Li Y., Ray P., Rao E.J., Shi C., Guo W., Chen X., Woodruff E.A., 3rd, Fushimi K., Wu J.Y. A drosophila model for TDP-43 proteinopathy. Proc. Natl. Acad. Sci. USA. 2010;107:3169–3174. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanson N.A., Jr., Maltare A., King H., Smith R., Kim J.H., Taylor J.P., Lloyd T.E., Pandey U.B. A drosophila model of FUS-related neurodegeneration reveals genetic interaction between fus and TDP-43. Hum. Mol. Genet. 2011;20:2510–2523. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rumpf S., Bagley J.A., Thompson-Peer K.L., Zhu S., Gorczyca D., Beckstead R.B., Jan L.Y., Jan Y.N. Drosophila valosin-containing protein is required for dendrite pruning through a regulatory role in mRNA metabolism. Proc. Natl. Acad. Sci. USA. 2014;111:7331–7336. doi: 10.1073/pnas.1406898111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritson G.P., Custer S.K., Freibaum B.D., Guinto J.B., Geffel D., Moore J., Tang W., Winton M.J., Neumann M., Trojanowski J.Q., et al. TDP-43 mediates degeneration in a novel drosophila model of disease caused by mutations in VCP/p97. J. Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J.-W., Brent J.R., Tomlinson A., Shneider N.A., McCabe B.D. The ALS-associated proteins FUS and TDP-43 function together to affect drosophila locomotion and life span. J. Clin. Investig. 2011;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azuma Y., Tokuda T., Shimamura M., Kyotani A., Sasayama H., Yoshida T., Mizuta I., Mizuno T., Nakagawa M., Fujikake N., et al. Identification of TER94, drosophila VCP, as a strong modulator of motor neuron degeneration induced by knockdown of CAZ, drosophila FUS. Hum. Mol. Genet. 2014;23:3467–3480. doi: 10.1093/hmg/ddu055. [DOI] [PubMed] [Google Scholar]
- 37.Freischmidt A., Wieland T., Richter B., Ruf W., Schaeffer V., Muller K., Marroquin N., Nordin F., Hubers A., Weydt P., et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 2015;18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 38.Sieben A., Van Mossevelde S., Wauters E., Engelborghs S., van der Zee J., Van Langenhove T., Santens P., Praet M., Boon P., Miatton M., et al. Extended FTLD pedigree segregating a belgian GRN-null mutation: Neuropathological heterogeneity in one family. Alzheimer Res. Ther. 2018;10:7. doi: 10.1186/s13195-017-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui R., Tuo M., Li P., Zhou C. Association between TBK1 mutations and risk of amyotrophic lateral sclerosis/frontotemporal dementia spectrum: A meta-analysis. Neurol. Sci. 2018;39:811–820. doi: 10.1007/s10072-018-3246-0. [DOI] [PubMed] [Google Scholar]
- 40.Oakes J.A., Davies M.C., Collins M.O. TBK1: A new player in als linking autophagy and neuroinflammation. Mol. Brain. 2017;10:5. doi: 10.1186/s13041-017-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West R.J., Lu Y., Marie B., Gao F.-B., Sweeney S.T. RAB8, POSH, and TAK1 regulate synaptic growth in a drosophila model of frontotemporal dementia. J. Cell Biol. 2015;208:931. doi: 10.1083/jcb.201404066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt O., Teis D. The escrt machinery. Curr. Biol. 2012;22:R116–R120. doi: 10.1016/j.cub.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henne W.M., Stenmark H., Emr S.D. Molecular mechanisms of the membrane sculpting escrt pathway. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katzmann D.J., Odorizzi G., Emr S.D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 45.Morita E., Sundquist W.I. Retrovirus budding. Ann. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 46.Carlton J.G., Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the escrt machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 47.Christ L., Raiborg C., Wenzel E.M., Campsteijn C., Stenmark H. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci. 2017;42:42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 48.Frankel E.B., Audhya A. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin. Cell Dev. Biol. 2018;74:4–10. doi: 10.1016/j.semcdb.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wollert T., Hurley J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurley J.H. The escrt complexes. Crit. Rev. Biochem. Mol. Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henne W.M., Buchkovich N.J., Emr S.D. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 52.Pineda-Molina E., Belrhali H., Piefer A.J., Akula I., Bates P., Weissenhorn W. The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic. 2006;7:1007–1016. doi: 10.1111/j.1600-0854.2006.00440.x. [DOI] [PubMed] [Google Scholar]
- 53.Im Y.J., Hurley J.H. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev. Cell. 2008;14:902–913. doi: 10.1016/j.devcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang B., Stjepanovic G., Shen Q., Martin A., Hurley J.H. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol. 2015;22:492–498. doi: 10.1038/nsmb.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCullough J., Colf L.A., Sundquist W.I. Membrane fission reactions of the mammalian ESCRT pathway. Ann. Rev. Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adell M.A.Y., Migliano S.M., Teis D. ESCRT-III and Vps4: A dynamic multipurpose tool for membrane budding and scission. FEBS J. 2016;283:3288–3302. doi: 10.1111/febs.13688. [DOI] [PubMed] [Google Scholar]
- 57.Truebestein L., Leonard T.A. Coiled-coils: The long and short of it. Bioessays. 2016;38:903–916. doi: 10.1002/bies.201600062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Obita T., Saksena S., Ghazi-Tabatabai S., Gill D.J., Perisic O., Emr S.D., Williams R.L. Structural basis for selective recognition of ESCRT-III by the AAA ATpase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 59.Stuchell-Brereton M.D., Skalicky J.J., Kieffer C., Karren M.A., Ghaffarian S., Sundquist W.I. ESCRT-III recognition by Vps4 ATpases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 60.Bodon G., Chassefeyre R., Pernet-Gallay K., Martinelli N., Effantin G., Hulsik D.L., Belly A., Goldberg Y., Chatellard-Causse C., Blot B. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J. Biol. Chem. 2011;286:40276–40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shim S., Kimpler L.A., Hanson P.I. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 62.Hanson P.I., Roth R., Lin Y., Heuser J.E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gőttlinger H.G., Weissenhorn W. Helical structures of ESCRT-III are disassembled by Vps4. Science (N. Y.) 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bajorek M., Schubert H.L., McCullough J., Langelier C., Eckert D.M., Stubblefield W.-M.B., Uter N.T., Myszka D.G., Hill C.P., Sundquist W.I. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buchkovich N.J., Henne W.M., Tang S., Emr S.D. Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev. Cell. 2013;27:201–214. doi: 10.1016/j.devcel.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 66.Malerød L., Stuffers S., Brech A., Stenmark H. Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic. 2007;8:1617–1629. doi: 10.1111/j.1600-0854.2007.00630.x. [DOI] [PubMed] [Google Scholar]
- 67.Bache K.G., Stuffers S., Malerød L., Slagsvold T., Raiborg C., Lechardeur D., Wälchli S., Lukacs G.L., Brech A., Stenmark H. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell. 2006;17:2513–2523. doi: 10.1091/mbc.e05-10-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doyotte A., Russell M.R.G., Hopkins C.R., Woodman P.G. Depletion of TSG101 forms a mammalian ‘class E′ compartment: A multicisternal early endosome with multiple sorting defects. J. Cell Sci. 2005;118:3003–3017. doi: 10.1242/jcs.02421. [DOI] [PubMed] [Google Scholar]
- 69.Verma R., Marchese A. The endosomal sorting complex required for transport pathway mediates chemokine receptor CXCR4-promoted lysosomal degradation of the mammalian target of rapamycin antagonist deptor. J. Biol. Chem. 2015;290:6810–6824. doi: 10.1074/jbc.M114.606699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lobert V.H., Brech A., Pedersen N.M., Wesche J., Oppelt A., Malerød L., Stenmark H. Ubiquitination of α5β1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev. Cell. 2010;19:148–159. doi: 10.1016/j.devcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 71.Mamińska A., Bartosik A., Banach-Orłowska M., Pilecka I., Jastrzębski K., Zdżalik-Bielecka D., Castanon I., Poulain M., Neyen C., Wolińska-Nizioł L., et al. Escrt proteins restrict constitutive NF-κb signaling by trafficking cytokine receptors. Sci. Signal. 2016;9:ra8. doi: 10.1126/scisignal.aad0848. [DOI] [PubMed] [Google Scholar]
- 72.Husebye H., Halaas Ø., Stenmark H., Tunheim G., Sandanger Ø., Bogen B., Brech A., Latz E., Espevik T. Endocytic pathways regulate toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 2006;25:683–692. doi: 10.1038/sj.emboj.7600991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Le Borgne R. Regulation of notch signalling by endocytosis and endosomal sorting. Curr. Opin. Cell Biol. 2006;18:213–222. doi: 10.1016/j.ceb.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 74.Szymanska E., Budick-Harmelin N., Miaczynska M. Endosomal “sort” of signaling control: The role of escrt machinery in regulation of receptor-mediated signaling pathways. Semin. Cell Dev. Biol. 2018;74:11–20. doi: 10.1016/j.semcdb.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 75.Thompson B.J., Mathieu J., Sung H.-H., Loeser E., Rørth P., Cohen S.M. Tumor suppressor properties of the ESCRT-II complex component Vps25 in drosophila. Dev. Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 76.Rodahl L.M., Haglund K., Sem-Jacobsen C., Wendler F., Vincent J.-P., Lindmo K., Rusten T.E., Stenmark H. Disruption of Vps4 and JNK function in drosophila causes tumour growth. PLoS ONE. 2009;4:e4354. doi: 10.1371/journal.pone.0004354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moberg K.H., Schelble S., Burdick S.K., Hariharan I.K. Mutations in erupted, the drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev. Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 78.Herz H.M., Bergmann A. Genetic analysis of escrt function in drosophila: A tumour model for human TSG101. Biochem. Soc. Trans. 2009;37:204–207. doi: 10.1042/BST0370204. [DOI] [PubMed] [Google Scholar]
- 79.Lee J.-A., Gao F.-B. Escrt, autophagy, and frontotemporal dementia. BMB Rep. 2008;41:827–832. doi: 10.5483/BMBRep.2008.41.12.827. [DOI] [PubMed] [Google Scholar]
- 80.Lefebvre C., Legouis R., Culetto E. ESCRT and Autophagies: Endosomal Functions and Beyond. Semin. Cell Dev. Biol. 2018;74:21–28. doi: 10.1016/j.semcdb.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 81.Lu Y., Zhang Z., Sun D., Sweeney S.T., Gao F.-B. Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol. Cell. 2013;52:264–271. doi: 10.1016/j.molcel.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sadoul R., Laporte M.H., Chassefeyre R., Chi K.I., Goldberg Y., Chatellard C., Hemming F.J., Fraboulet S. The role of escrt during development and functioning of the nervous system. Semin. Cell Dev. Biol. 2018;74:40–49. doi: 10.1016/j.semcdb.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 83.Chassefeyre R., Martínez-Hernández J., Bertaso F., Bouquier N., Blot B., Laporte M., Fraboulet S., Couté Y., Devoy A., Isaacs A.M. Regulation of postsynaptic function by the dementia-related ESCRT-III subunit CHMP2B. J. Neurosci. 2015;35:3155–3173. doi: 10.1523/JNEUROSCI.0586-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ahmad S.T., Sweeney S.T., Lee J.A., Sweeney N.T., Gao F.B. Genetic screen identifies SERPIN5 as a regulator of the toll pathway and CHMP2B toxicity associated with frontotemporal dementia. Proc. Natl. Acad. Sci. USA. 2009;106:12168–12173. doi: 10.1073/pnas.0903134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheruiyot A., Lee J.-A., Gao F.-B., Ahmad S.T. Expression of mutant CHMP2B, an ESCRT-III component involved in frontotemporal dementia, causes eye deformities due to notch misregulation in drosophila. FASEB J. 2014;28:667–675. doi: 10.1096/fj.13-234138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.West R.J., Ugbode C., Gao F.-B., Sweeney S.T. The pro-apoptotic JNK scaffold POSH/SH3RF1 mediates CHMP2Bintron5-associated toxicity in animal models of frontotemporal dementia. Hum. Mol. Genet. 2018;27:1382–1395. doi: 10.1093/hmg/ddy048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M.C., Isaacs A., Brech A., Stenmark H., Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee J.-A., Beigneux A., Ahmad S.T., Young S.G., Gao F.-B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 89.Vernay A., Therreau L., Blot B., Risson V., Dirrig-Grosch S., Waegaert R., Lequeu T., Sellal F., Schaeffer L., Sadoul R. A transgenic mouse expressing CHMP2Bintron5 mutant in neurons develops histological and behavioural features of amyotrophic lateral sclerosis and frontotemporal dementia. Hum. Mol. Genet. 2016;25:3341–3360. doi: 10.1093/hmg/ddw182. [DOI] [PubMed] [Google Scholar]
- 90.Ghazi-Noori S., Froud K.E., Mizielinska S., Powell C., Smidak M., Fernandez de Marco M., O’Malley C., Farmer M., Parkinson N., Fisher E.M.C., et al. Progressive neuronal inclusion formation and axonal degeneration in CHMP2B mutant transgenic mice. Brain. 2012;135:819–832. doi: 10.1093/brain/aws006. [DOI] [PubMed] [Google Scholar]
- 91.Urwin H., Authier A., Nielsen J.E., Metcalf D., Powell C., Froud K., Malcolm D.S., Holm I., Johannsen P., Brown J. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum. Mol. Genet. 2010;19:2228–2238. doi: 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee J.-A., Liu L., Javier R., Kreitzer A.C., Delaloy C., Gao F.-B. ESCRT-III subunits Snf7-1 and Snf7-2 differentially regulate transmembrane cargos in hESC-derived human neurons. Mol. Brain. 2011;4:37. doi: 10.1186/1756-6606-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y., Schmid B., Nielsen T.T., Nielsen J.E., Clausen C., Hyttel P., Holst B., Freude K.K. Generation of a human induced pluripotent stem cell line via CRISPR-CAS9 mediated integration of a site-specific homozygous mutation in CHMP2B. Stem Cell Res. 2016;17:151–153. doi: 10.1016/j.scr.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y., Schmid B., Nikolaisen N.K., Rasmussen M.A., Aldana B.I., Agger M., Calloe K., Stummann T.C., Larsen H.M., Nielsen T.T. Patient IPSC-derived neurons for disease modeling of frontotemporal dementia with mutation in CHMP2B. Stem Cell Rep. 2017;8:648–658. doi: 10.1016/j.stemcr.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clayton E.L., Mizielinska S., Edgar J.R., Nielsen T.T., Marshall S., Norona F.E., Robbins M., Damirji H., Holm I.E., Johannsen P., et al. Frontotemporal dementia caused by CHMP2B mutation is characterised by neuronal lysosomal storage pathology. Acta Neuropathol. 2015;130:511–523. doi: 10.1007/s00401-015-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gascon E., Lynch K., Ruan H., Almeida S., Verheyden J., Seeley W.W., Dickson D.W., Petrucelli L., Sun D., Jiao J., et al. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat. Med. 2014;20:1444–1451. doi: 10.1038/nm.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Y.-T., Lee Y.-C., Soong B.-W. What we have learned from the next-generation sequencing: Contributions to the genetic diagnoses and understanding of pathomechanisms of neurodegenerative diseases. J. Neurogenet. 2015;29:103–112. doi: 10.3109/01677063.2015.1060972. [DOI] [PubMed] [Google Scholar]
- 98.Wang Z., Gerstein M., Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]